Comparative Genomics of Lactobacillus crispatus from the Gut and Vagina Reveals Genetic Diversity and Lifestyle Adaptation
Abstract
:1. Introduction
2. Materials and Methods
2.1. L. crispatus Strain Isolation
2.2. Genome Sequencing, Assembly, and Annotation
2.3. Average Nucleotide Identity Calculation
2.4. Pangenome and Core Genome Analysis
2.5. Phylogenetic Analysis
2.6. Carbohydrate Active Enzyme Analysis
2.7. Statistical Analysis
3. Results
3.1. General Genome Features of the L. crispatus Strains
3.2. Pan-genome and Core Genome Analysis of L. crispatus
3.3. Phylogenetic Analysis of L. crispatus Strains
3.4. Evolution and Adaptation to Environment
3.5. Active Carbohydrate Enzymes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ruan, J. Bergey’s manual of systematic bacteriology (second edition) volume 5 and the study of Actinomycetes systematic in China. Wei Sheng Wu Xue Bao Acta Microbiol. Sin. 2013, 53, 521–530. [Google Scholar]
- Eslami, S.; Hadjati, J.; Motevaseli, E.; Mirzaei, R.; Bonab, S.F.; Ansaripour, B.; Khoramizadeh, M.R. Lactobacillus crispatus strain SJ-3C-US induces human dendritic cells (DCs) maturation and confers an anti-inflammatory phenotype to DCs. Apmis Acta Pathol. Microbiol. Immunol. Scand. 2016, 124, 697–710. [Google Scholar] [CrossRef]
- Tobita, K.; Yanaka, H.; Otani, H. Anti-allergic effects of Lactobacillus crispatus KT-11 strain on ovalbumin-sensitized BALB/c mice. Anim. Sci. J. 2010, 81, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Azam, R.; Ghafouri-Fard, S.; Tabrizi, M.; Modarressi, M.H.; Ebrahimzadeh-Vesal, R.; Daneshvar, M.; Mobasheri, M.B.; Motevaseli, E. Lactobacillus acidophilus and Lactobacillus crispatus culture supernatants downregulate expression of cancer-testis genes in the MDA-MB-231 cell line. Asian Pac. J. Cancer Prev. 2014, 15, 4255–4259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphries, C. Detecting diversity. Nature 2017, 550, S12–S14. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Q.Y.; Yang, E.C.; Yan, L.; Li, T.; Zhuang, H. Antimicrobial compounds produced by vaginal lactobacillus crispatus are able to strongly inhibit candida albicans growth, hyphal formation and regulate virulence-related gene expressions. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonio, M.A.; Petrina, M.A.; Meyn, L.A.; Hillier, S.L. Women colonised by lactobacillus crispatus have a lower risk of acquisition of bacterial vaginosis (Bv) than women colonised by other lactobacilli. Sex. Transm. Infect. 2013, 89, A232. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Liu, Z.; Zhang, X.; Chen, X.; Wang, S. Local probiotic lactobacillus crispatus and lactobacillus delbrueckii exhibit strong antifungal effects against vulvovaginal candidiasis in a rat model. Front. Microbiol. 2019, 10, 1033. [Google Scholar] [CrossRef]
- Bohbot, J.M.; Darai, E.; Bretelle, F.; Brami, G.; Daniel, C.; Cardot, J.M. Efficacy and safety of vaginally administered lyophilized Lactobacillus crispatus IP 174178 in the prevention of bacterial vaginosis recurrence. J. Gynecol. Obstet. Hum. Reprod. 2018, 47, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Stapleton, A.E.; Au-Yeung, M.; Hooton, T.M.; Fredricks, D.N.; Roberts, P.L.; Czaja, C.A.; Yarova-Yarovaya, Y.; Fiedler, T.; Cox, M.; Stamm, W.E. Randomized, placebo-controlled phase 2 trial of a Lactobacillus crispatus probiotic given intravaginally for prevention of recurrent urinary tract infection. Clin. Infect. Dis. 2011, 52, 1212–1217. [Google Scholar] [CrossRef]
- Binnewies, T.T.; Motro, Y.; Hallin, P.F.; Lund, O.; Dunn, D.; La, T.; Hampson, D.J.; Bellgard, M.; Wassenaar, T.M.; Ussery, D.W. Ten years of bacterial genome sequencing: Comparative-genomics-based discoveries. Funct. Integr. Genom. 2006, 6, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Wilson, G.; van der Gast, C. How do we compare hundreds of bacterial genomes? Curr. Opin. Microbiol. 2006, 9, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Ojala, T.; Kankainen, M.; Castro, J.; Cerca, N.; Edelman, S.; Westerlund-Wikstrom, B.; Paulin, L.; Holm, L.; Auvinen, P. Comparative genomics of Lactobacillus crispatus suggests novel mechanisms for the competitive exclusion of Gardnerella vaginalis. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Veer, C.; Hertzberger, R.Y.; Bruisten, S.M.; Tytgat, H.L.P.; Swanenburg, J.; Angelino-Bart, A.D.; Schuren, F.; Molenaar, D.; Reid, G.; de Vries, H.; et al. Comparative genomics of human Lactobacillus crispatus isolates reveals genes for glycosylation and glycogen degradation: Implications for in vivo dominance of the vaginal microbiota. Microbiome 2019, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Zhao, J.; Song, Y.; Zhang, J.; Yu, Z.; Zhang, H.; Sun, Z. Comparative genomics of the herbivore gut symbiont lactobacillus reuteri reveals genetic diversity and lifestyle adaptation. Front. Microbiol. 2018, 9, 1151. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Han, G.G.; Kim, E.B.; Choi, Y.J. Comparative genomics of Lactobacillus salivarius strains focusing on their host adaptation. Microbiol. Res. 2017, 205, 48–58. [Google Scholar] [CrossRef]
- Eisenbach, L.; Janssen, D.; Ehrmann, M.A.; Vogel, R.F. Comparative genomics of Lactobacillus curvatus enables prediction of traits relating to adaptation and strategies of assertiveness in sausage fermentation. Int. J. Food Microbiol. 2018, 286, 37–47. [Google Scholar] [CrossRef]
- Feyereisen, M.; Mahony, J.; Kelleher, P.; Roberts, R.J.; O’Sullivan, T.; Geertman, J.M.A.; van Sinderen, D. Comparative genome analysis of the Lactobacillus brevis species. BMC Genom. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Pan, M.C.; Hidalgo-Cantabrana, C.; Goh, Y.J.; Sanozky-Dawes, R.; Barrangou, R. Comparative analysis of lactobacillus gasseri and lactobacillus crispatus isolated from human urogenital and gastrointestinal tracts. Front. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Beasley, S.S.; Takala, T.M.; Reunanen, J.; Apajalahti, J.; Saris, P.E.J. Characterization and electrotransformation of Lactobacillus crispatus isolated from chicken crop and intestine. Poult. Sci. 2004, 83, 45–48. [Google Scholar] [CrossRef]
- Luo, R.B.; Liu, B.H.; Xie, Y.L.; Li, Z.Y.; Huang, W.H.; Yuan, J.Y.; He, G.Z.; Chen, Y.X.; Pan, Q.; Liu, Y.J.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Borodovsky, M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005, 33, W451–W454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Zhao, Y.B.; Wu, J.Y.; Yang, J.H.; Sun, S.X.; Xiao, J.F.; Yu, J. PGAP: Pan-genomes analysis pipeline. Bioinformatics 2012, 28, 416–418. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Gadagkar, S.R. Efficiency of the neighbor-joining method in reconstructing deep and shallow evolutionary relationships in large phylogenies. J. Mol. Evol. 2000, 51, 544–553. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Yang, B.; Ross, R.P.; Stanton, C.; Zhao, J.X.; Zhang, H.; Chen, W. Comparative genomics analysis of lactobacillus ruminis from different niches. Genes 2020, 11, 70. [Google Scholar] [CrossRef] [Green Version]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef]
- Medini, D.; Donati, C.; Tettelin, H.; Masignani, V.; Rappuoli, R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005, 15, 589–594. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, O.; O’Callaghan, J.; Sangrador-Vegas, A.; McAuliffe, O.; Slattery, L.; Kaleta, P.; Callanan, M.; Fitzgerald, G.F.; Ross, R.P.; Beresford, T. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol. 2009, 9, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broadbent, J.R.; Neeno-Eckwall, E.C.; Stahl, B.; Tandee, K.; Cai, H.; Morovic, W.; Horvath, P.; Heidenreich, J.; Perna, N.T.; Barrangou, R.; et al. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genom. 2012, 13, 533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, A.C.; Hill, J.E. Bifidobacteria isolated from vaginal and gut microbiomes are indistinguishable by comparative genomics. PLoS ONE 2018, 13, e0196290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabayek, S.; Spellerberg, B. Acid stress response mechanisms of group B streptococci. Front. Cell. Infect. Microbiol. 2017, 7, 395. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.A.; Beasley, D.E.; Dunn, R.R.; Archie, E.A. Lactobacilli dominance and vaginal pH: Why is the human vaginal microbiome unique? Front. Microbiol. 2016, 7, 1936. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C. Surviving the acid test: Responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. MMBR 2003, 67, 429–453. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.Y.; Tian, W.H.; Wan, C.X.; Jia, L.J.; Wang, L.Y.; Yuan, J.; Liu, C.M.; Zeng, M.; Wei, H. Antagonistic potential against pathogenic microorganisms and hydrogen peroxide production of indigenous lactobacilli isolated from vagina of Chinese pregnant women. Biomed. Environ. Sci. 2008, 21, 365–371. [Google Scholar] [CrossRef]
