Human ARF Specifically Inhibits Epimorphic Regeneration in the Zebrafish Heart
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish and Mice
2.2. Heat Shock Experiments
2.3. Cryoinjury
2.4. Mouse Anesthesia
2.5. Digit Amputation
2.6. Histology and Immunofluorescence
2.7. Quantitative Polymerase Chain Reaction (qPCR)
2.8. Statistical Analysis
3. Results
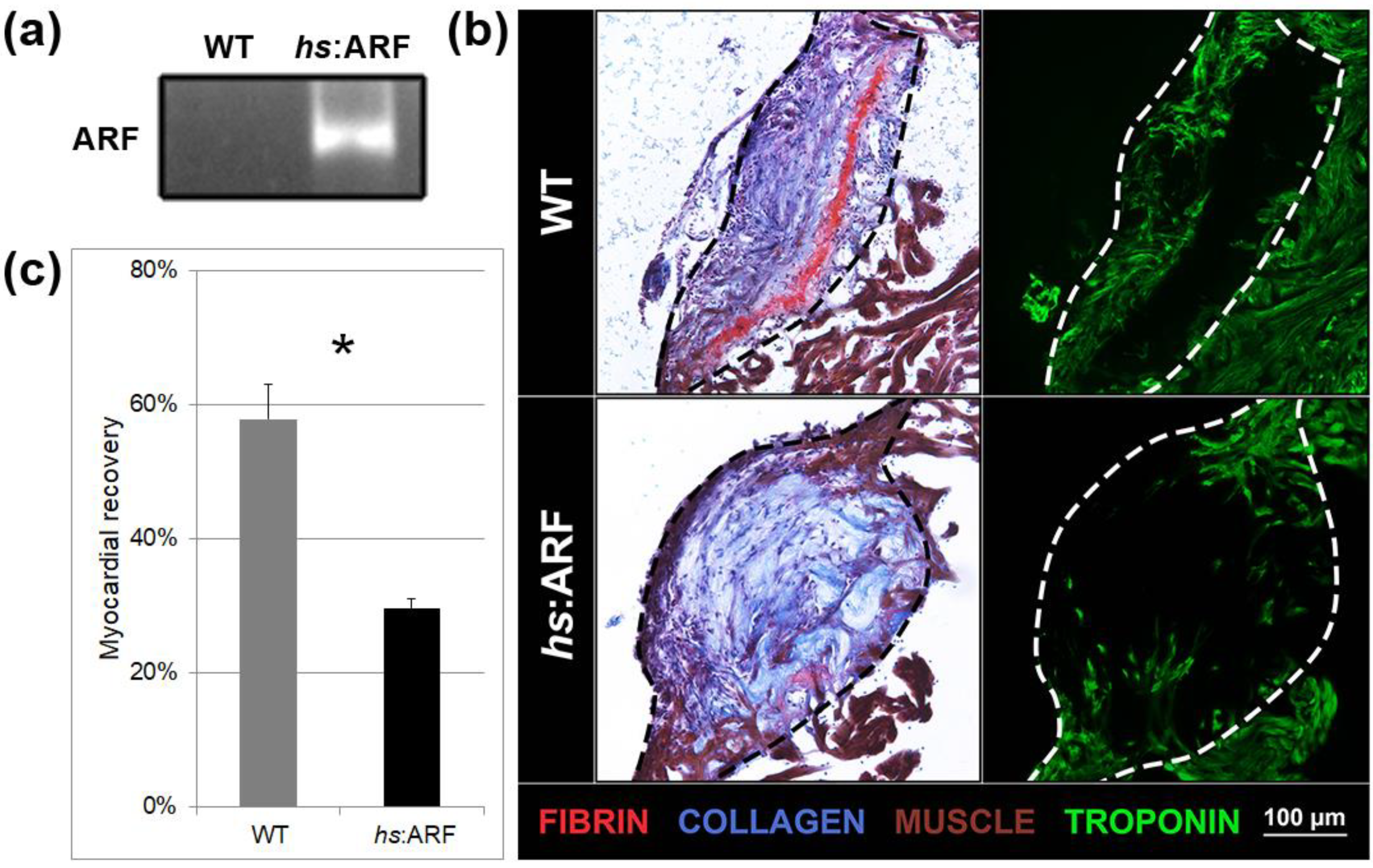
3.1. Induced Expression of ARF Suppresses Cardiac Regeneration
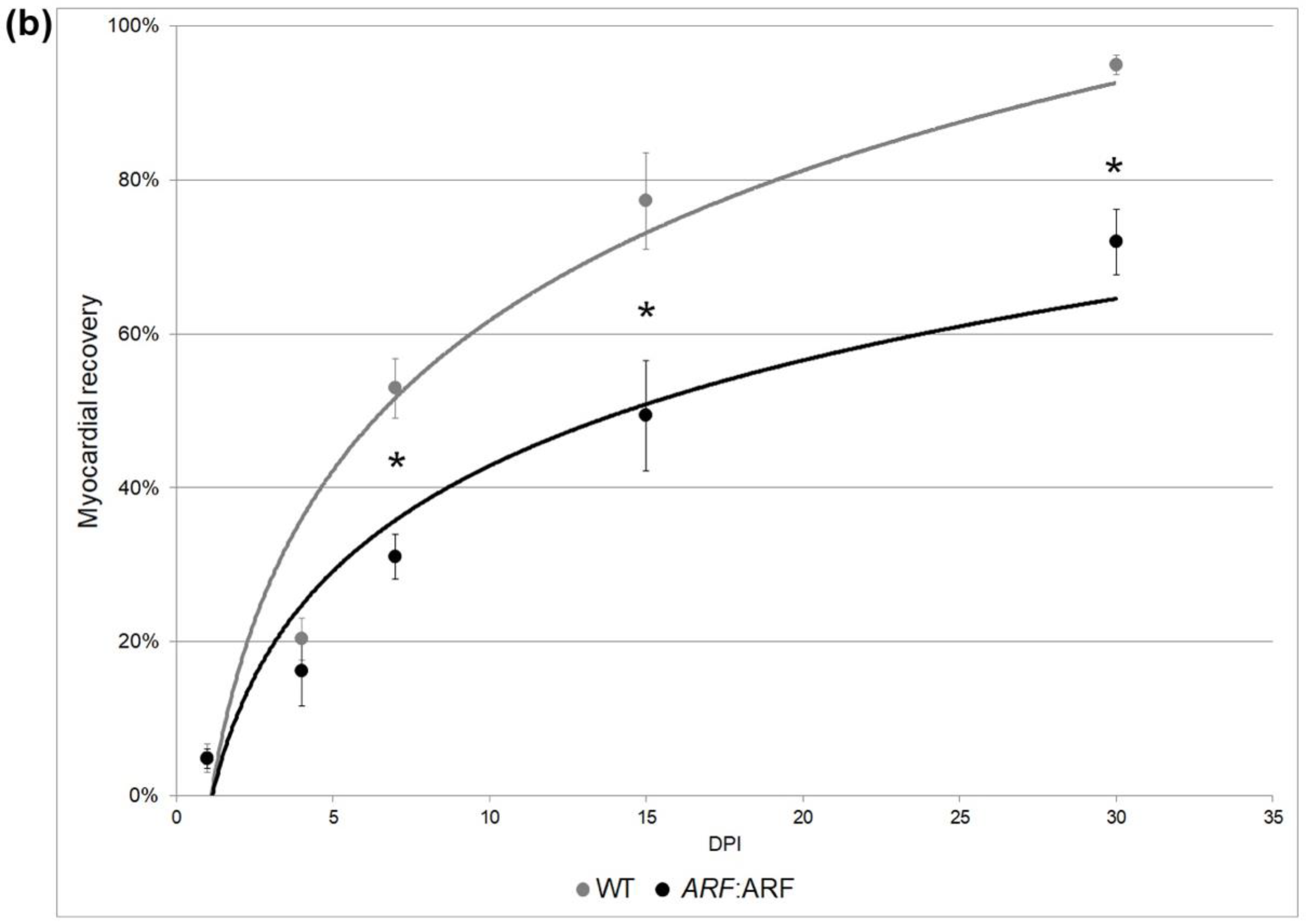
3.2. ARF Expressed under Control of the Endogenous Human ARF Promoter Suppresses Cardiac Regeneration
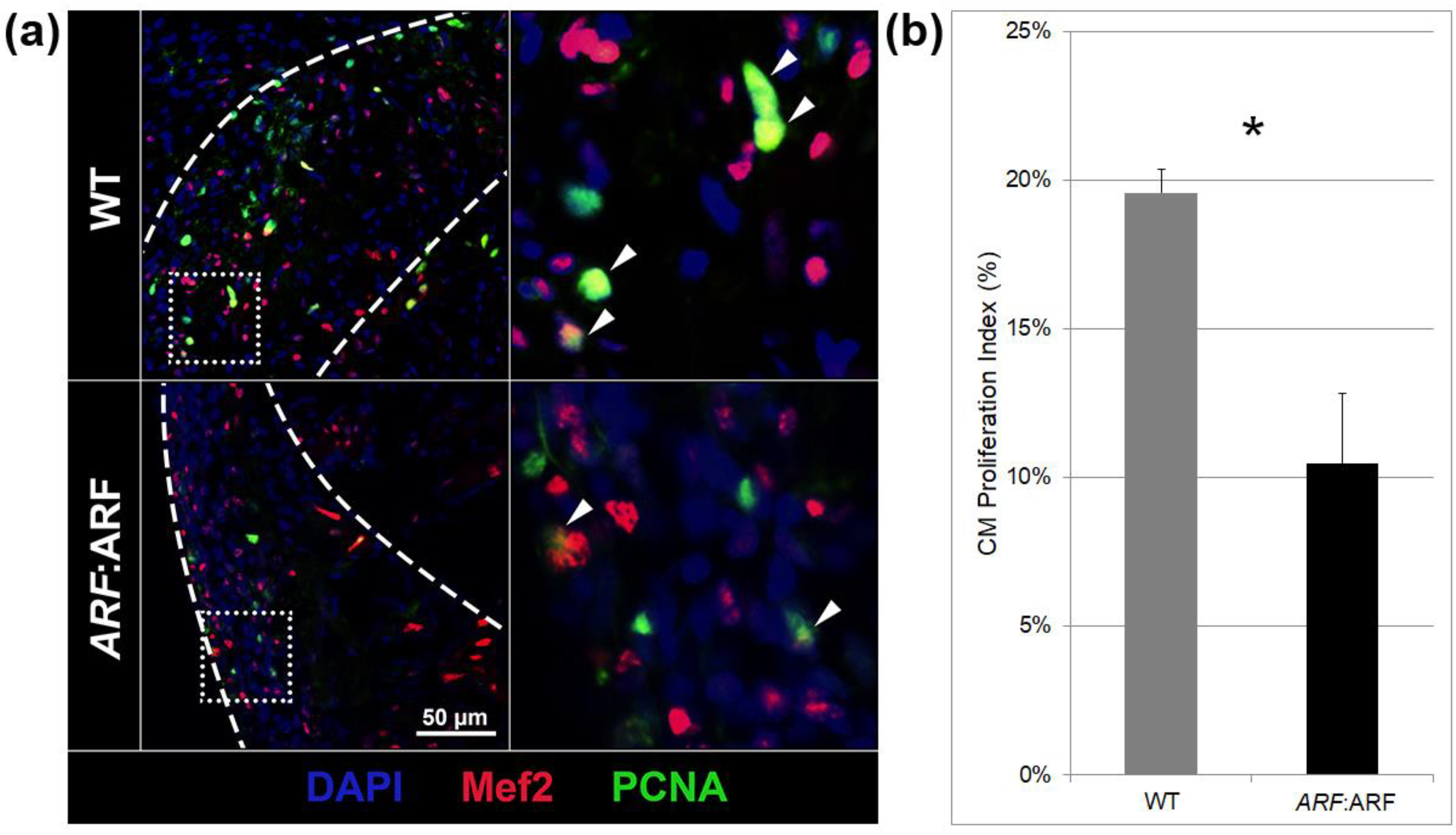
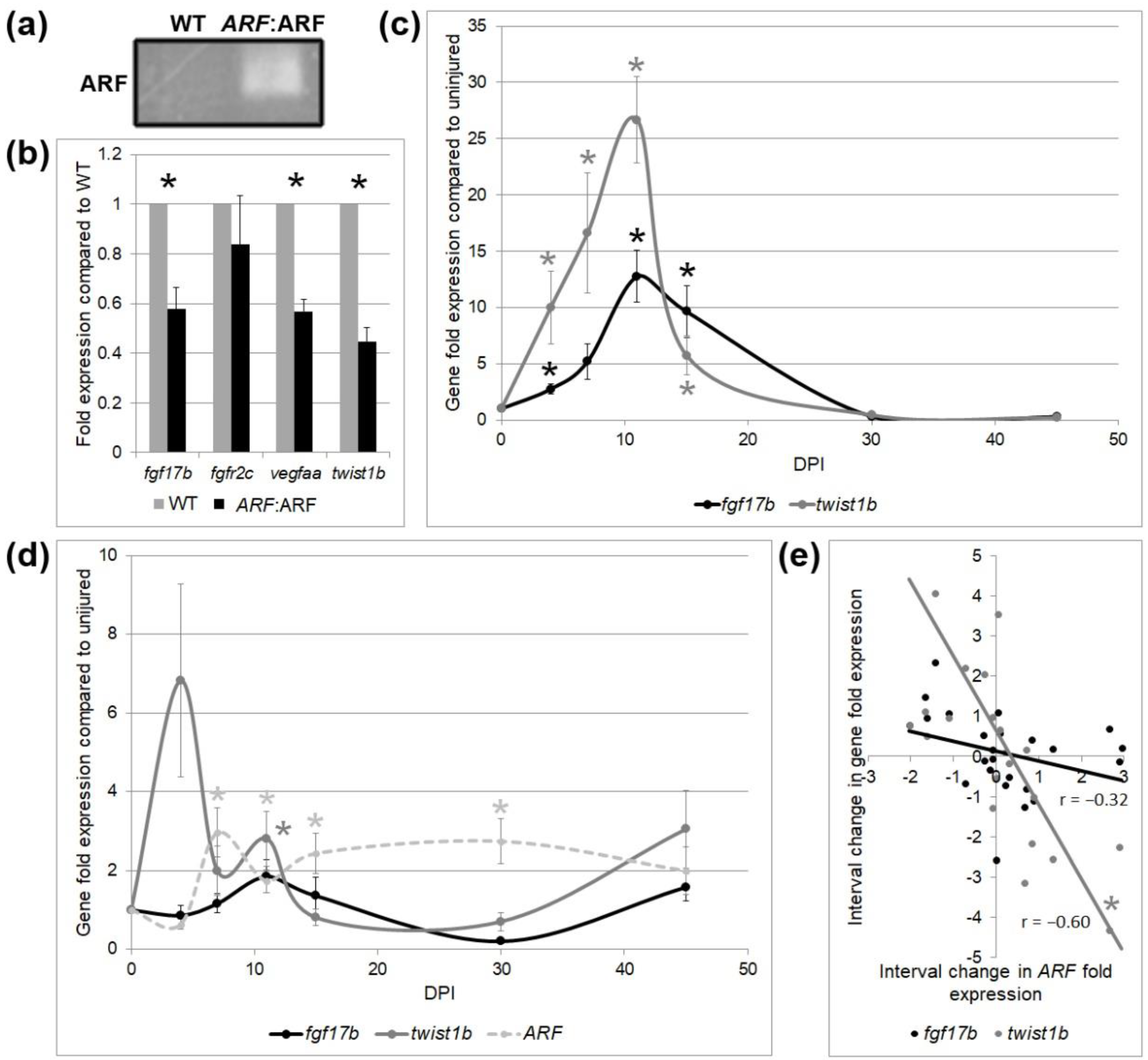
3.3. ARF Suppresses Cardiomyocyte Proliferation, Vascular Formation, and EMT
3.4. ARF Deficiency Is Insufficient to Permit Digit Regeneration in Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Antigen | Host Species | Company | Cat. No. | Dilution |
|---|---|---|---|---|
| Troponin | Mouse | Invitrogen | MA5-12960 | 1:1000 |
| Mef2 | Rabbit | Abcam | ab64644 | 1:50 |
| PCNA | Mouse | Cell Signaling | 2586T | 1:100 |
| Gene | NCBI ID | Forward | Reverse |
|---|---|---|---|
| B-Actin | NM_131031.1 | TTCACCACCACAGCCGAAAGA | TACCGCAAGATTCCATACCCA |
| ARF | NM_058195.3 | ATGGTGCGCAGGTTCTTGGTGA | CACCACCAGCGTGTCCAGGAAG |
| Fgf17b | NM_214808.1 | GCGGACGACGGAAACTCTTACG | CTCGAAGTGTTTTGGCTGCTCT |
| Fgfr2c | NM_001243004.1 | CGGCAGGTGTGAACACTACGG | CTCCGGCGAGTGGTGATTCTG |
| Vegfaa | NM_001110349.2 | GTGCAGGATGCTGTAATGATGAGG | AATTATGCTGCGATACGCGTTG |
| Twist1b | NM_001017820.1 | AGGTTCTACAGAGTGACGAGC | GCACAGGATTCGAACTAGAGG |
References
