Arginine Auxotrophy Affects Siderophore Biosynthesis and Attenuates Virulence of Aspergillus fumigatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
2.2. Deletion of argJ (AFUA_5G08120), argB (AFUA_4G07190) and Reconstitution of ∆argJ and ∆argB in A. fumigatus
2.3. Quantification of Biomass and Siderophore Production
2.4. Northern Blot Analysis
2.5. Galleria mellonella Infection Studies
2.6. Pulmonary Mouse Infection
3. Results
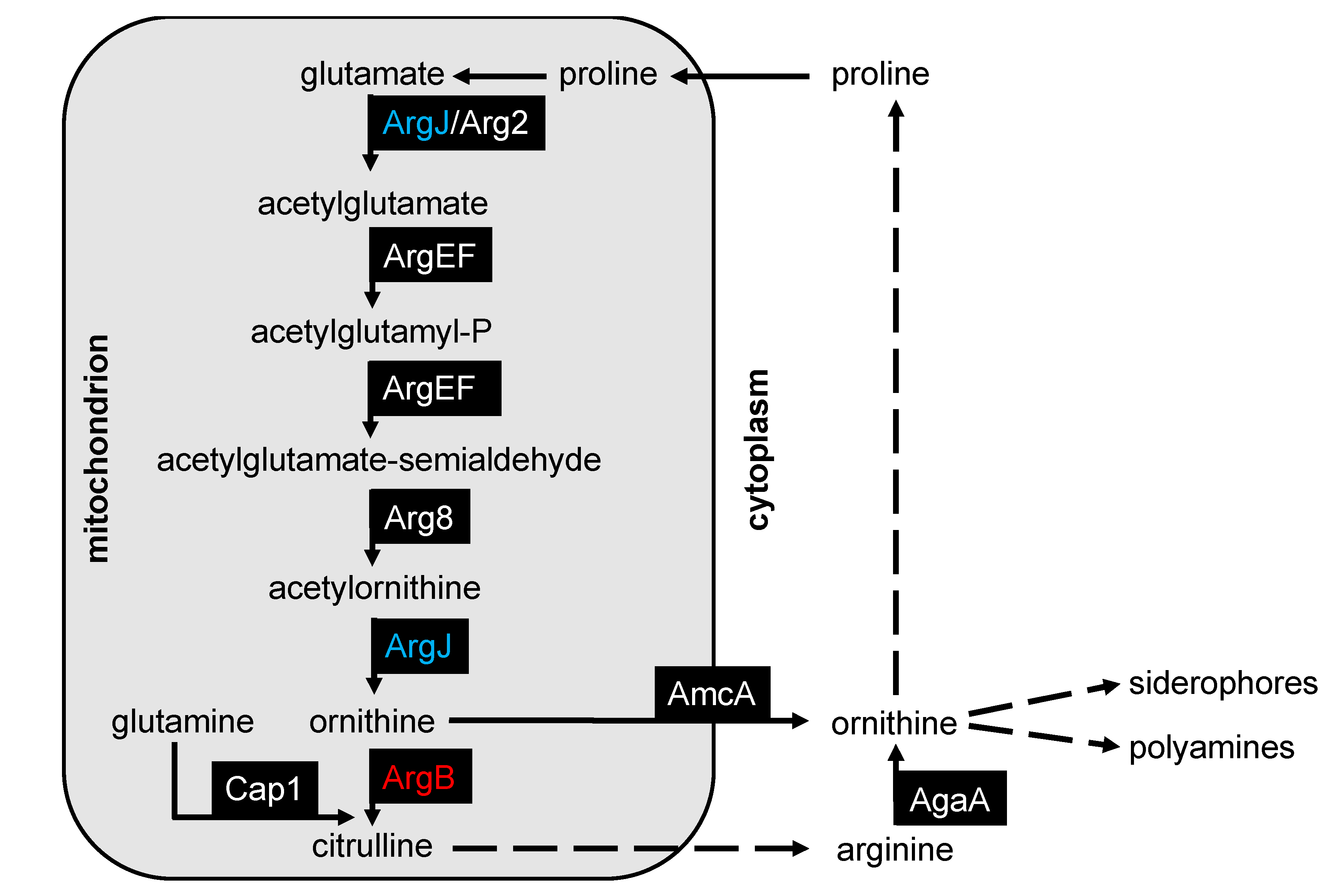
3.1. Generation of Arginine Auxotrophic Mutant Strains in A. fumigatus
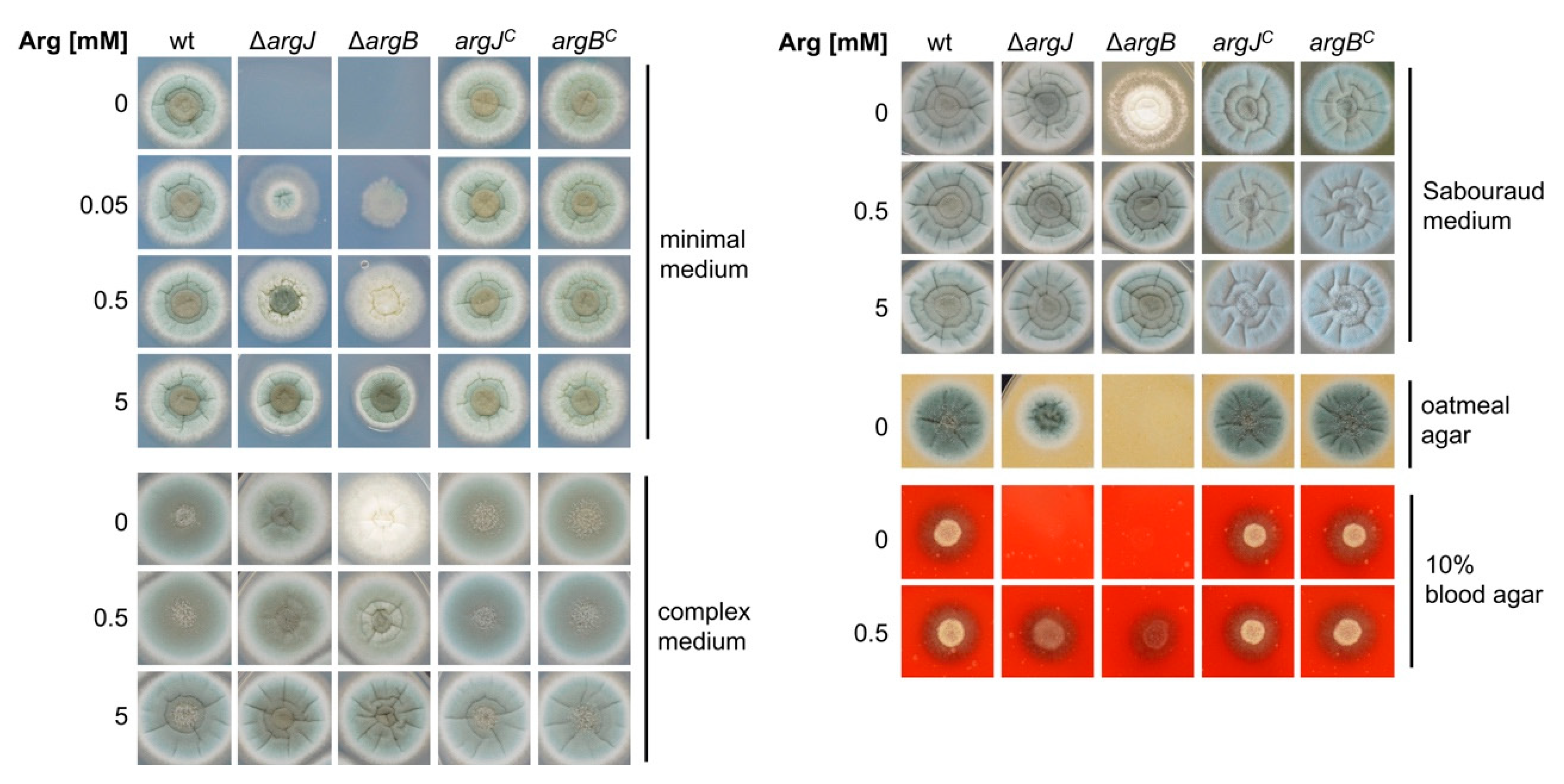
3.2. Deletion of argJ and argB Leads to Arginine Auxotrophy in A. fumigatus
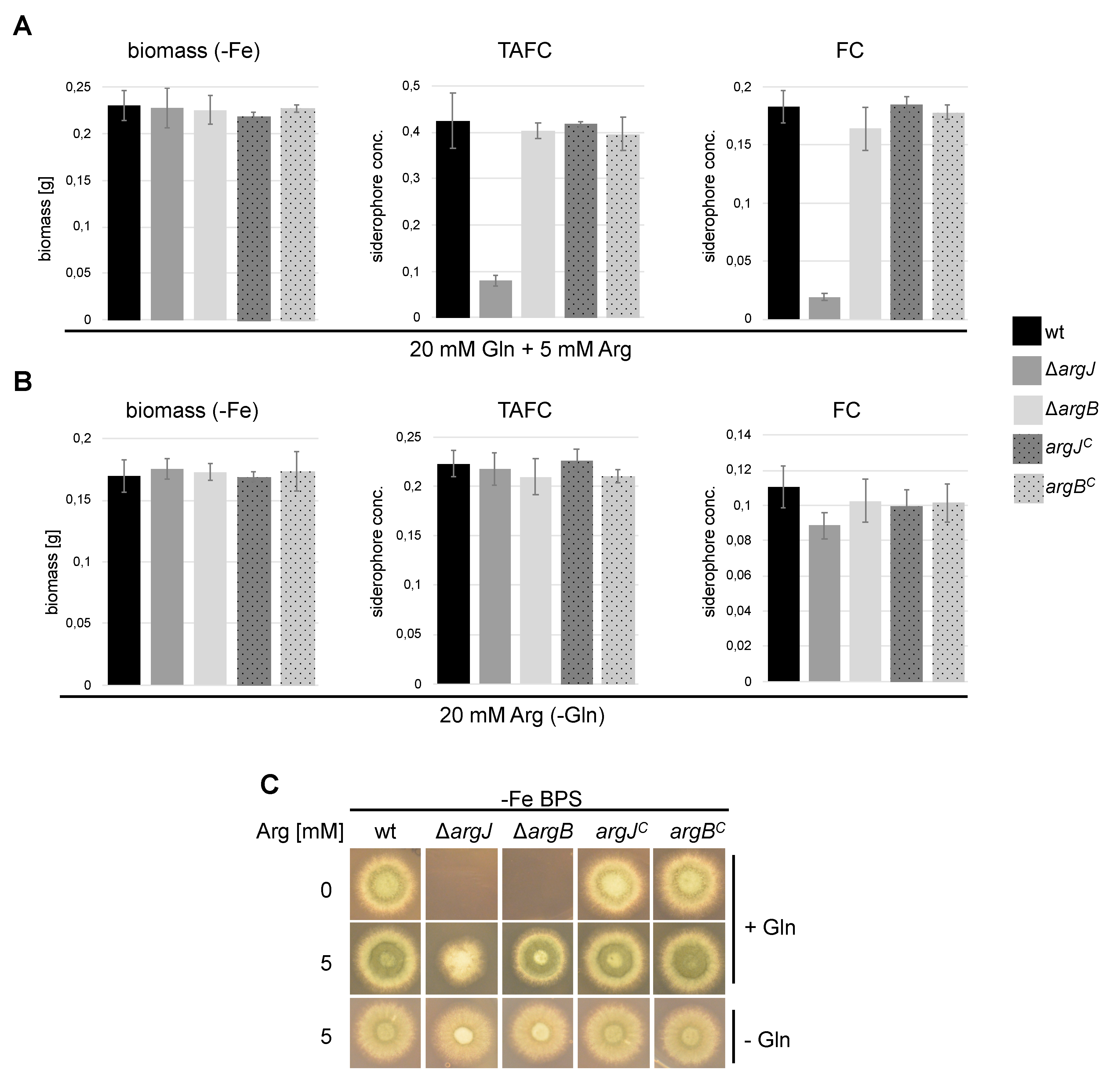
3.3. Deficiency of ArgJ, but not ArgB, Decreases Siderophore Production with Glutamine but Not with Arginine as Major Nitrogen Source
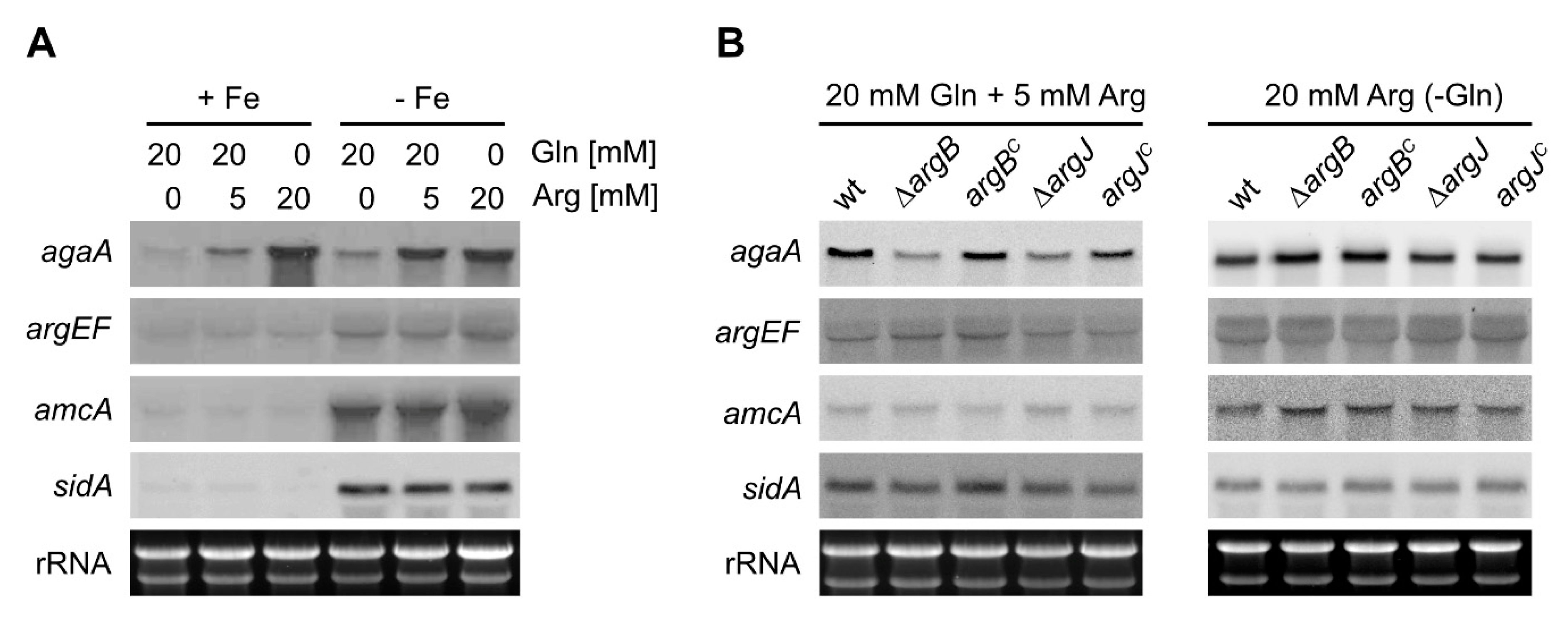
3.4. Arginase is Transcriptionally Repressed by Nitrogen Metabolite Repression in A. fumigatus
3.5. Deficiency of ArgJ, but Not ArgB, Impairs Growth during Iron-Limited Conditions with Glutamine as Nitrogen Source, When Reductive Iron Assimilation Is Blocked
3.6. Arginine Biosynthesis Is Crucial for Full Virulence of A. fumigatus in the Insect Model Galleria mellonella
3.7. Arginine Biosynthesis Is Crucial for Full Virulence of A. fumigatus in Murine Models for Invasive Aspergillosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Latge, J.P.; Chamilos, G. Aspergillus fumigatus and aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e0014018. [Google Scholar] [CrossRef]
- Amich, J.; Bignell, E. Amino acid biosynthetic routes as drug targets for pulmonary fungal pathogens: What is known and why do we need to know more? Curr. Opin. Microbiol. 2016, 32, 151–158. [Google Scholar] [CrossRef]
