The Polycomb Orthologues in Teleost Fishes and Their Expression in the Zebrafish Model
Abstract
1. Introduction
2. Materials and Methods
2.1. In Silico Identification and Analyses of Pc/CBX Sequences in Databases
2.2. Zebrafish Maintenance, Embryo Preparation and Animal Ethics Statements
2.3. Whole-Mount In Situ Hybridization
- ISH_cbx4a1_508F: 5′-TAATACGACTCACTATAGGGGTCGAGCAGCATATGAGCGG-3′
- ISH_cbx4a1_1921R: 5′-GATTTAGGTGACACTATAGAATTTGCCAGGGGCATGAAT-3′
- ISH_cbx6a2_1210F: 5′-TAATACGACTCACTATAGGGGTGGGTCGAGCTTCGGTCTGCC-3′
- ISH_cbx6a2_1460R: 5′-GATTTAGGTGACACTATAGGCAGTTGTGGTGGGAGAGTTTGGGT-3′
- ISH_cbx6b_1037F: 5′-TAATACGACTCACTATAGGGCCCCTTCGACTAAGGCCCCTGA-3′
- ISH_cbx6b_1633R: 5′-GATTTAGGTGACACTATAGGCTTGACCACATCCAAGCCCAACC-3′
- ISH_cbx7b_304F: 5′-TAATACGACTCACTATAGGGGCTCATCTGCCACTCTCGCTGGA-3′
- ISH_cbx7b_692R: 5′-GATTTAGGTGACACTATAGAAGCCTCTCGCAACCAAAGCCTCT-3′
2.4. RNA Extraction and RT-PCR
- cbx2_984F: 5′-TCAAAGGTCAGCATCTCTCCCGA-3′
- cbx2_1111R: 5′-TCAAATCAGTCCCGTTTCCCTGC-3′
- cbx4a1_656F: 5′-GGAACCTCCCTCCAGCCCTAC-3′
- cbx4a1_785R: 5′-AGTGTCCGCTCTGCTTTGTTAGC-3′
- cbx6a2_1295F: 5′-CCGATTGGCACCCTGAAATGGC-3′
- cbx6a2_1461R: 5′-TGCAGTTGTGGTGGGAGAGTTTGG-3′
- cbx6b_1275F: 5′-ACCAGAGGTGCCCGTGTCAGA-3′
- cbx6b_1437R: 5′-AGAAGAAGCAGAAGGCGGATGGC-3′
- cbx7_1691F: 5′-CCTCACCTGCTTGTCCTGTAACCA-3′
- cbx7_1933R: 5′-AGGCTCGGCTTTGATTTCTGCC-3′
- cbx7b_304F: 5′-GCTCATCTGCCACTCTCGCTGG-3′
- cbx7b_428R: 5′-TCCCTTCTGTCCCACTCTTCCTCA-3′
- cbx8a_831F: 5′-GGTTGGCTCTGGTGGTGCCATAA-3′
- cbx8a_977R: 5′-TTCAAACAAGGCAGCCAGGACTG-3′
- cbx8b_787F: 5′-AGCAACGTCATTCCAAGCAGGAGT -3′
- cbx8b_1045R: 5′-AGGGTCTCCAAGCATCTTCCACC -3′
- Dr_cDNA_ube2a_F: 5′-TGACTGTTGACCCACCTTACAG-3′
- Dr_cDNA_ube2a_R: 5′-CAAATAAAAGCAAGTAACCCCG-3′
- Dr_beta-actin_cDNA_5: 5′-CGTGACATCAAGGAGAAGCT-3′
- Dr_beta-actin_cDNA_3: 5′-ATCCACATCTGCTGGAAGGT-3′
3. Results
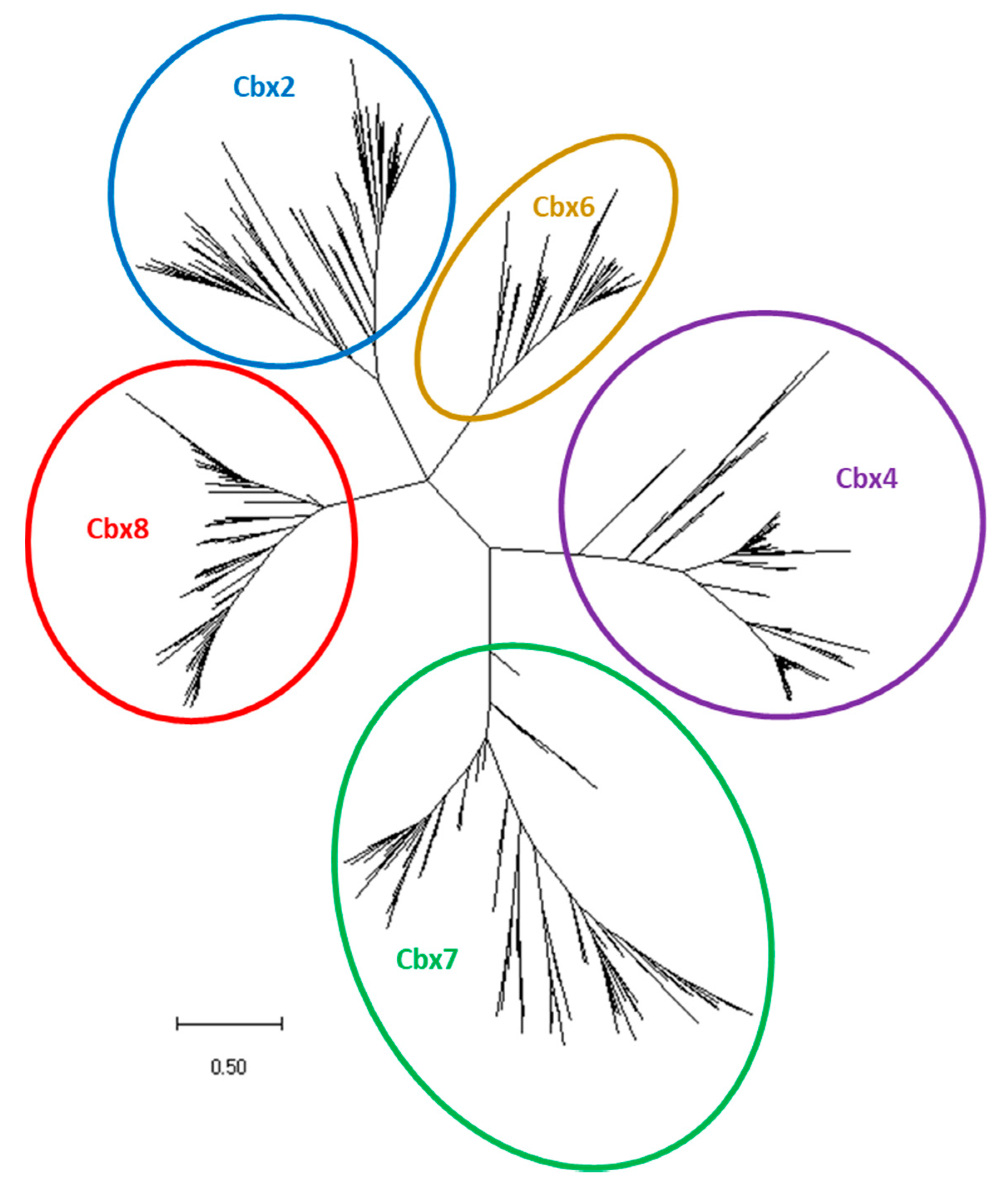
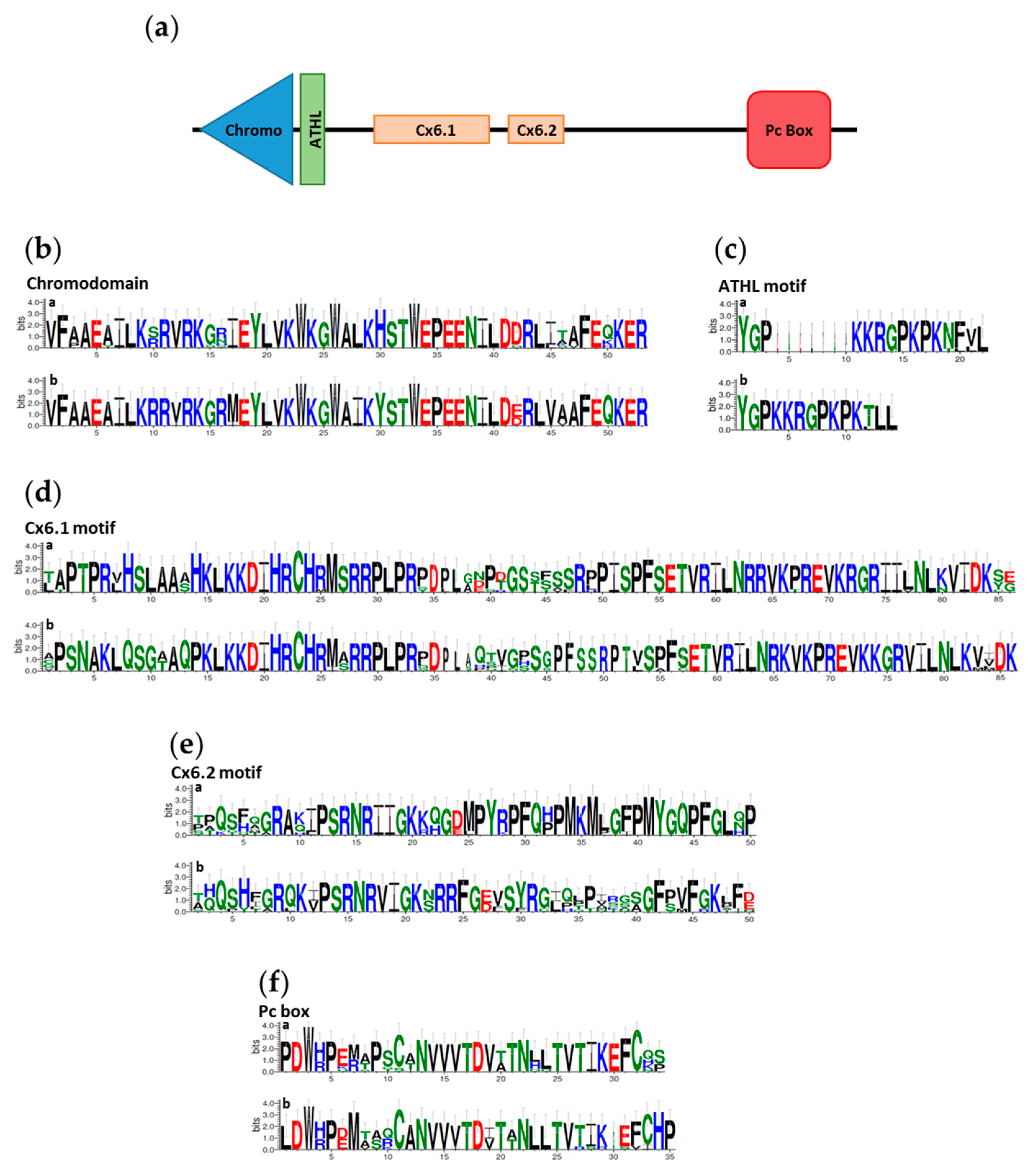
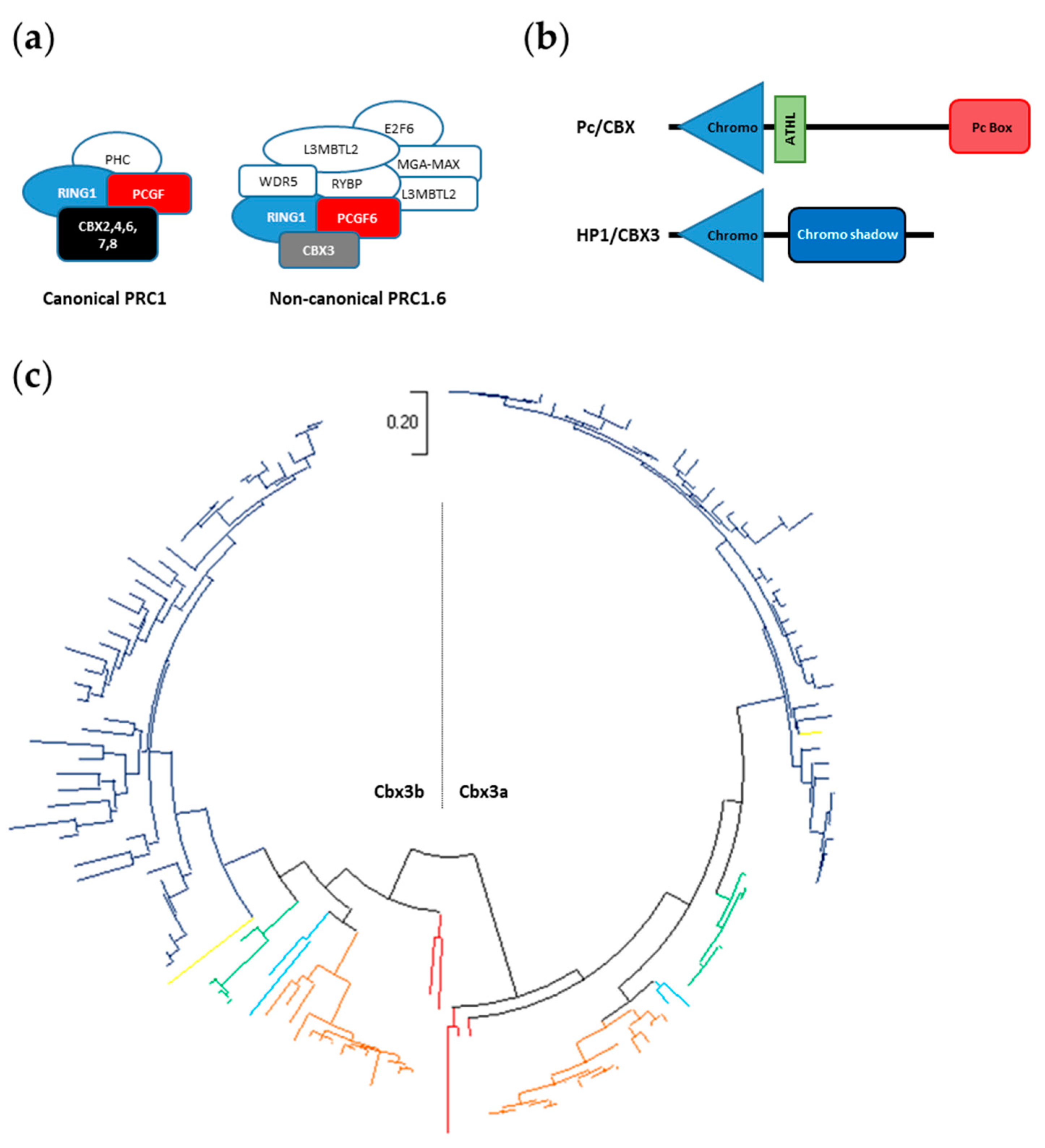
3.1. In Silico Identification of Teleost Pc Orthologues
3.2. The cbx6 Paralogue in Teleost Fishes
3.3. The cbx2 Paralogue in Teleost Fishes
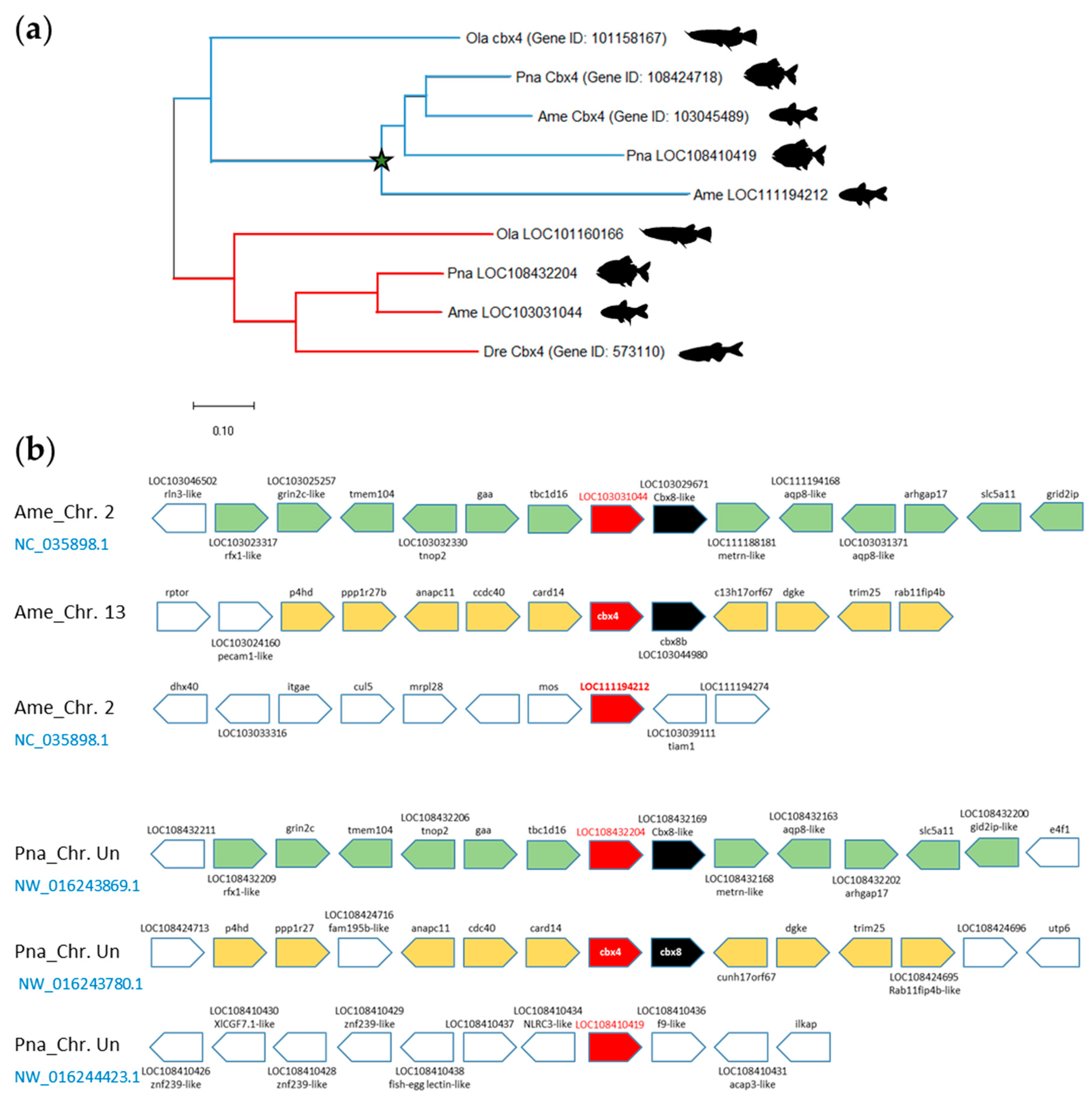
3.4. The cbx4, cbx7 and cbx8 Paralogues in Teleost Fishes
3.5. The cbx3 Genes in Teleost Fishes
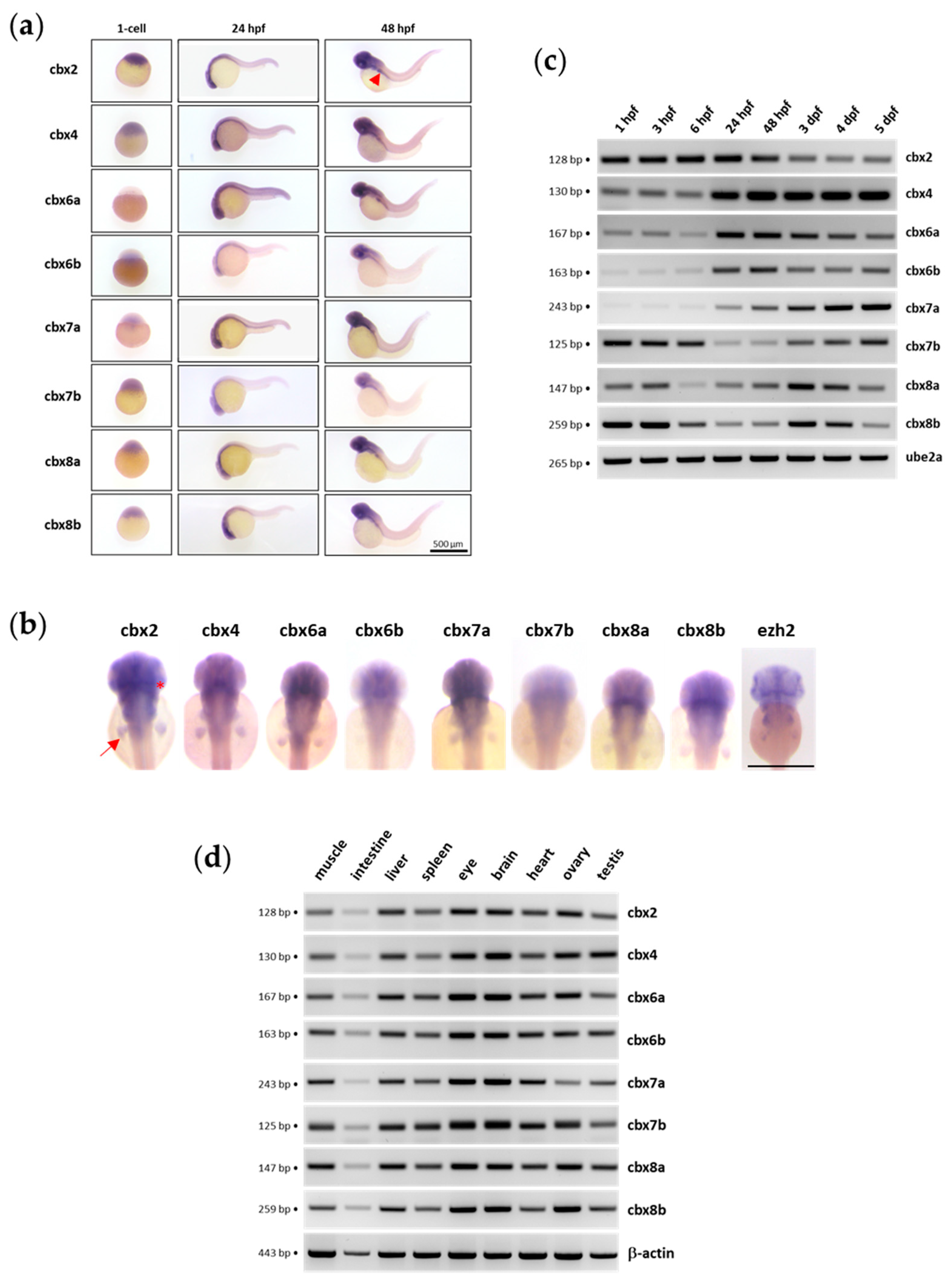
3.6. Pc Gene Expression in the Zebrafish Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Family | NCBI Genome | Ensembl ID | Assembly | ||
|---|---|---|---|---|---|
| Aca | Eastern happy (Astatotilapia calliptera) | Cichlidae | ID: 15018 | ENSACAG | fAstCal1.2 |
| Aci | Midas cichlid (Amphilophus citrinellus) | Cichlidae | ENSACIG | Midas_v5 | |
| Ame | Mexican cavefish (Astyanax mexicanus) | Characidae | ID: 13073 | ENSAMEG | Astyanax_mexicanus-2.0 |
| Aoc | Clown anemonefish (Amphiprion ocellaris) | Pomacentridae | ID: 8181 | ENSAOCG | AmpOce1.0 |
| Ape | Orange clownfish (Amphiprion percula) | Pomacentridae | ENSAPEG | Nemo_v1 | |
| Apo | Spiny chromis (Acanthochromis polyacanthus) | Pomacentridae | ID: 54316 | ENSAPOG | ASM210954v1 |
| Ate | Climbing perch (Anabas testudineus) | Anabantidae | ID: 33761 | ENSATEG | fAnaTes1.1 |
