Abstract
Drechslera teres (D. teres) is an ascomycete, responsible for net blotch, the most serious barley disease causing an important economic impact. The cell wall is a crucial structure for the growth and development of fungi. Thus, understanding cell wall structure, composition and biosynthesis can help in designing new strategies for pest management. Despite the severity and economic impact of net blotch, this is the first study analyzing the cell wall-related genes in D. teres. We have identified key genes involved in the synthesis/remodeling of cell wall polysaccharides, namely chitin, β-(1,3)-glucan and mixed-linkage glucan synthases, as well as endo/exoglucanases and a mitogen-activated protein kinase. We have also analyzed the differential expression of these genes in D. teres spores and in the mycelium after cultivation on different media, as well as in the presence of Paraburkholderia phytofirmans strain PsJN, a plant growth-promoting bacterium (PGPB). The targeted gene expression analysis shows higher gene expression in the spores and in the mycelium with the application of PGPB. Besides analyzing key cell-wall-related genes, this study also identifies the most suitable reference genes to normalize qPCR results in D. teres, thus serving as a basis for future molecular studies on this ascomycete.
1. Introduction
Barley is the fourth most-produced cereal in the world behind maize, wheat and rice. This crop is used mostly for animal feed (55%–60%) and by the malt industry (up to 35%) [1]. Globally, twenty million tons of malt are produced per year, and barley is intended for beer production in breweries (https://www.planetoscope.com). However, the production of this monocotyledon may be compromised by the phytopathogen ascomycete Drechslera teres [2,3]. This filamentous fungus is responsible for net blotch, easily recognizable by the occurrence of brown necrotic lesions on leaves [4,5]. This disease negatively impacts barley physiology and development and thus causes important agronomic and economic losses. Because of the use of chemical products to control D. teres, the emergence of fungicide resistance is a matter of growing concern.
The cell wall is a crucial structure for fungal development, constituting the first physical barrier of protection and is, therefore, a target for antifungal agents [6]. Although the fungal cell wall composition varies from one species to another [6,7], it includes the fundamental components chitin, glucans, mannans and/or galactomannans and glycoproteins [8].
Chitin is one of the main structural components of the fungal cell wall. More specifically, this β-(1,4)-linked homopolymer of N-acetylglucosamine (GlcNAc) is synthesized by chitin synthases, CHSs. Chitin plays a crucial role in fungal morphogenesis and in hyphal tip growth [9]. In addition to chitin, several glucans have been identified in fungal cell walls, including β-(1,3)-glucans, mixed-linkage glucans, β-(1,6)-glucans and α-(1,3)-glucans [10].
The FksA gene encodes a glycosyltransferase responsible for the synthesis of β-(1,3)-glucans. The inhibition of this gene disrupts the structural integrity of the cell wall and, therefore, impairs fungal development [6]. The majority of mannans and galactomannans decorate cell wall proteins, which are essential for cell wall formation [10]. Multiple classes of fungal cell wall proteins are present with diverse functions: glycosylphosphatidylinositol (GPI)-anchored proteins, transglycosidases, hydrolases including endo/exo-β-(1,3)-glucanase, chitinase, adhesins and hydrophobins. The roles of these proteins are to participate in cytokinesis, to remodel the cell wall and to ensure the adhesion to host cells and tissues [11].
In light of the crucial role played by the fungal cell walls as targets in the development of effective drugs to inhibit pathogens’ growth, we here mine the genome of D. teres to identify key genes involved in cell wall biosynthesis [12,13]. The goal is to analyze their expression pattern in the spores and the mycelium grown on different media and in the presence/absence of the plant growth-promoting bacterium (PGPB) Paraburkholderia phytofirmans, strain PsJN [14,15,16]. The co-cultivation with a PGPB was investigated because of the increasing attention that biocontrol methods are playing as environmentally friendly strategies to protect crops against pests.
According to many studies, PsJN is able to induce plant growth with an antagonistic effect on pathogens’ development. For instance, the bacterium restricts the spread of the gray mold disease caused by Botrytis cinerea [17,18]. PsJN-colonized tomato plants show an improved resistance against Verticillium sp. [19].
The remodeling of the cell wall has a crucial impact on the resistance of fungi to drugs [20]. Consequently, deciphering the regulation of cell wall-related genes in D. teres could inspire strategies aimed at controlling the spread of the disease. The present results constitute a resource for future molecular studies on the important phytopathogen D. teres. Despite the economic impact of the disease caused by D. teres, this is, to the best of our knowledge, the first study focusing on the cell wall-related genes of this pathogenic fungus.
2. Materials and Methods
2.1. Media and Cultivation Conditions
D. teres HE 019 (SAS BAYER Crop Lyon), the asexual form of the ascomycete fungus, was grown on different media, as hereafter described. Growing on these different media, D. teres presents several pathotypes. The abbreviations “+”, “−“ and “+/−“ are used to denote rich (+), middle (+/−) and poor (−) media for the whole paper.