- Amich, J.; Dumig, M.; O′Keeffe, G.; Binder, J.; Doyle, S.; Beilhack, A.; Krappmann, S. Exploration of sulfur assimilation of Aspergillus fumigatus reveals biosynthesis of sulfur-containing amino acids as a virulence determinant. Infect. Immun. 2016, 84, 917–929. [Google Scholar] [CrossRef]
- Dietl, A.M.; Amich, J.; Leal, S.; Beckmann, N.; Binder, U.; Beilhack, A.; Pearlman, E.; Haas, H. Histidine biosynthesis plays a crucial role in metal homeostasis and virulence of Aspergillus fumigatus. Virulence 2016, 7, 465–476. [Google Scholar] [CrossRef]
- Liebmann, B.; Muhleisen, T.W.; Muller, M.; Hecht, M.; Weidner, G.; Braun, A.; Brock, M.; Brakhage, A.A. Deletion of the Aspergillus fumigatus lysine biosynthesis gene lysf encoding homoaconitase leads to attenuated virulence in a low-dose mouse infection model of invasive aspergillosis. Arch. Microbiol. 2004, 181, 378–383. [Google Scholar] [CrossRef]
- Orasch, T.; Dietl, A.M.; Shadkchan, Y.; Binder, U.; Bauer, I.; Lass-Florl, C.; Osherov, N.; Haas, H. The leucine biosynthetic pathway is crucial for adaptation to iron starvation and virulence in Aspergillus fumigatus. Virulence 2019, 10, 925–934. [Google Scholar] [CrossRef]
- Sasse, A.; Hamer, S.N.; Amich, J.; Binder, J.; Krappmann, S. Mutant characterization and in vivo conditional repression identify aromatic amino acid biosynthesis to be essential for Aspergillus fumigatus virulence. Virulence 2016, 7, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Schobel, F.; Jacobsen, I.D.; Brock, M. Evaluation of lysine biosynthesis as an antifungal drug target: Biochemical characterization of Aspergillus fumigatus homocitrate synthase and virulence studies. Eukaryot. Cell 2010, 9, 878–893. [Google Scholar] [CrossRef] [PubMed]
- Namiki, F.; Matsunaga, M.; Okuda, M.; Inoue, I.; Nishi, K.; Fujita, Y.; Tsuge, T. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. Sp melonis. Mol. Plant Microbe Interact. 