| Bsp | Siamese fighting fish (Betta splendens) | Osphronemidae | ID: 36335 | fBetSpl5.2 | |
| Cau | Goldfish (Carassius auratus) | Cyprinidae | ID: 10773 | ASM336829v1 | |
| Cca | Common carp (Cyprinus carpio) | Cyprinidae | ID: 10839 | common carp genome | |
| Cgo | Channel bull blenny (Cottoperca gobio ) | Bovichtidae | ID: 76087 | CotGob3.1 | |
| Cha | Atlantic herring (Clupea harengus) | Clupeidae | ID: 15477 | Ch_v2.0.2 | |
| Cse | Tongue sole (Cynoglossus semilaevis) | Cynoglossidae | ID: 11788 | ENSSCEG | Cse_v1.0 |
| Cva | Sheepshead minnow (Cyprinodon variegatus) | Cyprinodontidae | ID: 13078 | ENSCVAG | C_variegatus-1.0 |
| Dcl | Denticle herring (Denticeps clupeoides) | Denticipitidae | ID: 7889 | fDenClu1.1 | |
| Dre | Zebrafish (Danio rerio) | Cyprinidae | ID: 50 | ENSDARG | GRCz11 |
| Eel | Electric eel (Electrophorus electricus) | Gymnotidae | ID: 72955 | ENSEELG | Ee_SOAP_WITH_SSPACE |
| Elu | Northern pike (Esox lucius) | Esocidae | ID: 22932 | ENSELUG | Eluc_v4 / Eluc_V3 |
| Ena | Live sharksucker (Echeneis naucrates) | Echeneidae | ID: 22910 | fEcheNa1.1 | |
| Fhe | Mummichog (Fundulus heteroclitus) | Fundulidae | ID: 743 | ENSFHEG | Fundulus_heteroclitus-3.0.2 |
| Gac | Stickleback (Gasterosteus aculeatus) | Gasterosteidae | ENSGACG | BROAD S1 | |
| Gaf | Western mosquitofish (Gambusia affinis) | Poeciliidae | ENSGAFG | ASM309773v1 | |
| Gmo | Cod (Gadus morhua) | Gadidae | ENSGMOG | gadMor1 | |
| Gwi | Blunt-snouted clingfish (Gouania willdenowi) | Gobiesocidae | ID: 76090 | ENSGWIG | fGouWil2.1 |
| Hbu | Burton’s mouthbrooder (Haplochromis burtoni) | Cichlidae | ID: 3328 | ENSHBUG | AstBur1.0 |
| Hco | Tiger tail seahorse (Hippocampus comes) | Syngnathidae | ID: 16754 | ENSHCOG | H_comes_QL1_v1 |
| Hhu | Huchen (Hucho hucho) | Salmonidae | ENSHHUG | ASM331708v1 | |
| Ipu | Channel catfish (Ictalurus punctatus) | Ictaluridae | ID: 198 | ENSIPUG | IpCoco_1.2 |
| Kma | Mangrove rivulus (Kryptolebias marmoratus) | Rivulinae | ID: 23212 | ENSKMAG | ASM164957v1 |
| Lbe | Ballan wrasse (Labrus bergylta) | Labridae | ID: 14767 | ENSLBEG | BallGen_V1 |
| Lca | Barramundi perch (Lates calcarifer) | Centropomidae | ID: 14180 | ENSLCAG | ASM164080v1 ASB_HGAPassembly_v1 |
| Lcr | Large yellow croaker (Larimichthys crocea) | Sciaenidae | ID: 12197 | L_crocea_2.0 | |
| Mal | Swamp eel (Monopterus albus) | Synbranchidae | ID: 24053 | ENSMALG | M_albus_1.0 |
| Mar | Zig-zag eel (Mastacembelus armatus) | Mastacembelidae | ID: 31700 | ENSMAMG | fMasArm1.1 |
| Mmo | Ocean sunfish (Mola mola) | Molidae | ENSMMOG | ASM169857v1 | |
| Mze | Zebra mbuna (Maylandia zebra) | Cichlidae | ID: 2640 | ENSMZEG | M_zebra_UMD2a |
| Nbr | Lyretail cichlid (Neolamprologus brichardi) | Cichlidae | ID: 3329 | ENSNBRG | NeoBri1.0 |
| Nco | Black rockcod (Notothenia coriiceps) | Nototheniidae | ID: 10753 | NC01 | |
| Nfu | Turquoise killifish (Nothobranchius furzeri) | Nothobranchiidae | ID: 2642 | Nfu_20140520 | |
| Oki | Coho salmon (Oncorhynchus kisutch) | Salmonidae | ID: 13127 | Okis_V1 | |
