A Stop-Gain Mutation within MLPH Is Responsible for the Lilac Dilution Observed in Jacob Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Whole Genome Sequencing
2.3. Candidate Variant Validation
3. Results
3.1. Whole-Genome Sequencing
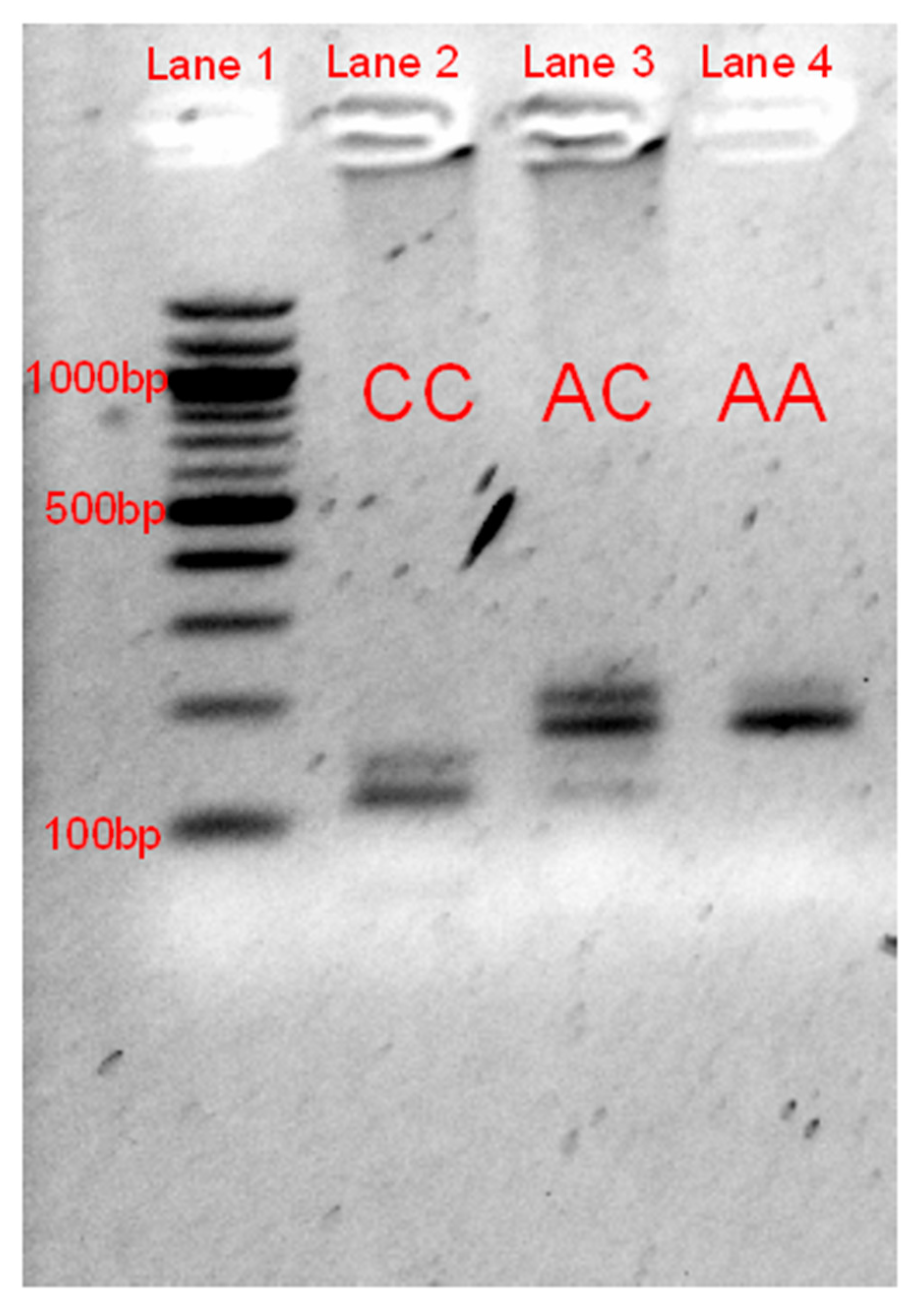
3.2. RFLP Validation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Anjola, O.; McEwan, N. Inheritance patterns of coat colouration and horn number in Jacob sheep. Open Agric. 2018, 3, 363–367. [Google Scholar]
- Nascimento, A.A.; Roland, J.T.; Gelfand, V.I. Pigment cells: A model for the study of organelle transport. Annu. Rev. Cell Dev. Biol. 2003, 19, 469–491. [Google Scholar] [CrossRef]
- Fukuda, M.; Kuroda, T.S.; Mikoshiba, K. Slac2-a/Melanophilin, the Missing Link between Rab27 and Myosin Va IMPLICations Of A Tripartite Protein Complex For Melanosome Transport. J. Biol. Chem. 2002, 277, 12432–12436. [Google Scholar] [CrossRef]
- Hume, A.N.; Ushakov, D.S.; Tarafder, A.K.; Ferenczi, M.A.; Seabra, M.C. Rab27a and MyoVa are the primary Mlph interactors regulating melanosome transport in melanocytes. J. Cell Sci. 2007, 120, 3111–3122. [Google Scholar] [CrossRef] [PubMed]
- Van Gele, M.; Dynoodt, P.; Lambert, J. Griscelli syndrome: A model system to study vesicular trafficking. Pigment Cell Melanoma Res. 2009, 22, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Vaez, M.; Follett, S.A.; Bed’Hom, B.; Gourichon, D.; Tixier-Boichard, M.; Burke, T. A single point-mutation within the melanophilin gene causes the lavender plumage colour dilution phenotype in the chicken. BMC Genet. 2008, 9, 7. [Google Scholar] [CrossRef]
- Philipp, U.; Hamann, H.; Mecklenburg, L.; Nishino, S.; Mignot, E.; Günzel-Apel, A.-R.; Schmutz, S.M.; Leeb, T. Polymorphisms within the canine MLPH gene are associated with dilute coat color in dogs. BMC Genet. 2005, 6, 34. [Google Scholar] [CrossRef]
- Drögemüller, C.; Philipp, U.; Haase, B.; Günzel-Apel, A.-R.; Leeb, T. A noncoding melanophilin gene (MLPH) SNP at the splice donor of exon 1 represents a candidate causal mutation for coat color dilution in dogs. J. Hered. 2007, 98, 468–473. [Google Scholar] [CrossRef]
