Genome-Wide Characterization of Snf1-Related Protein Kinases (SnRKs) and Expression Analysis of SnRK1.1 in Strawberry
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification and Characterization of FvSnRKs
2.3. Sequence Analysis of FvSnRKs
2.4. Gene Structure and Phylogenetic Tree of FvSnRKs
2.5. RNA Extraction and Gene Expression of FvSnRKs
3. Results
3.1. Identification and Properties of SnRKs in F. versa
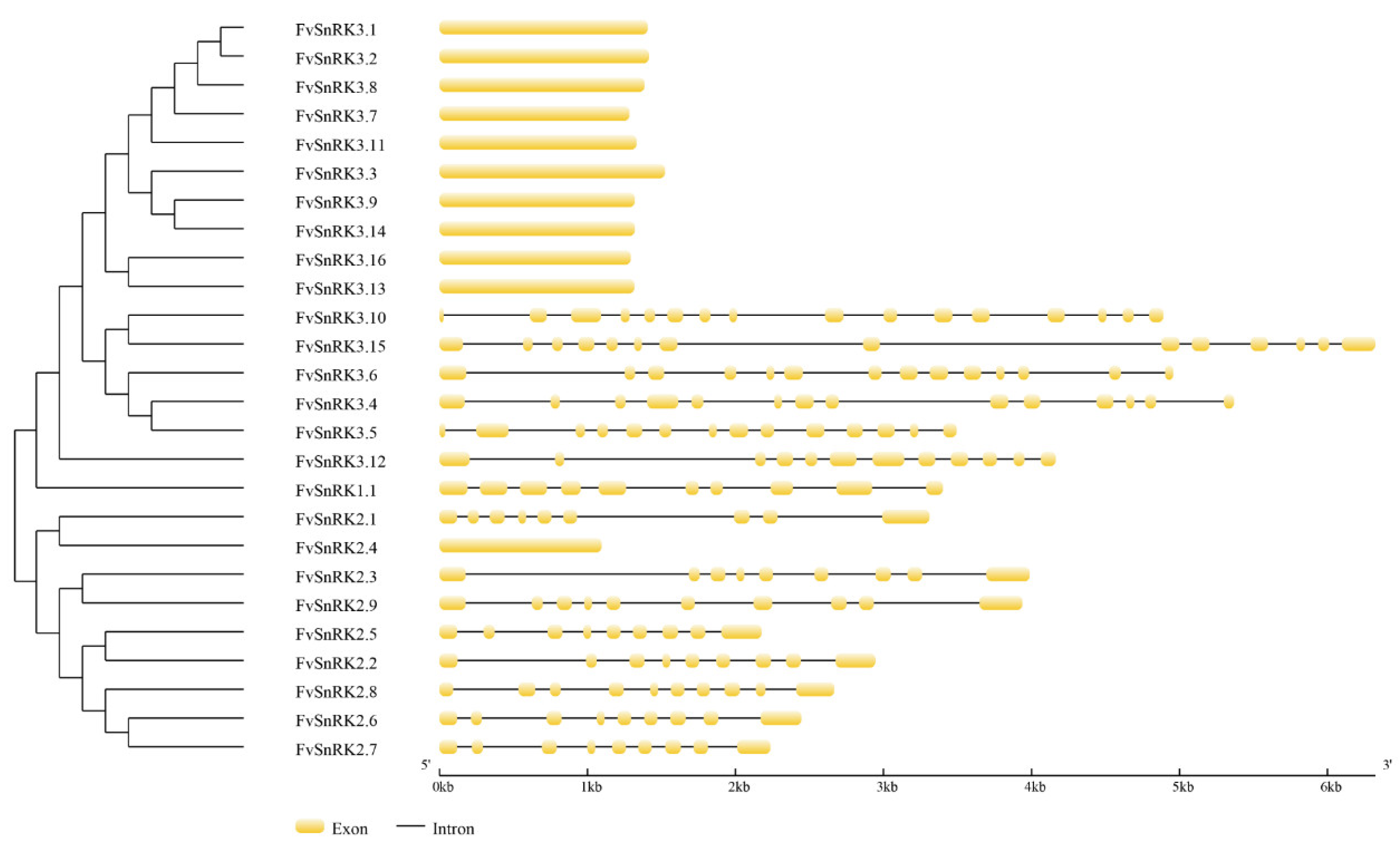
3.2. Analysis of Gene Structure
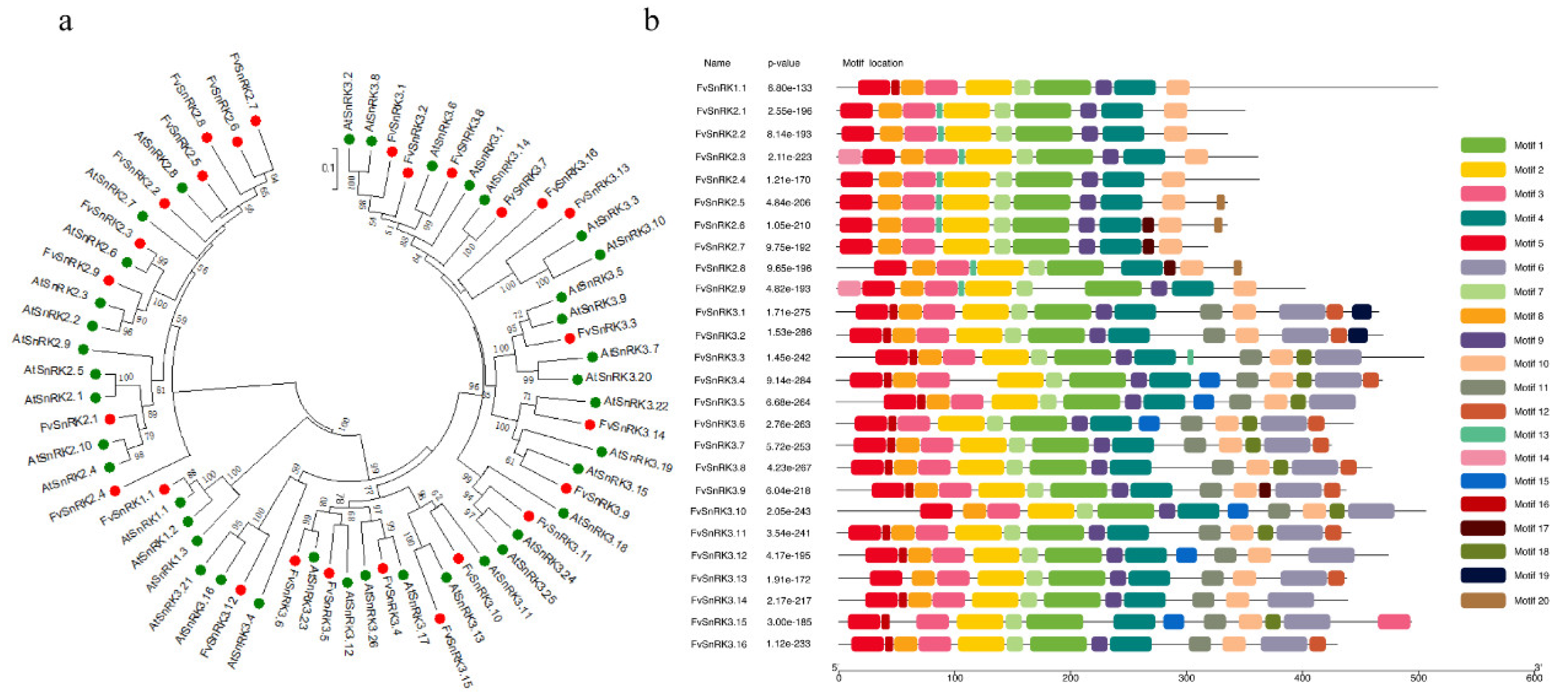
3.3. Phylogenetic Analysis and Conserved Motifs of FvSnRKs
3.4. Cis-element Analysis in the Promoter Region of FvSnRK1.1 Gene
3.5. Expression Pattern of FvSnRK1.1 in Different Tissues and During the Fruit Development
3.6. Expression Pattern of FvSnRK1.1 in Response to Different Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Demidchik, V.; Maathuis, F.; Voitsekhovskaja, O. Unravelling the plant signalling machinery: An update on the cellular and genetic basis of plant signal transduction. Funct. Plant Boil. 2018, 45, 1–8. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Boil. 2006, 9, 436–442. [Google Scholar] [CrossRef]
- Hashiguchi, A.; Komatsu, S. Impact of post-translational modifications of crop proteins under abiotic stress. Proteomes 2016, 4, 42. [Google Scholar] [CrossRef]
- Tena, G.; Boudsocq, M.; Sheen, J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Boil. 2011, 14, 519–529. [Google Scholar] [CrossRef]
- Coello, P.; Hey, S.J.; Halford, N.G. The sucrose non-fermenting-1-related (SnRK) family of protein kinases: Potential for manipulation to improve stress tolerance and increase yield. J. Exp. Bot. 2011, 62, 883–893. [Google Scholar] [CrossRef]
- Broeckx, T.; Hulsmans, S.; Rolland, F. The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 2016, 67, 6215–6252. [Google Scholar] [CrossRef]
- Hulsmans, S.; Rodriguez, M.; De Coninck, B.; Rolland, F. The SnRK1 energy sensor in plant biotic interactions. Trends Plant Sci. 2016, 21, 648–661. [Google Scholar] [CrossRef] [PubMed]
- Halford, N.G.; Hey, S.J. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 2009, 419, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Cui, N.; Wang, L.; Zhao, X.; Qu, B.; Li, T.; Zhang, G. The SnRK protein kinase family and the function of SnRK1 protein kinase. Int. J. Agric. Biol. 2012, 14, 196–200. [Google Scholar]