| Ola | Medaka (Oryzia latipes) | Adrianichthyidae | ID: 542 | ENSORLG | ASM223467v1 |
| Omy | Rainbow trout (Oncorhynchus mykiss) | Salmonidae | ID: 196 | Omyk_1.0 | |
| Oni | Tilapia (Oreochromis niloticus) | Cichlidae | ID: 197 | ENSONIG | O_niloticus_UMD_NMBU Orenil1.0 |
| Pfl | Yellow perch (Perca flavescens) | Percidae | ID: 15707 | PFLA_1.0 | |
| Pfo | Amazon molly (Poecilia formosa) | Poeciliidae | ID: 13072 | ENSPFOG | Poecilia_formosa-5.1.2 |
| Pki | Elephantfish (Paramormyrops kingsleyae) | Mormyridae | ID: 66590 | ENSPKIG | PKINGS_0.1 |
| Pla | Sailfin molly (Poecilia latipinna) | Poeciliidae | ID: 17477 | ENSPLAG | P_latipinna-1.0 |
| Pma | Mudskipper (Periophthalmus magnuspinnatus) | Oxudercidae | ENSPMG | PM.fa | |
| Pme | Shortfin molly (Poecilia mexicana) | Poeciliidae | ID: 14658 | ENSPMEG | P_mexicana-1.0 |
| Pna | Red-bellied piranha (Pygocentrus nattereri) | Serrasalmidae | ID: 10452 | ENSPNAG | Pygocentrus_nattereri-1.0.2 |
| Pny | Makobe Island cichlid (Pundamilia nyererei) | Cichlidae | ID: 3330 | ENSPNYG | PunNye1.0 |
| Pra | Indian glassy fish (Parambassis ranga) | Ambassidae | ID: 76088 | fParRan2.1 | |
| Pre | Guppy (Poecilia reticulata) | Poeciliidae | ID: 23338 | ENSPREG | Guppy_female_1.0_MT |
| Sal | Arctic char (Salvelinus alpinus) | Salmonidae | ID: 12179 | ASM291031v2 | |
| San | 安水金线鲃 (Sinocyclocheilus anshuiensis) | Cyprinidae | ID: 38362 | SAMN03320099.WGS_v1.1 | |
| Sdu | Greater amberjack (Seriola dumerili) | Carangidae | ID: 12614 | ENSSDUG | Sdu_1.0 |
| Sfa | Jewelled blenny (Salarias fasciatus) | Blenniidae | ID: 7248 | ENSSFAG | fSalaFa1.1 |
| Sfo | Asian bonytongue (Scleropages formosus) | Osteoglossidae | ID: 7635 | ENSSFOG | fSclFor1.1 / ASM162426v1 |
| Sgr | Golden-line barbel (Sinocyclocheilus grahami) | Cyprinidae | ID: 9142 | SAMN03320097.WGS_v1.1 | |
| Sla | Yellowtail amberjack (Seriola lalandi dorsalis) | Carangidae | ID: 12613 | ENSSLDG | Sedor1 |
| Sma | Turbot (Scophthalmus maximus) | Scophthalmidae | ENSSMAG | ASM318616v1 | |
| Spa | Bicolor damselfish (Stegastes partitus) | Pomacentridae | ID: 13077 | ENSSPAG | Stegastes_partitus-1.0.2 |
| Srh | 犀角金线鲃 (Sinocyclocheilus rhinocerous) | Cyprinidae | ID: 38394 | SAMN03320098_v1.1 | |
| Ssa | Atlantic salmon (Salmo salar) | Salmonidae | ID: 369 | ICSASG_v2 | |
| Tni | Tetraodon (Tetraodon nigroviridis) | Tetraodontidae | ENSTNIG | TETRAODON 8.0 | |
| Tru | Japanese puffer (Takifugu rubripes) | Tetraodontidae | ID: 63 | ENSTRUG | fTakRub1.2 / FUGU5 |
| Xco | Monterrey platyfish (Xiphophorus couchianus) | Poeciliidae | ID: 16825 | ENSXCOG | X_couchianus-1.0 Xiphophorus_couchianus-4.0.1 |
| Xma | Platyfish (Xiphophorus maculatus) | Poeciliidae | ID: 10764 | ENSXMAG | X_maculatus-5.0-male |
References
- Nelson, J.S. Fishes of the World, 4th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Faircloth, B.C.; Sorenson, L.; Santini, F.; Alfaro, M.E. A Phylogenomic Perspective on the Radiation of Ray-Finned Fishes Based upon Targeted Sequencing of Ultraconserved Elements (UCEs). PLoS ONE 2013, 8, e65923. [Google Scholar] [CrossRef]