- Bauer, A.; Kehl, A.; Jagannathan, V.; Leeb, T. A novel MLPH variant in dogs with coat colour dilution. Anim. Genet. 2018, 49, 94–97. [Google Scholar] [CrossRef]
- Lehner, S.; Gähle, M.; Dierks, C.; Stelter, R.; Gerber, J.; Brehm, R.; Distl, O. Two-exon skipping within MLPH is associated with coat color dilution in rabbits. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Fontanesi, L.; Scotti, E.; Allain, D.; Dall’olio, S. A frameshift mutation in the melanophilin gene causes the dilute coat colour in rabbit (Oryctolagus cuniculus) breeds. Anim. Genet. 2014, 45, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Demars, J.; Iannuccelli, N.; Utzeri, V.J.; Auvinet, G.; Riquet, J.; Fontanesi, L.; Allain, D. New Insights into the Melanophilin (MLPH) Gene Affecting Coat Color Dilution in Rabbits. Genes 2018, 9, 430. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; David, V.A.; Eizirik, E.; Schäffer, A.A.; Neelam, B.A.; Roelke, M.E.; Hannah, S.S.; O’Brien, S.J.; Menotti-Raymond, M. A homozygous single-base deletion in MLPH causes the dilute coat color phenotype in the domestic cat. Genomics 2006, 88, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Cirera, S.; Markakis, M.N.; Christensen, K.; Anistoroaei, R. New insights into the melanophilin (MLPH) gene controlling coat color phenotypes in American mink. Gene 2013, 527, 48–54. [Google Scholar] [CrossRef]
- Manakhov, A.D.; Andreeva, T.V.; Trapezov, O.V.; Kolchanov, N.A.; Rogaev, E.I. Genome analysis identifies the mutant genes for common industrial Silverblue and Hedlund white coat colours in American mink. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Li, W.; Sartelet, A.; Tamma, N.; Coppieters, W.; Georges, M.; Charlier, C. Reverse genetic screen for loss-of-function mutations uncovers a frameshifting deletion in the melanophilin gene accountable for a distinctive coat color in Belgian Blue cattle. Anim. Genet. 2016, 47, 110–113. [Google Scholar] [CrossRef]
- Jacob Sheep Pedigree Search. Available online: https://jsba.org/search.html (accessed on 20 April 2018).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Goleft Github Repository. Available online: https://github.com/brentp/goleft (accessed on 1 February 2020).