- Saha, J.; Chatterjee, C.; Sengupta, A.; Gupta, K.; Gupta, B. Genome-wide analysis and evolutionary study of sucrose non-fermenting 1-related protein kinase 2 (SnRK2) gene family members in Arabidopsis and Oryza. Comput. Boil. Chem. 2014, 49, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Anderberg, R.J.; Walker-Simmons, M.K. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc. Natl. Acad. Sci. USA. 1992, 89, 10183–10187. [Google Scholar] [CrossRef]
- Holappa, L.D.; Walker-Simmons, M.K. The wheat abscisic acid-responsive protein kinase mRNA, PKABA1, is up-regulated by dehydration, cold temperature, and osmotic stress. Plant Physiol. 1995, 108, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Li, Y.; Rehman, S.U.; Miao, L.; Zhang, Y.; Chen, X.; Yu, C.; Wang, J.; Li, C.; Jing, R. The sucrose non-fermenting 1-related protein kinase 2 (SnRK2) genes are multifaceted players in plant growth, development and response to environmental stimuli. Plant Cell Physiol. 2020, 61, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Halfter, U.; Ishitani, M.; Zhu, J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 2001, 13, 1383–1400. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Pandey, G.K.; Grant, J.J.; Batistic, O.; Li, L.; Kim, B.G.; Lee, S.C.; Kudla, J.; Luan, S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007, 52, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.J.; Wang, C.; Li, K.; Luan, S. The CBL–CIPK calcium signaling network: Unified paradigm from 20 years of discoveries. Trends Plant Sci. 2020. [Google Scholar] [CrossRef]
- Nietzsche, M.; Landgraf, R.; Tohge, T.; Börnke, F. A protein–protein interaction network linking the energy-sensor kinase SnRK1 to multiple signaling pathways in Arabidopsis thaliana. Curr. Plant Boil. 2016, 5, 36–44. [Google Scholar] [CrossRef]
- Crozet, P.; Margalha, L.; Confraria, A.; Rodrigues, A.; Martinho, C.; Adamo, M.; Elias, C.A.; Baena-González, E. Mechanisms of regulation of SNF1/AMPK/SNRK1 protein kinases. Front. Plant Sci. 2014, 5, 190. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; He, Y.; Xia, R. Tbtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Chen, Q.; Yu, H.; Wang, X.; Xie, X.; Yue, X.; Tang, H. An alternative cetyltrimethylammonium bromide-based protocol for RNA isolation from blackberry (Rubus L.). Genet. Mol. Res. 2012, 11, 1773–1782. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.G.; Kudla, J.; Luan, S.; Nimmo, H.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Kolukisaoglu, Ü.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Weinl, S.; Kudla, J. The CBL-CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytol. 2009, 184, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Lyzenga, W.J.; Liu, H.; Schofield, A.; Muise-Hennessey, A.; Stone, S.L. Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J. Exp. Bot. 2013, 64, 2779–2791. [Google Scholar] [CrossRef]
- Wang, L.; Hu, W.; Sun, J.; Liang, X.; Yang, X.; Wei, S.; Wang, X.; Zhou, Y.; Xiao, Q.; Yang, G.; et al. Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9. Plant Sci. 2015, 237, 33–45. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, H.; Qiu, Z.; Hu, B.; Zeng, B.; Zhong, C.; Fan, C. Comprehensive analysis of SnRK gene family and their responses to salt stress in Eucalyptus grandis. Int. J. Mol. Sci. 2019, 20, 2786. [Google Scholar] [CrossRef]
- Huai, J.; Wang, M.; He, J.; Zheng, J.; Dong, Z.; Lv, H.; Zhao, J.; Wang, G. Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep. 2008, 27, 1861–1868. [Google Scholar] [CrossRef]
- Li, L.B.; Zhang, Y.R.; Liu, K.C.; Ni, Z.F.; Fang, Z.J.; Sun, Q.X.; Gao, J.W. Identification and bioinformatics analysis of SnRK2 and CIPK family genes in sorghum. Agric. Sci. China 2010, 9, 19–30. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Wan, S.Q.; Wang, W.D.; Chen, J.F.; Huang, L.L.; Duan, M.S.; Yu, Y.B. Genome-wide identification and characterization of the CsSnRK2 family in Camellia sinensis. Plant Physiol. Biochem. 2018, 132, 287–296. [Google Scholar] [CrossRef]
- Liu, J.Y.; Chen, N.N.; Cheng, Z.M.; Xiong, J.S. Genome-wide identification, annotation and expression profile analysis of SnRK2 gene family in grapevine. Aust. J. Grape Wine Res. 2016, 22, 478–488. [Google Scholar] [CrossRef]
- Crepin, N.; Rolland, F. SnRK1 activation, signaling, and networking for energy homeostasis. Curr. Opin. Plant Boil. 2019, 51, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Feng, P.; Yu, H.; Yu, X.; Sun, Q.; Liu, S.; Minh, T.N.; Chen, J.; Wang, D.; Zhang, Q.; et al. GsSnRK1 interplays with transcription factor GsERF7 from wild soybean to regulate soybean stress resistance. Plant Cell Environ. 2020, 1–20. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Micallef, B.J.; Tetlow, I.J.; Mullen, R.T.; Feil, R.; Lunn, J.E.; Emes, M.J. AKINβ1, a subunit of SnRK1, regulates organic acid metabolism and acts as a global modulator of genes involved in carbon, lipid, and nitrogen metabolism. J. Exp. Bot. 2020, 71, 1010–1028. [Google Scholar] [PubMed]