- Sallan, L. Major issues in the origins of ray-finned fish (Actinopterygii) biodiversity. Biol. Rev. Camb. Phylos. Soc. 2014, 89, 950–971. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Force, A.; Yan, Y.-L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.-L.; et al. Zebrafish hox Clusters and Vertebrate Genome Evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.S.; Van De Peer, Y.; Braasch, I.; Meyer, A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 1661–1679. [Google Scholar] [CrossRef] [PubMed]
- Hoegg, S.; Brinkmann, H.; Taylor, J.S.; Meyer, A. Phylogenetic Timing of the Fish-Specific Genome Duplication Correlates with the Diversification of Teleost Fish. J. Mol. Evol. 2004, 59, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Sémon, M.; Wolfe, K.H. Reciprocal gene loss between Tetraodon and zebrafish after whole genome duplication in their ancestor. Trends Genet. 2007, 23, 108–112. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, J. Rapid Subfunctionalization Accompanied by Prolonged and Substantial Neofunctionalization in Duplicate Gene Evolution. Genetics 2005, 169, 1157–1164. [Google Scholar] [CrossRef]
- Brunet, F.; Crollius, H.R.; Paris, M.; Aury, J.-M.; Gibert, P.; Jaillon, O.; Laudet, V.; Robinson-Rechavi, M. Gene Loss and Evolutionary Rates Following Whole-Genome Duplication in Teleost Fishes. Mol. Biol. Evol. 2006, 23, 1808–1816. [Google Scholar] [CrossRef]
- Sémon, M.; Wolfe, K.H. Rearrangement Rate following the Whole-Genome Duplication in Teleosts. Mol. Biol. Evol. 2006, 24, 860–867. [Google Scholar]
- Hufton, A.; Groth, D.; Vingron, M.; Lehrach, H.; Poustka, A.J.; Panopoulou, G. Early vertebrate whole genome duplications were predated by a period of intense genome rearrangement. Genome Res. 2008, 18, 1582–1591. [Google Scholar] [CrossRef]
- Robinson-Rechavi, M.; Laudet, V. Evolutionary rates of duplicate genes in fish and mammals. Mol. Biol. Evol. 2001, 18, 681–683. [Google Scholar] [CrossRef] [PubMed]
- Di Croce, L.; Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013, 20, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Sauvageau, G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010, 7, 299–313. [Google Scholar] [CrossRef]
- Surface, L.E.; Thornton, S.R.; Boyer, L.A. Polycomb Group Proteins Set the Stage for Early Lineage Commitment. Cell Stem Cell 2010, 7, 288–298. [Google Scholar] [CrossRef]
- Shao, Z.; Raible, F.; Mollaaghababa, R.; Guyon, J.R.; Wu, C.-T.; Bender, W.; Kingston, R.E. Stabilization of Chromatin Structure by PRC1, a Polycomb Complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genome Res. 2002, 16, 2893–2905. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef]
- Ringrose, L.; Paro, R. Epigenetic Regulation of Cellular Memory by the Polycomb and Trithorax Group Proteins. Annu. Rev. Genet. 2004, 38, 413–443. [Google Scholar] [CrossRef]
- Völkel, P.; Angrand, P.-O. The control of histone lysine methylation in epigenetic regulation. Biochimie 2007, 89, 1–20. [Google Scholar] [CrossRef]
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.; Kingston, R.E.; A Simon, J. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef]