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic. Acids. Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Desai, U.J.; Pfaffle, P.K. Single-step purification of a thermostable DNA polymerase expressed in Escherichia coli. BioTechniques 1995, 19, 780–782, 784. [Google Scholar]
- Norris, B.J.; Whan, V.A. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res. 2008, 18, 1282–1293. [Google Scholar] [CrossRef]
- Våge, D.I.; Klungland, H.; Lu, D.; Cone, R.D. Molecular and pharmacological characterization of dominant black coat color in sheep. Mammalian Genome Off. J. Int. Mamm. Genome Soc. 1999, 10, 39–43. [Google Scholar] [CrossRef]
- Våge, D.I.; Fleet, M.R.; Ponz, R.; Olsen, R.T.; Monteagudo, L.V.; Tejedor, M.T.; Arruga, M.V.; Gagliardi, R.; Postiglioni, A.; Nattrass, G.S.; et al. Mapping and characterization of the dominant black colour locus in sheep. Pigment cell Res./Spons. Eur. Soc. Pigment Cell Res. Int. Pigment Cell Soc. 2003, 16, 693–697. [Google Scholar] [CrossRef]
- Gratten, J.; Beraldi, D.; Lowder, B.V.; McRae, A.F.; Visscher, P.M.; Pemberton, J.M.; Slate, J. Compelling evidence that a single nucleotide substitution in TYRP1 is responsible for coat-colour polymorphism in a free-living population of Soay sheep. Proc. R. Soc. Lond. B Biol. Sci. 2007, 274, 619–626. [Google Scholar] [CrossRef]
- Posbergh, C.; Huson, H. Making Moorit: Mutations in TYRP1 are responsible for brown coat color in different United States sheep breeds. In Proceedings of the 11th World Congress on Genetics Applied to Livestock Production, Auckland, New Zealand, 11–16 February 2018. [Google Scholar]
- Ramalho, J.S.; Lopes, V.S.; Tarafder, A.K.; Seabra, M.C.; Hume, A.N. Myrip uses distinct domains in the cellular activation of myosin VA and myosin VIIA in melanosome transport. Pigment Cell Melanoma Res. 2009, 22, 461–473. [Google Scholar] [CrossRef]


| Breed | Dilute Status | N | NC_019458.2:g.3451931C>A Genotype | ||
|---|---|---|---|---|---|
| AA | AC | CC | |||
| Jacob | Non-dilute | 39 | 0 | 13 | 26 |
| Dilute | 22 | 22 | 0 | 0 | |
| Non-Jacob | Non-dilute | 163 | 0 | 0 | 163 |
| Total | 224 | 22 | 13 | 189 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Posbergh, C.J.; Staiger, E.A.; Huson, H.J. A Stop-Gain Mutation within MLPH Is Responsible for the Lilac Dilution Observed in Jacob Sheep. Genes 2020, 11, 618. https://doi.org/10.3390/genes11060618
Posbergh CJ, Staiger EA, Huson HJ. A Stop-Gain Mutation within MLPH Is Responsible for the Lilac Dilution Observed in Jacob Sheep. Genes. 2020; 11(6):618. https://doi.org/10.3390/genes11060618
Chicago/Turabian StylePosbergh, Christian J., Elizabeth A. Staiger, and Heather J. Huson. 2020. "A Stop-Gain Mutation within MLPH Is Responsible for the Lilac Dilution Observed in Jacob Sheep" Genes 11, no. 6: 618. https://doi.org/10.3390/genes11060618
APA StylePosbergh, C. J., Staiger, E. A., & Huson, H. J. (2020). A Stop-Gain Mutation within MLPH Is Responsible for the Lilac Dilution Observed in Jacob Sheep. Genes, 11(6), 618. https://doi.org/10.3390/genes11060618