2001, 14, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shi, H.; Liang, S.; Ning, G.; Xu, N.; Lu, J.; Liu, X.; Lin, F. Moarg1, moarg5,6 and moarg7 involved in arginine biosynthesis are essential for growth, conidiogenesis, sexual reproduction, and pathogenicity in Magnaporthe oryzae. Microbiol. Res. 2015, 180, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Purnell, D.M. The effects of specific auxotrophic mutations on the virulence of Aspergillus nidulans for mice. Mycopathol. Mycol. Appl. 1973, 50, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Andrew, P.J.; Mayer, B. Enzymatic function of nitric oxide synthases. Cardiovasc. Res. 1999, 43, 521–531. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.W.; Davis, T.A.; Kim, S.W.; Li, P.; Marc Rhoads, J.; Carey Satterfield, M.; Smith, S.B.; Spencer, T.E.; Yin, Y. Arginine metabolism and nutrition in growth, health and disease. Amino Acids 2009, 37, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.H. Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol. Rev. 1986, 50, 280–313. [Google Scholar] [CrossRef] [PubMed]
- Messenguy, F. Multiplicity of regulatory mechanisms controlling amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Sci. 1987, 4, 150–153. [Google Scholar]
- Crabeel, M.; Abadjieva, A.; Hilven, P.; Desimpelaere, J.; Soetens, O. Characterization of the Saccharomyces cerevisiae arg7 gene encoding ornithine acetyltransferase, an enzyme also endowed with acetylglutamate synthase activity. Eur. J. Biochem. 1997, 250, 232–241. [Google Scholar] [CrossRef]
- Negredo, A.; Monteoliva, L.; Gil, C.; Pla, J.; Nombela, C. Cloning, analysis and one-step disruption of the arg5,6 gene of Candida albicans. Microbiology 1997, 143, 297–302. [Google Scholar] [CrossRef]
- Boonchird, C.; Messenguy, F.; Dubois, E. Determination of amino acid sequences involved in the processing of the arg5/arg6 precursor in Saccharomyces cerevisiae. Eur. J. Biochem. 1991, 199, 325–335. [Google Scholar] [CrossRef]