- Poss, K.D. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genet. 2010, 11, 710–722. [Google Scholar] [CrossRef] [Green Version]
- Poss, K.D.; Wilson, L.G.; Keating, M.T. Heart regeneration in zebrafish. Science 2002, 298, 2188–2190. [Google Scholar] [CrossRef]
- Raya, A.; Koth, C.M.; Büscher, D.; Kawakami, Y.; Itoh, T.; Raya, R.M.; Sternik, G.; Tsai, H.-J.; Rodríguez-Esteban, C.; Izpisua Belmonte, J.C. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. USA 2003, 100, 11889–11895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Panáková, D.; Kikuchi, K.; Holdway, J.E.; Gemberling, M.; Burris, J.S.; Singh, S.P.; Dickson, A.L.; Lin, Y.F.; Sabeh, M.K.; et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 2011, 138, 3421–3430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parente, V.; Balasso, S.; Pompilio, G.; Verduci, L.; Colombo, G.I.; Milano, G.; Guerrini, U.; Squadroni, L.; Cotelli, F.; Pozzoli, O.; et al. Hypoxia/reoxygenation cardiac injury and regeneration in zebrafish adult heart. PLoS ONE. 2013, 8, e53748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chablais, F.; Veit, J.; Rainer, G.; Jaźwińska, A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev. Biol. 2011, 11, 21. [Google Scholar] [CrossRef] [Green Version]
- González-Rosa, J.M.; Martín, V.; Peralta, M.; Torres, M.; Mercader, N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 2011, 138, 1663–1674. [Google Scholar] [CrossRef] [Green Version]