2.1.1. Malt Extract Agar (MP)
MP (−), a poor medium, is composed of 10 g/L bacto malt extract and 15 g/L agar. A volume of 150 µL of spores suspension of D. teres at a concentration of 50,000 spores/mL is spread on MP (−) medium. After 15 days at 20 °C, D. teres produces a small feather duster of approximately 1 cm of height and white in color on this medium. At this stage of development, a transfer from MP (−) media onto BOA (+) media is carried out to maintain D. teres and its pathogenicity.
2.1.2. Barley Oat Meal Agar (BOA)
The medium composition is as follows: 18 g/L meal agar, 50 g/L milled leaves of barley and 17 g/L agar. This enriched medium brings all nutritive elements to the growth and spore production of D. teres. The fungus was grown for 15 days on this medium with 12 h in darkness and 12 h under blue light near UV emission at 20 °C. Onesirosan and Banttari (1969) demonstrated that spore production was greatest when the fungal cultures were exposed to this wavelength [21]. Two inoculation conditions were tested including the mycelium of the fungus containing spores (first condition) or spore suspension (second condition). For the first type of inoculation, sterile water was deposited on the surface of the fungal mycelium to facilitate its harvest with a rod. For the second inoculation condition, mycelium and spores were filtered through sterile gauze tissues. The filtrate consisted mainly of spores. The concentration of the suspension was adjusted at 105 spores/mL using a Malassez counting chamber (Marienfeld, Lauda-Königshofen, Germany).
2.1.3. Potato Dextrose Agar (PDA)
Co-culture of the fungus and PsJN was carried out on PDA (+/−) medium (39 g/L PDA, pH 4.5 ± 0.2), a medium allowing both the development of fungi and bacteria. After 15 days of growth, a rod was passed on the whole mycelium surface together with sterile water.
2.2. Bioinformatics
The maximum likelihood phylogenetic analysis of CHS (protein sequences) was obtained with several CHSs from the following fungi: D. teres, Aspergillus nidulans [22], Alternaria alternata, Botrytis cinerea [23], Blumeria graminis [24], Fusarium graminearum, Tuber melanosporum [25] and Magnaporthe grisea [9]. The tree was generated with PhyML [26] and available at http://www.phylogeny.fr. The tree was visualized with the online software iTOL (http://itol.embl.de). The CHS sequences from D. teres, A. alternata, B. graminis, F. graminearum, T. melanosporum and M. grisea were obtained by blasting the A. nidulans CHSs (National Center for Biotechnology Information (NCBI) [27]. The identification of CHS domains was carried out with Motif Scan [28]. Sequences were aligned with Clustal Omega [29]. The prediction of the transmembrane domain was performed using the online programs TMHMM (v. 2.0) [30] and Phobius [31]. Protein identifications and corresponding accession numbers from NCBI are indicated in Table 1.

Table 1.
List of the chitin synthase (CHS) protein accession numbers from the species used in this study.
2.3. RNA Extraction and cDNA Synthesis
D. teres mycelium was crushed with liquid nitrogen using a mortar and a pestle. Spore suspensions, stored at −80 °C, were lyophilized (Freeze Dryer Alpha 1/ 2-4 Christ) for 12 h at −55 °C, then milled using a ball mill MM400 (Retsch) for 2 min at 20 Hz. The extraction of total RNA was carried out using the RNeasy Plant Mini Kit including the DNase I on-column digestion (Qiagen, Leusden, The Netherlands). The integrity of the obtained RNA was evaluated with an Agilent Bioanalyzer (Agilent, Santa Clara, CA, USA). RNA Integrity Numbers (RINs) were >7. The RNA quality and quantity were checked using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Villebon-sur-Yvette, France) (A 260/280 and A 260/230 ratios between 1.9 and 2.2). In the case of contamination (ratio 260/230 < 2), samples were precipitated with ammonium acetate (NH4OAc) and washed in ethanol [32]. Subsequently, 1 µg of extracted RNA was retro-transcribed using the Superscript II cDNA Synthesis kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions.
2.4. Selection and Primer Design of Reference and Target Genes
Eleven putative genes were chosen from the available literature as candidate reference genes [33,34,35]. These genes are actin (EFQ93811.1), aminopeptidase C ApsC (EFQ89971.1), cytochrome C oxidase Cos4 (EFQ89754.1), glyceraldehyde-3-phosphate dehydrogenase GAPDH (EFQ89753.1), glucokinase GlkA (EFQ96576.1), phosphofructokinase PfkA (EFQ89228.1), phosphoglucose isomerase PgiA (EFQ88542.1), secretion-associated GTP-binding protein SarA (EFQ89474.1), isocitrate dehydrogenase precursor IsdA (EFQ94718.1), histone H2B (EFQ87126.1) and ribosomal protein S14 RS14 (EFQ94228.1).