- Schafferer, L.; Beckmann, N.; Binder, U.; Brosch, G.; Haas, H. Amca-a putative mitochondrial ornithine transporter supporting fungal siderophore biosynthesis. Front. Microbiol. 2015, 6, 252. [Google Scholar] [CrossRef]
- Jadoun, J.; Shadkchan, Y.; Osherov, N. Disruption of the Aspergillus fumigatus argb gene using a novel in vitro transposon-based mutagenesis approach. Curr. Genet. 2004, 45, 235–241. [Google Scholar] [CrossRef]
- Crabeel, M.; Messenguy, F.; Lacroute, F.; Glansdorff, N. Cloning arg3, the gene for ornithine carbamoyltransferase from Saccharomyces-cerevisiae - expression in escherichia-coli requires secondary mutations - production of plasmid beta-lactamase in yeast. Proc. Natl. Acad. Sci. USA 1981, 78, 5026–5030. [Google Scholar] [CrossRef] [PubMed]
- Borsuk, P.; Dzikowska, A.; Empel, J.; Grzelak, A.; Grzeskowiak, R.; Weglenski, P. Structure of the arginase coding gene and its transcript in aspergillus nidulans. Acta Biochim. Pol. 1999, 46, 391–403. [Google Scholar] [CrossRef]
- Beckmann, N.; Schafferer, L.; Schrettl, M.; Binder, U.; Talasz, H.; Lindner, H.; Haas, H. Characterization of the link between ornithine, arginine, polyamine and siderophore metabolism in Aspergillus fumigatus. PLoS ONE 2013, 8, e67426. [Google Scholar] [CrossRef] [PubMed]
- Schrettl, M.; Beckmann, N.; Varga, J.; Heinekamp, T.; Jacobsen, I.D.; Jochl, C.; Moussa, T.A.; Wang, S.; Gsaller, F.; Blatzer, M.; et al. Hapx-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010, 6, e1001124. [Google Scholar] [CrossRef] [PubMed]
- Tapiero, H.; Mathe, G.; Couvreur, P.; Tew, K.D.I. Arginine. Biomed. Pharmacother. 2002, 56, 439–445. [Google Scholar] [CrossRef]
- Pieper, G.M.; Dondlinger, L.A. Plasma and vascular tissue arginine are decreased in diabetes: Acute arginine supplementation restores endothelium-dependent relaxation by augmenting cgmp production. J. Pharmacol. Exp. Ther. 1997, 283, 684–691. [Google Scholar]