- Balkus, J.E.; Mitchell, C.; Agnew, K.; Liu, C.; Fiedler, T.; Cohn, S.E.; Luque, A.; Coombs, R.; Fredricks, D.N.; Hitti, J. Detection of hydrogen peroxide-producing Lactobacillus species in the vagina: A comparison of culture and quantitative PCR among HIV-1 seropositive women. BMC Infect. Dis. 2012, 12, 188. [Google Scholar] [CrossRef] [Green Version]
- Pascual, L.M.; Daniele, M.B.; Pajaro, C.; Barberis, L. Lactobacillus species isolated from the vagina: Identification, hydrogen peroxide production and nonoxynol-9 resistance. Contraception 2006, 73, 78–81. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Peng, T.; Oh, J.I.; Glaeser, J.; Weber, L.; Li, Q.; Klug, G. A response regulator of the OmpR family is part of the regulatory network controlling the oxidative stress response of Rhodobacter sphaeroides. Environ. Microbiol. Rep. 2019, 11, 118–128. [Google Scholar] [CrossRef]
- Kiley, P.J.; Beinert, H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 2003, 6, 181–185. [Google Scholar] [CrossRef]
- Selbach, B.P.; Chung, A.H.; Scott, A.D.; George, S.J.; Cramer, S.P.; Dos Santos, P.C. Fe-S cluster biogenesis in Gram-positive bacteria: SufU is a zinc-dependent sulfur transfer protein. Biochemistry 2014, 53, 152–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachin, L.; Loiseau, L.; Expert, D.; Barras, F. SufC: An unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 2003, 22, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Maresca, D.; Zotta, T.; Mauriello, G. Adaptation to aerobic environment of lactobacillus johnsonii/gasseri strains. Front. Microbiol. 2018, 9, 157. [Google Scholar] [CrossRef]
- Mirmonsef, P.; Hotton, A.L.; Gilbert, D.; Gioia, C.J.; Maric, D.; Hope, T.J.; Landay, A.L.; Spear, G.T. Glycogen levels in undiluted genital fluid and their relationship to vaginal ph, estrogen, and progesterone. PLoS ONE 2016, 11, e0153553. [Google Scholar] [CrossRef] [Green Version]
- Tachedjian, G.; Aldunate, M.; Bradshaw, C.S.; Cone, R.A. The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res. Microbiol. 2017, 168, 782–792. [Google Scholar] [CrossRef]