- Schnabel, K.; Wu, C.-C.; Kurth, T.; Weidinger, G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicar-dial activation and cardiomyocyte proliferation. PLoS ONE 2011, 6, e18503. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, J.H.; Blau, H.M. Tumor suppressors: Enhancers or suppressors of regeneration? Development 2013, 140, 2502–2512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flicek, P.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2014. Nucleic Acids Res. 2014, 42, D749–D755. [Google Scholar] [CrossRef] [PubMed]
- Karolchik, D.; Barber, G.P.; Casper, J.; Clawson, H.; Cline, M.S.; Diekhans, M.; Dreszer, T.R.; Fujita, P.A.; Guruvadoo, L.; Haeussler, M.; et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014, 42, D764–D770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, L.; Pomerantz, J.H.; DePinho, R.A. The INK4a/ARF tumor suppressor: One gene—two products—two pathways. Trends Biochem. Sci. 1998, 23, 291–296. [Google Scholar] [CrossRef]
- Lowe, S.W.; Sherr, C.J. Tumor suppression by Ink4a–arf: Progress and puzzles. Current Opinion in Genetics & Development. Curr. Opin. Genet. Dev. 2003, 13, 77–83. [Google Scholar] [PubMed]
- Pomerantz, J.H.; Schreiber-Agus, N.; Liégeois, N.J.; Silverman, A.; Alland, L.; Chin, L.; Potes, J.; Chen, K.; Orlow, I.; Lee, H.W.; et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 1998, 92, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.D.; Taylor, L.J.; Roussel, M.F.; Sherr, C.J.; Bar-Sagi, D. Nucleolar arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1999, 1, 20–26. [Google Scholar] [CrossRef]
- Pajcini, K.V.; Corbel, S.Y.; Pomerantz, J.H.; Blau, H.M.; Sage, J.; Pomerantz, J.H.; Blau, H.M. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell 2010, 7, 198–213. [Google Scholar] [CrossRef] [Green Version]