- Wu, G.; Morris, S.M., Jr. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336, 1–17. [Google Scholar] [CrossRef]
- Wu, G.; Davis, P.K.; Flynn, N.E.; Knabe, D.A.; Davidson, J.T. Endogenous synthesis of arginine plays an important role in maintaining arginine homeostasis in postweaning growing pigs. J. Nutr. 1997, 127, 2342–2349. [Google Scholar] [CrossRef]
- Pontecorvo, G.; Roper, J.A.; Hemmons, L.M.; Macdonald, K.D.; Bufton, A.W. The genetics of Aspergillus nidulans. Adv. Genet. 1953, 5, 141–238. [Google Scholar]
- Dietl, A.M.; Meir, Z.; Shadkchan, Y.; Osherov, N.; Haas, H. Riboflavin and pantothenic acid biosynthesis are crucial for iron homeostasis and virulence in the pathogenic mold Aspergillus fumigatus. Virulence 2018, 9, 1036–1049. [Google Scholar] [CrossRef]
- Nielsen, M.L.; Albertsen, L.; Lettier, G.; Nielsen, J.B.; Mortensen, U.H. Efficient pcr-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal. Genet. Biol. 2006, 43, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Krappmann, S.; Sasse, C.; Braus, G.H. Gene targeting in aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot. Cell 2006, 5, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Oberegger, H.; Schoeser, M.; Zadra, I.; Abt, B.; Haas, H. Srea is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 2001, 41, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Maurer, E.; Browne, N.; Surlis, C.; Jukic, E.; Moser, P.; Kavanagh, K.; Lass-Florl, C.; Binder, U. Galleria mellonella as a host model to study aspergillus terreus virulence and amphotericin b resistance. Virulence 2015, 6, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Mowlds, P.; Kavanagh, K. Effect of pre-incubation temperature on susceptibility of Galleria mellonella larvae to infection by candida albicans. Mycopathologia 2008, 165, 5–12. [Google Scholar] [CrossRef]
- Bergstrom, J.; Furst, P.; Noree, L.O.; Vinnars, E. Intracellular free amino acid concentration in human muscle tissue. J. Appl. Physiol. 1974, 36, 693–697. [Google Scholar] [CrossRef]