- Stern, A.; Mick, E.; Tirosh, I.; Sagy, O.; Sorek, R. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res. 2012, 22, 1985–1994. [Google Scholar] [CrossRef] [Green Version]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Gao, J.; Wang, B.; Huo, D.; Wang, Z.; Zhang, J.; Shao, Y. Whole-genome sequencing reveals the mechanisms for evolution of streptomycin resistance in Lactobacillus plantarum. J. Dairy Sci. 2018, 101, 2867–2874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKeegan, K.S.; Borges-Walmsley, M.I.; Walmsley, A.R. The structure and function of drug pumps: An update. Trends Microbiol. 2003, 11, 21–29. [Google Scholar] [CrossRef]
- Miguez Amil, S.; Jimenez-Ortega, E.; Ramirez-Escudero, M.; Talens-Perales, D.; Marin-Navarro, J.; Polaina, J.; Sanz-Aparicio, J.; Fernandez-Leiro, R. The cryo-EM structure of thermotoga maritima beta-galactosidase: Quaternary structure guides protein engineering. ACS Chem. Biol. 2020, 15, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Novy, V.; Aissa, K.; Nielsen, F.; Straus, S.K.; Ciesielski, P.; Hunt, C.G.; Saddler, J. Quantifying cellulose accessibility during enzyme-mediated deconstruction using 2 fluorescence-tagged carbohydrate-binding modules. Proc. Natl. Acad. Sci. USA 2019, 116, 22545–22551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, C.J.; Huang, M.Y.; Pang, H.; Zhao, J.; Wu, C.X.; Feng, J.X. Characterization of a novel theme C glycoside hydrolase family 9 cellulase and its CBM-chimeric enzymes. Appl. Microbiol. Biotechnol. 2017, 101, 5723–5737. [Google Scholar] [CrossRef] [PubMed]
- O’Riordan, N.; O’Callaghan, J.; Butto, L.F.; Kilcoyne, M.; Joshi, L.; Hickey, R.M. Bovine glycomacropeptide promotes the growth of Bifidobacterium longum ssp. infantis and modulates its gene expression. J. Dairy Sci. 2018, 101, 6730–6741. [Google Scholar] [CrossRef]
- Zaloba, P.; Bailey-Elkin, B.A.; Derksen, M.; Mark, B.L. Structural and biochemical insights into the peptidoglycan hydrolase domain of FlgJ from Salmonella typhimurium. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Morais, S.; Cockburn, D.W.; Ben-David, Y.; Koropatkin, N.M.; Martens, E.C.; Duncan, S.H.; Flint, H.J.; Mizrahi, I.; Bayer, E.A. Lysozyme activity of the Ruminococcus champanellensis cellulosome. Environ. Microbiol. 2016, 18, 5112–5122. [Google Scholar] [CrossRef]