- Hesse, R.G.; Kouklis, G.K.; Ahituv, N.; Pomerantz, J.H. The human ARF tumor suppressor senses blastema activity and suppresses epimorphic tissue regeneration. Elife 2015, 4, e07702. [Google Scholar] [CrossRef]
- Knopf, F.; Hammond, C.; Chekuru, A.; Kurth, T.; Hans, S.; Weber, C.W.; Mahatma, G.; Fisher, S.; Brand, M.; Schulte-Merker, S.; et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 2011, 20, 713–724. [Google Scholar] [CrossRef] [Green Version]
- Tu, S.; Johnson, S.L. Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell 2011, 20, 725–732. [Google Scholar] [CrossRef]
- Bertero, A.; Murry, C.E. Hallmarks of cardiac regeneration. Nat. Rev. Cardiol. 2018, 15, 579–580. [Google Scholar] [CrossRef]
- Kikuchi, K.; Holdway, J.E.; Werdich, A.A.; Anderson, R.M.; Fang, Y.; Egnaczyk, G.F.; Evans, T.; Macrae, C.A.; Stainier, D.Y.; Poss, K.D. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010, 464, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Lepilina, A.; Coon, A.N.; Kikuchi, K.; Holdway, J.E.; Roberts, R.W.; Burns, C.G.; Poss, K.D. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 2006, 127, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Poss, K.D. The epicardium as a hub for heart regeneration. Nat. Rev. Cardiol. 2018, 15, 631–647. [Google Scholar] [CrossRef]
- Zhao, L.; Borikova, A.L.; Ben-Yair, R.; Guner-Ataman, B.; MacRae, C.A.; Lee, R.T.; Burns, C.G.; Burns, C.E. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2014, 111, 1403–1408. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Honor, L.B.; He, H.; Ma, Q.; Oh, J.H.; Butterfield, C.; Lin, R.Z.; Melero-Martin, J.M.; Dolmatova, E.; Duffy, H.S.; et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Investig. 