- Schrettl, M.; Kim, H.S.; Eisendle, M.; Kragl, C.; Nierman, W.C.; Heinekamp, T.; Werner, E.R.; Jacobsen, I.; Illmer, P.; Yi, H.; et al. Srea-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 2008, 70, 27–43. [Google Scholar] [CrossRef]
- Macios, M.; Caddick, M.X.; Weglenski, P.; Scazzocchio, C.; Dzikowska, A. The gata factors area and areb together with the co-repressor nmra, negatively regulate arginine catabolism in Aspergillus nidulans in response to nitrogen and carbon source. Fungal. Genet. Biol. 2012, 49, 189–198. [Google Scholar] [CrossRef]
- Schrettl, M.; Bignell, E.; Kragl, C.; Joechl, C.; Rogers, T.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 2004, 200, 1213–1219. [Google Scholar] [CrossRef]
- Xue, T.; Nguyen, C.K.; Romans, A.; Kontoyiannis, D.P.; May, G.S. Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain af293. Arch. Microbiol. 2004, 182, 346–353. [Google Scholar] [CrossRef]
- Punt, P.J.; Greaves, P.A.; Kuyvenhoven, A.; van Deutekom, J.C.; Kinghorn, J.R.; Pouwels, P.H.; van den Hondel, C.A. A twin-reporter vector for simultaneous analysis of expression signals of divergently transcribed, contiguous genes in filamentous fungi. Gene 1991, 104, 119–122. [Google Scholar] [CrossRef]
- Slater, J.L.; Gregson, L.; Denning, D.W.; Warn, P.A. Pathogenicity of Aspergillus fumigatus mutants assessed in galleria mellonella matches that in mice. Med. Mycol. 2011, 49 (Suppl. 1), S107–S113. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; Monzon, S.; Alcazar-Fuoli, L.; Cuesta, I.; Mellado, E. Genome-wide comparative analysis of Aspergillus fumigatus strains: The reference genome as a matter of concern. Genes 2018, 9, 363. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Heterogeneity confounds establishment of "a" model microbial strain. Mbio 2017, 8, e00135-17. [Google Scholar] [CrossRef]