- Nekrasov, M.; Klymenko, T.; Fraterman, S.; Papp, B.; Oktaba, K.; Köcher, T.; Cohen, A.; Stunnenberg, H.G.; Wilm, M.; Müller, J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007, 26, 4078–4088. [Google Scholar] [CrossRef] [PubMed]
- Fischle, W.; Wang, Y.; Jacobs, S.A.; Kim, Y.; Allis, C.D.; Khorasanizadeh, S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003, 17, 1870–1881. [Google Scholar] [CrossRef] [PubMed]
- Min, J.; Zhang, Y.; Xu, R.-M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genome Res. 2003, 17, 1823–1828. [Google Scholar] [CrossRef] [PubMed]
- Levine, S.S.; Weiss, A.; Erdjument-Bromage, H.; Shao, Z.; Tempst, P.; Kingston, R.E. The Core of the Polycomb Repressive Complex Is Compositionally and Functionally Conserved in Flies and Humans. Mol. Cell. Biol. 2002, 22, 6070–6078. [Google Scholar] [CrossRef]
- Connelly, K.E.; Dykhuizen, E.C. Compositional and functional diversity of canonical PRC1 complexes in mammals. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2017, 1860, 233–245. [Google Scholar] [CrossRef]
- Maertens, G.; El Messaoudi-Aubert, S.; Raček, T.; Stock, J.K.; Nicholls, J.; Rodríguez-Niedenfuhr, M.; Gil, J.; Peters, G. Several Distinct Polycomb Complexes Regulate and Co-Localize on the INK4a Tumor Suppressor Locus. PLOS ONE 2009, 4, e6380. [Google Scholar] [CrossRef]
- Vandamme, J.; Völkel, P.; Rosnoblet, C.; Le Faou, P.; Angrand, P.-O. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol. Cell. Proteomics 2011, 10. [Google Scholar] [CrossRef]
- Ren, X.; Kerppola, T. REST Interacts with Cbx Proteins and Regulates Polycomb Repressive Complex 1 Occupancy at RE1 Elements. Mol. Cell. Biol. 2011, 31, 2100–2110. [Google Scholar] [CrossRef][Green Version]
- Trojer, P.; Cao, A.R.; Gao, Z.; Li, Y.; Zhang, J.; Xu, X.; Li, G.; Losson, R.; Erdjument-Bromage, H.; Tempst, P.; et al. L3MBTL2 Protein Acts in Concert with PcG Protein-Mediated Monoubiquitination of H2A to Establish a Repressive Chromatin Structure. Mol. Cell 2011, 42, 438–450. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF Homologs, CBX Proteins, and RYBP Define Functionally Distinct PRC1 Family Complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef]
- O’Loghlen, A.; Muñoz-Cabello, A.M.; Gaspar-Maia, A.; Wu, H.-A.; Banito, A.; Kunowska, N.; Racek, T.; Pemberton, H.N.; Beolchi, P.; Lavial, F.; et al. MicroRNA Regulation of Cbx7 Mediates a Switch of Polycomb Orthologs during ESC Differentiation. Cell Stem Cell 2012, 10, 33–46. [Google Scholar] [CrossRef]
- Bernstein, E.; Duncan, E.M.; Masui, O.; Gil, J.; Heard, E.; Allis, C.D. Mouse Polycomb Proteins Bind Differentially to Methylated Histone H3 and RNA and Are Enriched in Facultative Heterochromatin. Mol. Cell. Biol. 2006, 26, 2560–2569. [Google Scholar] [CrossRef] [PubMed]