- Yu, C.; Song, L.; Song, J.; Ouyang, B.; Guo, L.; Shang, L.; Wang, T.; Li, H.X.; Zhang, J.; Ye, Z. ShCIGT, a Trihelix family gene, mediates cold and drought tolerance by interacting with SnRK1 in tomato. Plant Sci. 2018, 270, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Peng, F.; Zhang, L.; Shi, X.; Wang, Z. Cloning and characterization of a SnRK1-encoding gene from Malus hupehensis Rehd and heterologous expression in tomato. Mol. Boil. Rep. 2010, 37, 947–954. [Google Scholar] [CrossRef]
- Wang, X.; Peng, F.; Li, M.; Yang, L.; Li, G. Expression of a heterologous SnRK1 in tomato increases carbon assimilation, nitrogen uptake and modifies fruit development. J. Plant Physiol. 2012, 169, 1173–1182. [Google Scholar] [CrossRef]
- Yu, W.; Peng, F.; Xiao, Y.; Wang, G.; Luo, J. Overexpression of PpSnRK1α in Tomato Promotes Fruit Ripening by Enhancing RIPENING INHIBITOR Regulation Pathway. Front. Plant Sci. 2018, 9, 1856. [Google Scholar] [CrossRef]


| Gene Name | Gene Id | Chr. | Locus | ORF (bp) | Amino Acid (aa) | MW (kDa) | pI | Instability Index | Aliphatic Index | GRAVY | SignalP | TMH | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FvSnRK1.1 | gene04397 | chr6 | 32622471..32625872+ | 1557 | 518 | 59.19 | 8.68 | 48.23 (unstable) | 88.78 | −0.313 | no | 0 | a, b |
| FvSnRK2.1 | gene11031 | chr5 | 16681534..16684843+ | 1059 | 352 | 40.40 | 6.12 | 44.84 (unstable) | 82.78 | −0.511 | no | 0 | a, b |
| FvSnRK2.2 | gene10769 | chr5 | 23595280..23598224+ | 1014 | 337 | 38.04 | 5.85 | 35.42 (stable) | 86.20 | −0.399 | no | 0 | a, b |
| FvSnRK2.3 | gene31902 | chr2 | 12574424..12578412+ | 1092 | 363 | 41.23 | 4.88 | 41.24 (unstable) | 88.35 | −0.316 | no | 0 | a, b |
| FvSnRK2.4 | gene06595 | chr4 | 14755025..14756119+ | 1095 | 364 | 41.48 | 9.10 | 45.92 (unstable) | 82.17 | −0.429 | no | 0 | a, b |
| FvSnRK2.5 | gene16244 | chr1 | 13571546..13573722+ | 1014 | 337 | 38.38 | 5.18 | 33.76 (stable) | 90.50 | −0.286 | no | 0 | a, b |
| FvSnRK2.6 | gene11971 | chr5 | 12304605..12307049− | 1014 | 337 | 38.10 | 5.76 | 36.88 (stable) | 89.91 | −0.242 | no | 0 | a, b |