2011, 121, 1894–1904. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, K.; Holdway, J.E.; Major, R.J.; Blum, N.; Dahn, R.D.; Begemann, G.; Poss, K.D. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 2011, 20, 397–404. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Han, P.; Yang, H.; Ouyang, K.; Lee, D.; Lin, Y.F.; Ocorr, K.; Kang, G.; Chen, J.; Stainier, D.Y.; et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 2013, 498, 497–501. [Google Scholar] [CrossRef]
- Kikuchi, K. Dedifferentiation, Transdifferentiation, and Proliferation: Mechanisms Underlying Cardiac Muscle Regeneration in Zebrafish. Curr. Pathobiol. Rep. 2015, 3, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Chablais, F.; Jazwinska, A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development 2012, 139, 1921–1930. [Google Scholar] [CrossRef] [Green Version]
- Lien, C.L.; Schebesta, M.; Makino, S.; Weber, G.J.; Keating, M.T. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006, 4, e260. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.Y.; Gemberling, M.; Wang, J.; Holdway, J.E.; Shen, M.C.; Karlstrom, R.O.; Poss, K.D. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 2013, 140, 660–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Harrison, M.R.; Osorio, A.; Kim, J.; Baugh, A.; Duan, C.; Sucov, H.M.; Lien, C.L. Igf Signaling is Required for Cardiomyocyte Proliferation during Zebrafish Heart Development and Regeneration. PLoS ONE 2013, 8, e67266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jopling, C.; Sleep, E.; Raya, M.; Marti, M.; Raya, A.; Belmonte, J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef]