- Morey, L.; Pascual, G.; Cozzuto, L.; Roma, G.; Wutz, A.; Benitah, S.A.; Di Croce, L. Nonoverlapping Functions of the Polycomb Group Cbx Family of Proteins in Embryonic Stem Cells. Cell Stem Cell 2012, 10, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Klauke, K.; Radulović, V.; Broekhuis, M.; Weersing, E.; Zwart, E.; Olthof, S.; Ritsema, M.; Bruggeman, S.; Wu, X.; Helin, K.; et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nature 2013, 15, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Le Faou, P.; Völkel, P.; Angrand, P.-O. The zebrafish genes encoding the Polycomb repressive complex (PRC) 1. Gene 2011, 475, 10–21. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; López, R.; McWilliam, H.; Remmert, M.; Soeding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Boil. Evol. 1987, 4, 406–425. [Google Scholar]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, USA, 2000. [Google Scholar]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2007, 3, 59–69. [Google Scholar] [CrossRef]
- Dupret, B.; Völkel, P.; Follet, P.; Le Bourhis, X.; Angrand, P.-O. Combining genotypic and phenotypic analyses on single mutant zebrafish larvae. MethodsX 2018, 5, 244–256. [Google Scholar] [CrossRef]
- Wang, R.; Taylor, A.B.; Leal, B.Z.; Chadwell, L.V.; Ilangovan, U.; Robinson, A.K.; Schirf, V.; Hart, P.J.; Lafer, E.; Demeler, B.; et al. Polycomb Group Targeting through Different Binding Partners of RING1B C-Terminal Domain. Structure 2010, 18, 966–975. [Google Scholar] [CrossRef]
- Senthilkumar, R.; Mishra, R.K. Novel motifs distinguish multiple homologues of Polycomb in vertebrates: Expansion and diversification of the epigenetic toolkit. BMC Genomics 2009, 10, 549. [Google Scholar] [CrossRef]
- Vincenz, C.; Kerppola, T. Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 16572–16577. [Google Scholar] [CrossRef]
- Near, T.J.; Eytan, R.I.; Dornburg, A.; Kuhn, K.L.; Moore, J.; Davis, M.P.; Wainwright, P.C.; Friedman, M.; Smith, W.L. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA 2012, 109, 13698–13703. [Google Scholar] [CrossRef] [PubMed]
- Betancur-R, R.; Broughton, R.E.; Wiley, E.O.; Carpenter, K.; López, J.A.; Li, C.; Holcroft, N.I.; Arcila, D.; Sanciangco, M.; Cureton, J.C., II; et al. The Tree of Life and a New Classification of Bony Fishes. PLoS Curr. 2013, 5, 5. [Google Scholar]
- Betancur-R, R.; Wiley, E.O.; Arratia, G.; Acero, A.; Bailly, N.; Miya, M.; Lecointre, G.; Ortí, G. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017, 17, 162. [Google Scholar] [CrossRef] [PubMed]
- Hughes, L.C.; Orti, G.; Huang, Y.; Sun, Y.; Baldwin, C.C.; Thompson, A.W.; Arcila, D.; Betancur-R, R.; Li, C.; Becker, L.; et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. USA 2018, 115, 6249–6254. [Google Scholar] [CrossRef]
- Woods, I.G.; Kelly, P.D.; Chu, F.; Ngo-Hazelett, P.; Yan, Y.-L.; Huang, H.; Postlethwait, J.H.; Talbot, W.S. A Comparative Map of the Zebrafish Genome. Genome Res. 2000, 10, 1903–1914. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.-M.; Brunet, F.; Petit, J.-L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef]
- Gallant, J.R.; Losilla, M.; Tomlinson, C.; Warren, W.C. The Genome and Adult Somatic Transcriptome of the Mormyrid Electric Fish Paramormyrops kingsleyae. Genome Biol. Evol. 2017, 9, 3525–3530. [Google Scholar] [CrossRef]