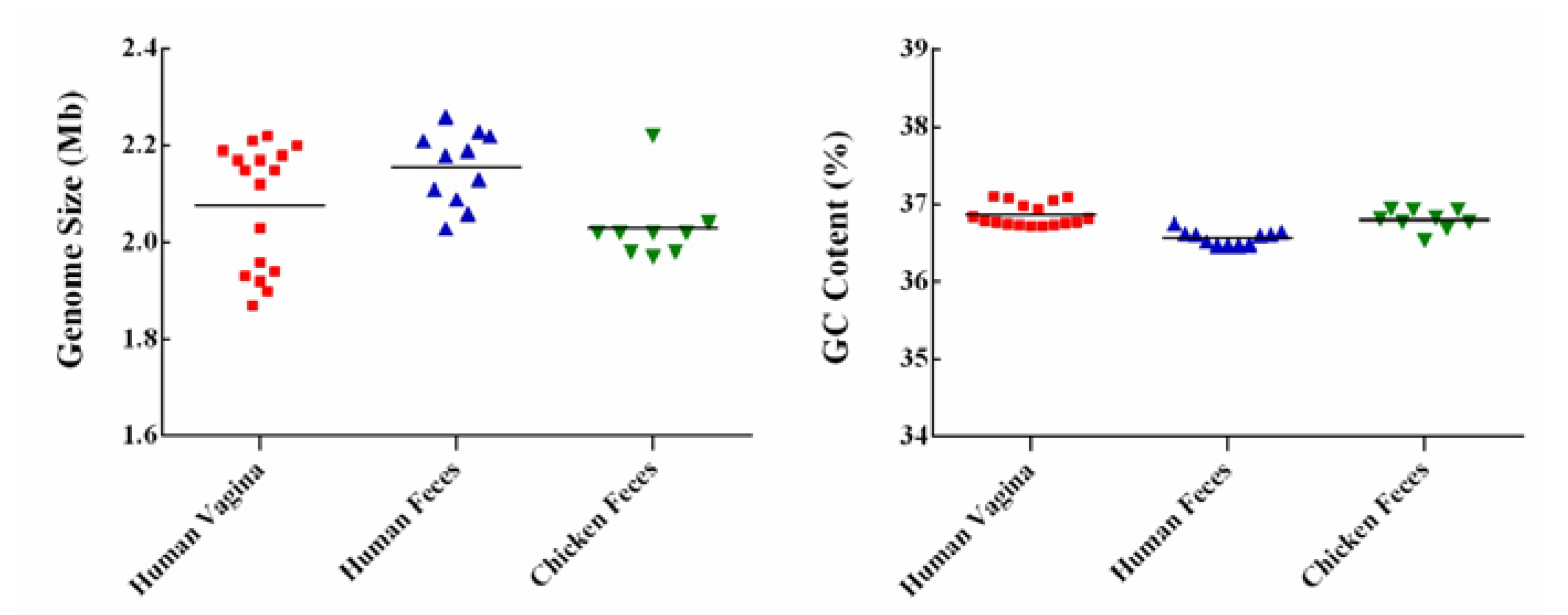
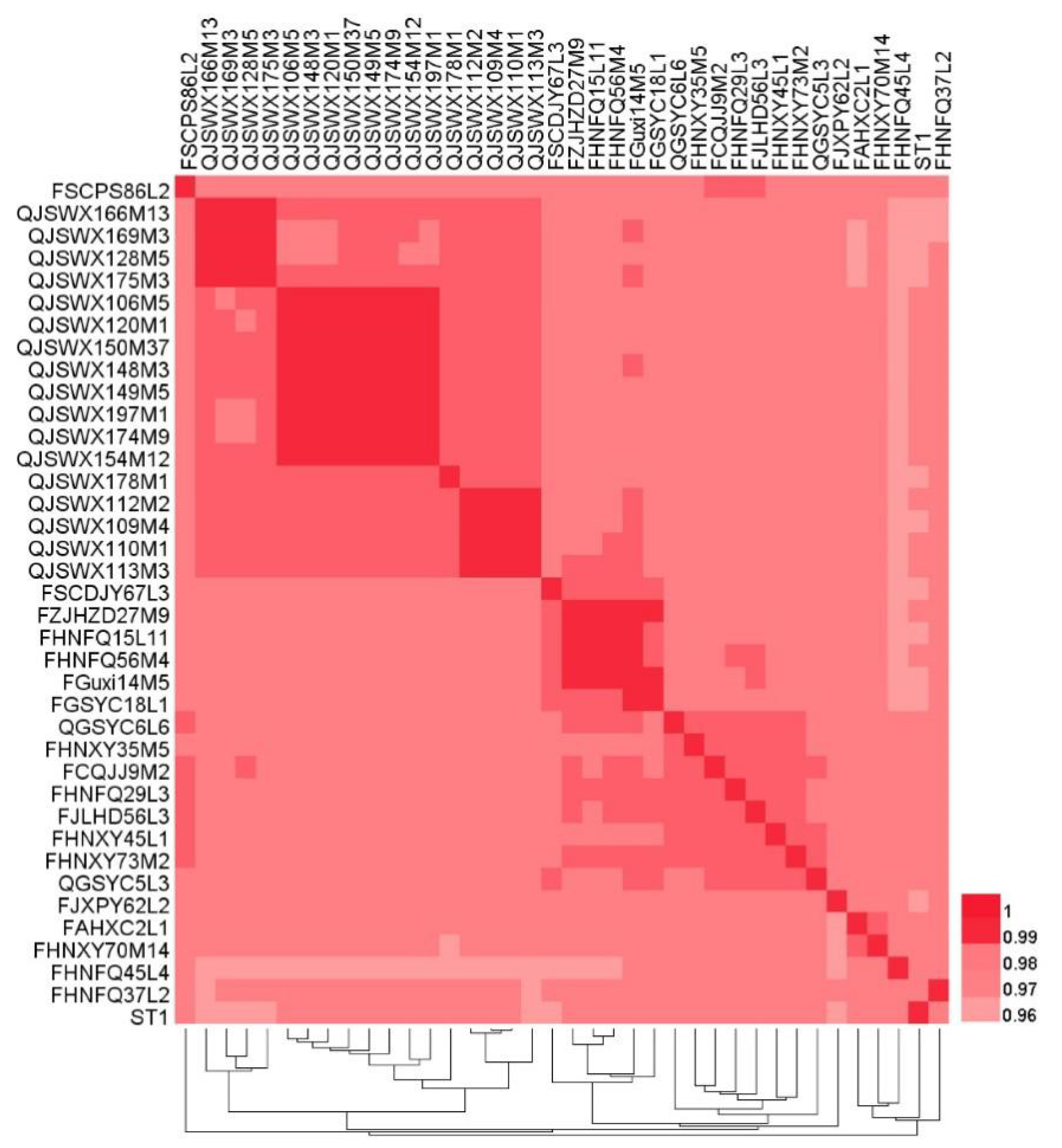

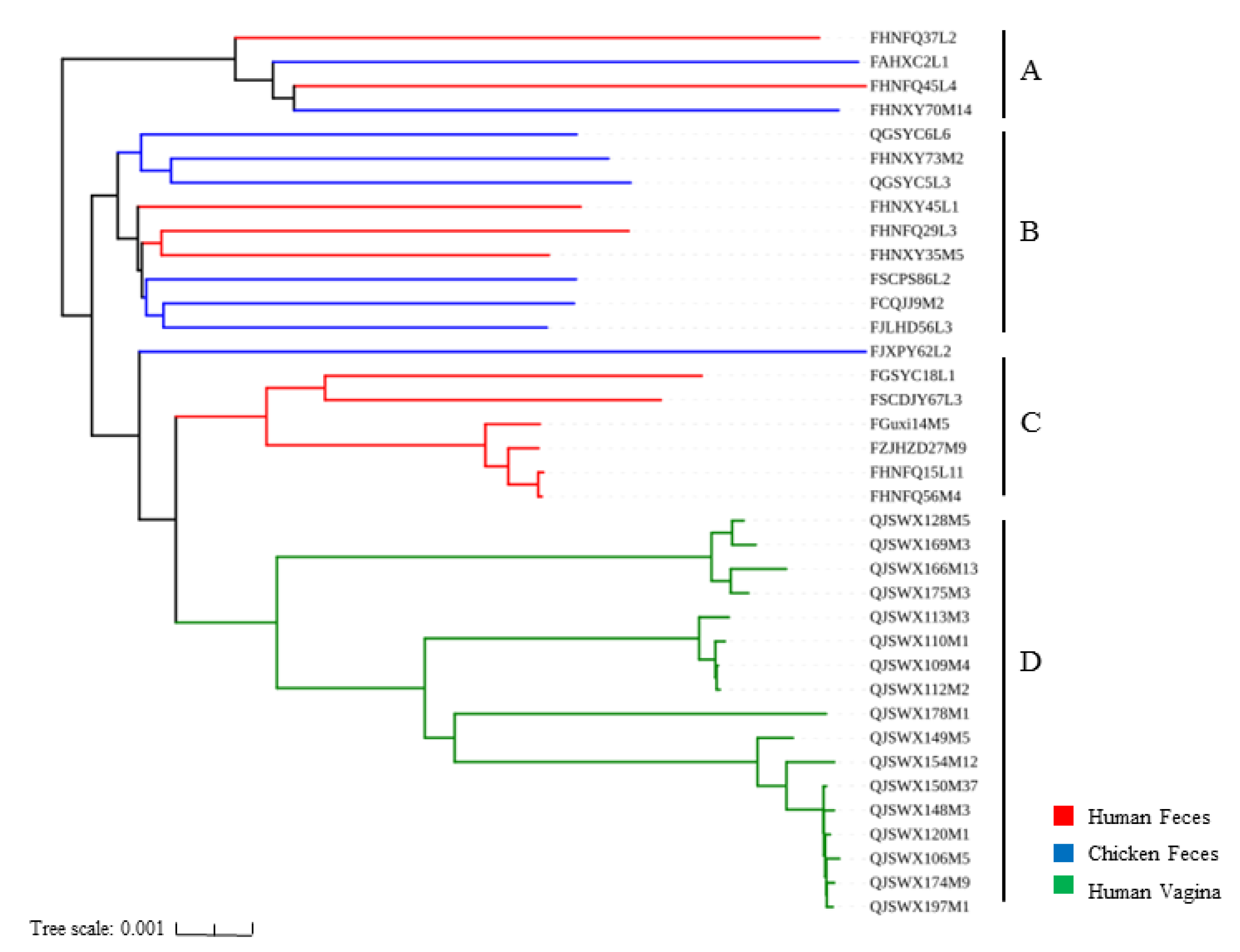

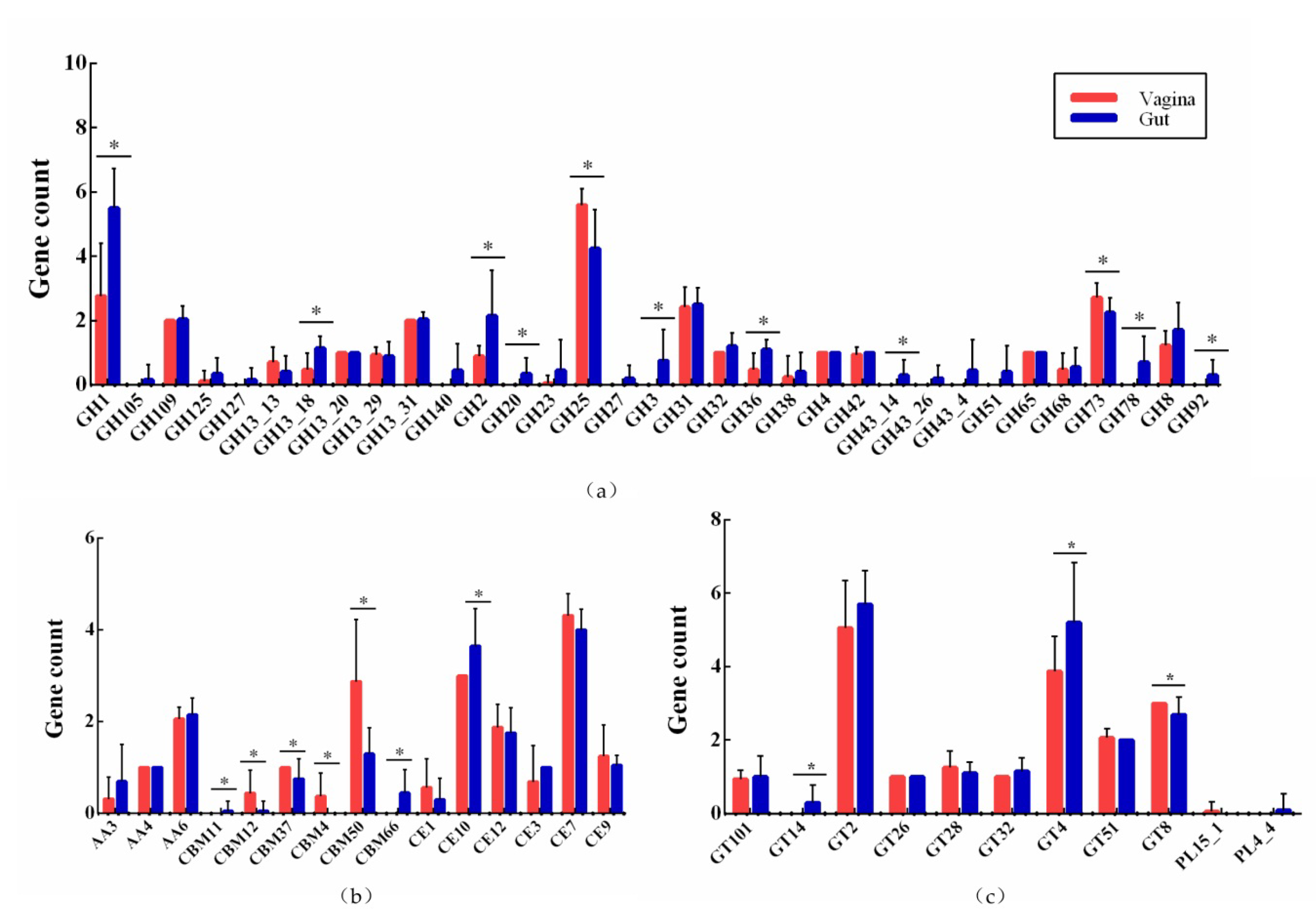
| Strains | Source | Genome Size (Mb) | GC Content (%) | CDS No. | tRNA No. | rRNA No. | Accession No. |
|---|---|---|---|---|---|---|---|
| ST1 | Chicken Feces | 2.04 | 36.9 | 2023 | 64 | 12 | SAMEA2272191 |
| FGSYC18L1 | Human Feces | 2.18 | 36.47 | 2177 | 37 | 3 | SAMN12869300 |
| FGuxi14M5 | Human Feces | 2.19 | 36.47 | 2191 | 42 | 1 | SAMN12869301 |
| FHNFQ15L11 | Human Feces | 2.26 | 36.48 | 2241 | 44 | 3 | SAMN12869302 |
| FHNFQ29L3 | Human Feces | 2.03 | 36.75 | 2053 | 56 | 3 | SAMN12869303 |
| FHNFQ37L2 | Human Feces | 2.09 | 36.60 | 2091 | 45 | 1 | SAMN12869304 |
| FHNFQ45L4 | Human Feces | 2.13 | 36.64 | 2190 | 44 | 3 | SAMN12869305 |
| FHNFQ56M4 | Human Feces | 2.23 | 36.52 | 2218 | 40 | 3 | SAMN12869306 |
| FHNXY35M5 | Human Feces | 2.11 | 36.61 | 2121 | 40 | 3 | SAMN12869307 |
| FHNXY45L1 | Human Feces | 2.06 | 36.62 | 2066 | 35 | 0 | SAMN12869308 |
| FSCDJY67L3 | Human Feces | 2.21 | 36.61 | 2190 | 45 | 1 | SAMN12869309 |
| FZJHZD27M9 | Human Feces | 2.22 | 36.47 | 2243 | 41 | 1 | SAMN12869310 |
| FAHXC2L1 | Chicken Feces | 2.04 | 36.81 | 2023 | 46 | 4 | SAMN12869311 |
| FCQJJ9M2 | Chicken Feces | 1.97 | 36.93 | 1988 | 44 | 3 | SAMN12869312 |
| FHNXY70M14 | Chicken Feces | 1.98 | 36.92 | 1985 | 45 | 3 | SAMN12869313 |
| FHNXY73M2 | Chicken Feces | 2.02 | 36.69 | 1989 | 49 | 3 | SAMN12869314 |
| FJLHD56L3 | Chicken Feces | 2.02 | 36.77 | 1970 | 38 | 3 | SAMN12869315 |
| FJXPY62L2 | Chicken Feces | 2.02 | 36.92 | 1977 | 41 | 3 | SAMN12869316 |
| FSCPS86L2 | Chicken Feces | 2.22 | 36.53 | 2203 | 49 | 3 | SAMN12869317 |
| QGSYC5L3 | Chicken Feces | 2.02 | 36.82 | 2003 | 45 | 3 | SAMN12869318 |
| QGSYC6L6 | Chicken Feces | 1.98 | 36.77 | 2004 | 43 | 3 | SAMN12869319 |
| QJSWX106M5 | Human Vagina | 2.15 | 36.82 | 2194 | 72 | 4 | SAMN12869320 |
| QJSWX109M4 | Human Vagina | 2.18 | 36.72 | 2231 | 53 | 3 | SAMN12869321 |