| FvSnRK2.7 | gene11970 | chr5 | 12295362..12297597− | 963 | 320 | 36.30 | 8.76 | 37.85 (stable) | 91.34 | −0.253 | no | 0 | a, b |
| FvSnRK2.8 | gene11969 | chr5 | 12290219..12292886− | 1050 | 349 | 39.43 | 6.76 | 41.81 (unstable) | 81.20 | −0.342 | no | 0 | a, b |
| FvSnRK2.9 | gene24096 | chr1 | 11643335..11647273+ | 1215 | 404 | 45.72 | 4.82 | 44.09 (unstable) | 90.20 | −0.170 | no | 0 | a, b |
| FvSnRK3.1 | gene28136 | chr3 | 21779588..21780994− | 1407 | 468 | 53.65 | 8.77 | 40.89 (unstable) | 88.12 | −0.491 | no | 0 | c, d |
| FvSnRK3.2 | gene13841 | chr6 | 7167764..7169179+ | 1416 | 471 | 53.28 | 9.00 | 32.77 (stable) | 89.36 | −0.414 | no | 0 | c, d |
| FvSnRK3.3 | gene29682 | chr3 | 7803967..7805490− | 1524 | 507 | 56.29 | 7.64 | 39.44 (stable) | 88.22 | −0.219 | no | 0 | c, d |
| FvSnRK3.4 | gene15372 | chr2 | 32504181..32509548+ | 1416 | 471 | 53.98 | 6.03 | 32.57 (stable) | 88.58 | −0.334 | no | 0 | a, b |
| FvSnRK3.5 | gene18646 | chr7 | 3032869..3036362− | 1347 | 448 | 51.53 | 9.13 | 40.50 (unstable) | 77.46 | −0.539 | no | 0 | a, b |
| FvSnRK3.6 | gene30382 | chr3 | 2915121..2920078+ | 1341 | 446 | 49.55 | 8.83 | 30.14 (stable) | 83.30 | −0.314 | no | 0 | a, b |
| FvSnRK3.7 | gene10067 | chr1 | 842428..843711− | 1284 | 427 | 47.96 | 9.34 | 28.34 (stable) | 83.58 | −0.326 | no | 0 | c, d |
| FvSnRK3.8 | gene29681 | chr3 | 7799332..7800717+ | 1386 | 461 | 51.94 | 9.11 | 31.99 (stable) | 78.87 | −0.429 | no | 0 | c, d |
| FvSnRK3.9 | gene13849 | chr6 | 7132323..7133642− | 1320 | 439 | 49.58 | 7.58 | 31.35 (stable) | 91.69 | −0.266 | no | 0 | a, b, d |
| FvSnRK3.10 | gene31049 | chr1 | 3172739..3177629− | 1524 | 507 | 57.42 | 9.24 | 43.34 (unstable) | 86.37 | −0.286 | no | 0 | a, b, d |
| FvSnRK3.11 | gene15015 | chr2 | 35278943..35280274− | 1332 | 443 | 49.74 | 8.94 | 38.64 (stable) | 77.24 | −0.346 | no | 1 | c, d |
| FvSnRK3.12 | gene22806 | chr4 | 15674061..15678224+ | 1425 | 474 | 53.06 | 8.06 | 44.47 (unstable) | 93.54 | −0.368 | no | 0 | a, b |
| FvSnRK3.13 | gene29066 | chr4 | 6439716..6441032+ | 1317 | 438 | 47.51 | 8.87 | 35.28 (stable) | 94.86 | −0.080 | no | 0 | a, b |
| FvSnRK3.14 | gene28132 | chr3 | 21757318..21758637+ | 1320 | 439 | 49.48 | 8.52 | 38.61 (stable) | 87.40 | −0.298 | no | 0 | a, b, d |
| FvSnRK3.15 | gene26443 | chr1 | 4822218..4828540− | 1485 | 494 | 55.46 | 5.67 | 37.29 (stable) | 93.30 | −0.074 | no | 0 | a, b, d |