- Bian, C.; Hu, Y.; Ravi, V.; Kuznetsova, I.S.; Shen, X.; Mu, X.; Sun, Y.; You, X.; Li, J.; Li, X.; et al. The Asian arowana (Scleropages formosus) genome provides new insights into the evolution of an early lineage of teleosts. Sci. Rep. 2016, 6, 24501. [Google Scholar] [CrossRef]
- Stout, C.C.; Tan, M.; Lemmon, A.R.; Lemmon, E.M.; Armbruster, J.W. Resolving Cypriniformes relationships using an anchored enrichment approach. BMC Evol. Biol. 2016, 16, 244. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, X.; Wang, X.; Li, J.; Liu, G.; Kuang, Y.-Y.; Xu, J.; Zheng, X.; Ren, L.; Wang, G.; et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef]
- Chen, Z.; Omori, Y.; Koren, S.; Shirokiya, T.; Kuroda, T.; Miyamoto, A.; Wada, H.; Fujiyama, A.; Toyoda, A.; Zhang, S.; et al. De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication. Sci. Adv. 2019, 5, eaav0547. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, X.; Bai, J.; Fang, D.; Qiu, Y.; Jiang, W.; Yuan, H.; Bian, C.; Lu, J.; He, S.; et al. The Sinocyclocheilus cavefish genome provides insights into cave adaptation. BMC Biol. 2016, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Rondeau, E.B.; Minkley, D.R.; Leong, J.S.; Messmer, A.M.; Jantzen, J.R.; Von Schalburg, K.R.; Lemon, C.; Bird, N.H.; Koop, B.F. The Genome and Linkage Map of the Northern Pike (Esox lucius): Conserved Synteny Revealed between the Salmonid Sister Group and the Neoteleostei. PLoS ONE 2014, 9, e102089. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Brunet, F.; Chalopin, M.; Juanchich, A.; Bernard, M.; Noel, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef]
- Vidal, M.; Starowicz, K.; Starowicz, K. Polycomb complexes PRC1 and their function in hematopoiesis. Exp. Hematol. 2017, 48, 12–31. [Google Scholar] [CrossRef]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef]
- Ogawa, H.; Ishiguro, K.-I.; Gaubatz, S.; Livingston, D.M.; Nakatani, Y. A Complex with Chromatin Modifiers That Occupies E2F- and Myc-Responsive Genes in G0 Cells. Science 2002, 296, 1132–1136. [Google Scholar] [CrossRef]
- Attwooll, C.; Oddi, S.; Cartwright, P.; Prosperini, E.; Agger, K.; Steensgaard, P.; Wagener, C.; Sardet, C.; Moroni, M.C.; Helin, K. A Novel Repressive E2F6 Complex Containing the Polycomb Group Protein, EPC1, That Interacts with EZH2 in a Proliferation-specific Manner. J. Biol. Chem. 2004, 280, 1199–1208. [Google Scholar] [CrossRef]
- Pelegri, F. Maternal factors in zebrafish development. Dev. Dyn. 2003, 228, 535–554. [Google Scholar] [CrossRef] [PubMed]
- Dosch, R.; Wagner, D.S.; A Mintzer, K.; Runke, G.; Wiemelt, A.P.; Mullins, M.C. Maternal Control of Vertebrate Development before the Midblastula Transition. Dev. Cell 2004, 6, 771–780. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; E Collins, J.; Sealy, I.M.; Wali, N.; Dooley, C.M.; Digby, Z.; Stemple, D.L.; Murphy, D.N.; Billis, K.; Hourlier, T.; et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Kuraku, S.; Holt, C.; Sauka-Spengler, T.; Jiang, N.; Campbell, M.S.; Yandell, M.D.; Manousaki, T.; Meyer, A.; Bloom, O.; et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 2013, 45, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, Y.; Takeda, H.; Kohara, Y.; Morishita, S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007, 17, 1254–1265. [Google Scholar] [CrossRef] [PubMed]