- Marín-Juez, R.; Marass, M.; Gauvrit, S.; Rossi, A.; Lai, S.; Materna, S.C.; Black, B.L.; Stainier, D. Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2016, 40, 11237–11242. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Wu, Q.; Zhang, Y.; Wiens, K.M.; Huang, Y.; Rubin, N.; Shimada, H.; Handin, R.I.; Chao, M.Y.; Tuan, T.L.; et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc. Natl. Acad. Sci. USA 2010, 40, 17206–17210. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; Weinberg, R. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Sharpless, N.E.; Depinho, R.A. How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell Biol. 2007, 8, 703–713. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Hesse, R.; Tamaki, S.; Garland, C.; Pomerantz, J.H. Human ARF Specifically Inhibits Epimorphic Regeneration in the Zebrafish Heart. Genes 2020, 11, 666. https://doi.org/10.3390/genes11060666
Lee S, Hesse R, Tamaki S, Garland C, Pomerantz JH. Human ARF Specifically Inhibits Epimorphic Regeneration in the Zebrafish Heart. Genes. 2020; 11(6):666. https://doi.org/10.3390/genes11060666
Chicago/Turabian StyleLee, Solomon, Robert Hesse, Stanley Tamaki, Catharine Garland, and Jason H. Pomerantz. 2020. "Human ARF Specifically Inhibits Epimorphic Regeneration in the Zebrafish Heart" Genes 11, no. 6: 666. https://doi.org/10.3390/genes11060666
APA StyleLee, S., Hesse, R., Tamaki, S., Garland, C., & Pomerantz, J. H. (2020). Human ARF Specifically Inhibits Epimorphic Regeneration in the Zebrafish Heart. Genes, 11(6), 666. https://doi.org/10.3390/genes11060666