- Kurucz, V.; Kiss, B.; Szigeti, Z.M.; Nagy, G.; Orosz, E.; Hargitai, Z.; Harangi, S.; Wiebenga, A.; de Vries, R.P.; Pocsi, I.; et al. Physiological background of the remarkably high cd2+ tolerance of the Aspergillus fumigatus Af293 strain. J. Basic Microb. 2018, 58, 957–967. [Google Scholar] [CrossRef]
- Sugui, J.A.; Pardo, J.; Chang, Y.C.; Muellbacher, A.; Zarember, K.A.; Galvez, E.M.; Brinster, L.; Zerfas, P.; Gallin, J.I.; Simon, M.M.; et al. Role of laea in the regulation of alb1, glip, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot. Cell 2007, 6, 1552–1561. [Google Scholar] [CrossRef]
- Watkins, T.N.; Liu, H.; Chung, M.; Hazen, T.H.; Hotopp, J.C.D.; Filler, S.G.; Bruno, V.M. Comparative transcriptomics of Aspergillus fumigatus strains upon exposure to human airway epithelial cells. Microb. Genom. 2018, 4. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dietl, A.-M.; Binder, U.; Bauer, I.; Shadkchan, Y.; Osherov, N.; Haas, H. Arginine Auxotrophy Affects Siderophore Biosynthesis and Attenuates Virulence of Aspergillus fumigatus. Genes 2020, 11, 423. https://doi.org/10.3390/genes11040423
Dietl A-M, Binder U, Bauer I, Shadkchan Y, Osherov N, Haas H. Arginine Auxotrophy Affects Siderophore Biosynthesis and Attenuates Virulence of Aspergillus fumigatus. Genes. 2020; 11(4):423. https://doi.org/10.3390/genes11040423
Chicago/Turabian StyleDietl, Anna-Maria, Ulrike Binder, Ingo Bauer, Yana Shadkchan, Nir Osherov, and Hubertus Haas. 2020. "Arginine Auxotrophy Affects Siderophore Biosynthesis and Attenuates Virulence of Aspergillus fumigatus" Genes 11, no. 4: 423. https://doi.org/10.3390/genes11040423
APA StyleDietl, A.-M., Binder, U., Bauer, I., Shadkchan, Y., Osherov, N., & Haas, H. (2020). Arginine Auxotrophy Affects Siderophore Biosynthesis and Attenuates Virulence of Aspergillus fumigatus. Genes, 11(4), 423. https://doi.org/10.3390/genes11040423