- Dupret, B.; Völkel, P.; Le Bourhis, X.; Angrand, P.-O. The Polycomb Group Protein Pcgf1 Is Dispensable in Zebrafish but Involved in Early Growth and Aging. PLoS ONE 2016, 11, e0158700. [Google Scholar] [CrossRef] [PubMed]
- Bajusz, I.; Henry, S.; Sutus, E.; Kovács, G.; Pirity, M. Evolving Role of RING1 and YY1 Binding Protein in the Regulation of Germ-Cell-Specific Transcription. Genes 2019, 10, 941. [Google Scholar] [CrossRef]
- Fursova, N.A.; Blackledge, N.P.; Nakayama, M.; Ito, S.; Koseki, Y.; Farcas, A.M.; King, H.W.; Koseki, H.; Klose, R.J. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Mol. Cell 2019, 74, 1020–1036.e8. [Google Scholar] [CrossRef]
- Van Der Velden, Y.U.; Wang, L.; Van Lohuizen, M.; Haramis, A.-P.G. The Polycomb group protein Ring1b is essential for pectoral fin development. Development 2012, 139, 2210–2220. [Google Scholar] [CrossRef]
- San, B.; Chrispijn, N.; Wittkopp, N.; Van Heeringen, S.J.; Lagendijk, A.K.; Aben, M.; Bakkers, J.; Ketting, R.; Kamminga, L.M. Normal formation of a vertebrate body plan and loss of tissue maintenance in the absence of ezh2. Sci. Rep. 2016, 6, 24658. [Google Scholar] [CrossRef]
- Dupret, B.; Völkel, P.; Vennin, C.; Toillon, R.-A.; Le Bourhis, X.; Angrand, P.-O. The histone lysine methyltransferase Ezh2 is required for maintenance of the intestine integrity and for caudal fin regeneration in zebrafish. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2017, 1860, 1079–1093. [Google Scholar] [CrossRef] [PubMed]
- Völkel, P.; Bary, A.; Raby, L.; Chapart, A.; Dupret, B.; Le Bourhis, X.; Angrand, P.-O. Ezh1 arises from Ezh2 gene duplication but its function is not required for zebrafish development. Sci. Rep. 2019, 9, 4319. [Google Scholar] [CrossRef] [PubMed]
- Chrispijn, N.; Andralojc, K.M.; Castenmiller, C.; Kamminga, L.M. Gene expression profile of a selection of Polycomb Group genes during zebrafish embryonic and germ line development. PLoS ONE 2018, 13, e0200316. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raby, L.; Völkel, P.; Le Bourhis, X.; Angrand, P.-O. The Polycomb Orthologues in Teleost Fishes and Their Expression in the Zebrafish Model. Genes 2020, 11, 362. https://doi.org/10.3390/genes11040362
Raby L, Völkel P, Le Bourhis X, Angrand P-O. The Polycomb Orthologues in Teleost Fishes and Their Expression in the Zebrafish Model. Genes. 2020; 11(4):362. https://doi.org/10.3390/genes11040362
Chicago/Turabian StyleRaby, Ludivine, Pamela Völkel, Xuefen Le Bourhis, and Pierre-Olivier Angrand. 2020. "The Polycomb Orthologues in Teleost Fishes and Their Expression in the Zebrafish Model" Genes 11, no. 4: 362. https://doi.org/10.3390/genes11040362
APA StyleRaby, L., Völkel, P., Le Bourhis, X., & Angrand, P.-O. (2020). The Polycomb Orthologues in Teleost Fishes and Their Expression in the Zebrafish Model. Genes, 11(4), 362. https://doi.org/10.3390/genes11040362