| QJSWX110M1 | Human Vagina | 2.17 | 36.72 | 2227 | 41 | 4 | SAMN12869322 |
| QJSWX112M2 | Human Vagina | 2.17 | 36.73 | 2241 | 53 | 3 | SAMN12869323 |
| QJSWX113M3 | Human Vagina | 2.22 | 36.77 | 2309 | 46 | 3 | SAMN12869324 |
| QJSWX120M1 | Human Vagina | 2.15 | 36.77 | 2206 | 68 | 4 | SAMN12869325 |
| QJSWX128M5 | Human Vagina | 1.94 | 36.73 | 1972 | 41 | 0 | SAMN12869326 |
| QJSWX148M3 | Human Vagina | 1.87 | 37.05 | 1869 | 43 | 2 | SAMN12869327 |
| QJSWX149M5 | Human Vagina | 1.96 | 37.08 | 1981 | 53 | 3 | SAMN12869328 |
| QJSWX150M37 | Human Vagina | 1.90 | 36.98 | 1914 | 40 | 3 | SAMN12869329 |
| QJSWX154M12 | Human Vagina | 2.03 | 37.1 | 2108 | 42 | 2 | SAMN12869330 |
| QJSWX166M13 | Human Vagina | 1.92 | 36.75 | 1962 | 42 | 1 | SAMN12869331 |
| QJSWX169M3 | Human Vagina | 2.21 | 36.74 | 2325 | 66 | 7 | SAMN12869332 |
| QJSWX174M9 | Human Vagina | 1.93 | 37.09 | 1969 | 49 | 3 | SAMN12869333 |
| QJSWX175M3 | Human Vagina | 2.12 | 36.79 | 2112 | 50 | 2 | SAMN12869334 |
| QJSWX178M1 | Human Vagina | 2.19 | 36.84 | 2243 | 71 | 6 | SAMN12869335 |
| QJSWX197M1 | Human Vagina | 2.20 | 36.94 | 2275 | 43 | 3 | SAMN12869336 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Zhang, L.; Ross, P.; Zhao, J.; Zhang, H.; Chen, W. Comparative Genomics of Lactobacillus crispatus from the Gut and Vagina Reveals Genetic Diversity and Lifestyle Adaptation. Genes 2020, 11, 360. https://doi.org/10.3390/genes11040360
Zhang Q, Zhang L, Ross P, Zhao J, Zhang H, Chen W. Comparative Genomics of Lactobacillus crispatus from the Gut and Vagina Reveals Genetic Diversity and Lifestyle Adaptation. Genes. 2020; 11(4):360. https://doi.org/10.3390/genes11040360
Chicago/Turabian StyleZhang, Qiuxiang, Lili Zhang, Paul Ross, Jianxin Zhao, Hao Zhang, and Wei Chen. 2020. "Comparative Genomics of Lactobacillus crispatus from the Gut and Vagina Reveals Genetic Diversity and Lifestyle Adaptation" Genes 11, no. 4: 360. https://doi.org/10.3390/genes11040360
APA StyleZhang, Q., Zhang, L., Ross, P., Zhao, J., Zhang, H., & Chen, W. (2020). Comparative Genomics of Lactobacillus crispatus from the Gut and Vagina Reveals Genetic Diversity and Lifestyle Adaptation. Genes, 11(4), 360. https://doi.org/10.3390/genes11040360