| FvSnRK3.16 | gene21319 | chr7 | 19497585..19498877+ | 1293 | 430 | 48.84 | 8.99 | 30.50 (stable) | 87.09 | −0.328 | no | 0 | c, d |
| Function Class | Cis-Elements | Amount | Function |
|---|---|---|---|
| Stress | ARE | 3 | essential for anaerobic induction |
| LTR | 1 | low-temperature responsiveness | |
| MBS | 1 | MYB binding site involved in drought-inducibility | |
| TC-rich repeats | 3 | defense and stress responsiveness | |
| Hormone | ABRE | 1 | abscisic acid responsiveness |
| CGTCA-motif | 3 | MeJA-responsiveness | |
| P-box | 1 | gibberellin-responsive element | |
| TCA-element | 1 | salicylic acid responsiveness | |
| TGACG-motif | 3 | MeJA-responsiveness | |
| Others | AE-box | 3 | part of a module for light response |
| Box 4 | 2 | light responsiveness | |
| G-box | 3 | light responsiveness | |
| GATA-motif | 2 | part of a light responsive element | |
| GT1-motif | 3 | light responsive element | |
| L-box | 1 | part of a light responsive element | |
| TCT-motif | 2 | part of a light responsive element | |
| MRE | 3 | MYB binding site involved in light responsiveness | |
| GCN4_motif | 1 | endosperm expression | |
| O2-site | 1 | zein metabolism regulation |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ye, Y.; Jiang, L.; Lin, Y.; Gu, X.; Chen, Q.; Sun, B.; Zhang, Y.; Luo, Y.; Wang, Y.; et al. Genome-Wide Characterization of Snf1-Related Protein Kinases (SnRKs) and Expression Analysis of SnRK1.1 in Strawberry. Genes 2020, 11, 427. https://doi.org/10.3390/genes11040427
Zhang Y, Ye Y, Jiang L, Lin Y, Gu X, Chen Q, Sun B, Zhang Y, Luo Y, Wang Y, et al. Genome-Wide Characterization of Snf1-Related Protein Kinases (SnRKs) and Expression Analysis of SnRK1.1 in Strawberry. Genes. 2020; 11(4):427. https://doi.org/10.3390/genes11040427
Chicago/Turabian StyleZhang, Yunting, Yuyun Ye, Leiyu Jiang, Yuanxiu Lin, Xianjie Gu, Qing Chen, Bo Sun, Yong Zhang, Ya Luo, Yan Wang, and et al. 2020. "Genome-Wide Characterization of Snf1-Related Protein Kinases (SnRKs) and Expression Analysis of SnRK1.1 in Strawberry" Genes 11, no. 4: 427. https://doi.org/10.3390/genes11040427
APA StyleZhang, Y., Ye, Y., Jiang, L., Lin, Y., Gu, X., Chen, Q., Sun, B., Zhang, Y., Luo, Y., Wang, Y., Wang, X., & Tang, H. (2020). Genome-Wide Characterization of Snf1-Related Protein Kinases (SnRKs) and Expression Analysis of SnRK1.1 in Strawberry. Genes, 11(4), 427. https://doi.org/10.3390/genes11040427