Thirteen target genes were chosen: mitogen-activated-protein kinase PTK1 (AF272831.1) [36], six isoforms of CHSs, i.e., CHS1, CHS2, CHS3, CHS4, CHS5, CHS7 (EFQ95838.1, EFQ92549.1, EFQ88914.1, EFQ93986.1, EFQ92060.1, EFQ96223.1, respectively), β-(1,3)-glucan synthase FksA (EFQ90969.1), endo-β-(1-3)-glucanase EngA (EFQ96294.1), three isoforms of putative exo-β-(1,3)-glucanases ExgB, ExgC and ExgD (respectively, EFQ93528.1, EFQ89593.1, EFQ92573.1) and the mixed-linkage glucan synthase celA (EFQ94510.1) [22]. All sequences, except PTK1, were retrieved by blasting the A. nidulans corresponding genes. Primers for qPCR amplification were designed with “Primer3Plus” (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer 3plus.cgi) and analyzed using the OligoAnalyzer tool from Integrated DNA Technologies (http://eu.idtdna.com/calc/analyzer). Primer efficiencies and specificities were calculated and checked using serial dilutions of cDNA with a factor 5 (10, 2, 0.4, 0.08, 0.016, 0.0032 ng/µL). The characteristics of the primers of both reference and target genes are reported in Table 2.

Table 2.
Description of the primers used for the qPCR analyses with details on the sequences, amplicons’ length, Tm, amplification efficiency and regression coefficient.
2.5. qPCR: Analysis of the Results and Statistics
A liquid handling robot (epMotion 5073, Eppendorf, Hamburg, Germany) was used to dispense the reaction mixture and cDNA in 384-well plates. The Takyon Low ROX SYBR MasterMix dTTP Blue Kit (Eurogentec, Liège, Belgium) was used for qPCR. The reactions were run in technical triplicates and repeated on four independent biological replicates. PCR was performed on a ViiA 7 Real PCR System (Thermo Scientific, Villebon-sur-Yvette, France) using the following conditions: initial denaturation at 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 15 s, primer annealing at 60 °C for 60 s. At the end of the experiment, the specificity of the amplified products was checked by the analysis of the melting curve. qBasePLUS (version 3.2, Biogazelle, Ghent, Belgium) was used to calculate gene expression. Cos4 and PgiA were identified as the most stable genes in the experimental set-up chosen and as sufficient for normalization. A one-way ANOVA with Tukey’s post-hoc test was performed with IBM SPSS Statistics v19 (IBM SPSS, Chicago, IL, USA) to determine the statistically significant differences among groups.
A hierarchical clustering using uncentered absolute correlation and complete linkage was performed with Cluster 3.0 (Pearson correlation coefficient threshold = 0.83) [37]. The heat map was visualized with Java TreeView (http://jtreeview.sourceforge.net/) [38].
3. Results and Discussion
3.1. Analysis of D. teres Phenotypes on Several Media
D. teres shows varying phenotypes according to the culture media used (Figure 1). On MP (−) medium, the fungus develops a white structure, similar to a feather duster (Figure 1a) as described in the literature [39]. This structure appears when the fungus is searching for nutritive resources. On this medium, spore production is not possible since their formation and subsequent germination require, in most filamentous fungi, the availability of nutrients in the culture medium, such as sugars, amino acids and inorganic salts [40].
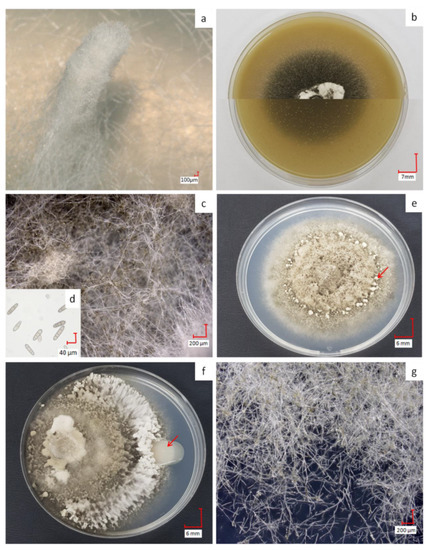
Figure 1.
Phenotypes of D. teres on malt extract agar (MP) (−) with the typical feather duster (a); aspect of D. teres on BOA (+) with top and bottom views of the Petri dish in the two halves of the image (b); mycelium of D. teres on BOA (+) (c); transversely septate conidia produced by D. teres on BOA (+) (d) and D. teres phenotype on PDA (+/−) medium with fruiting bodies (red arrow in e), aspect in the presence of PsJN (f) and mycelium on PDA (+/−) (g). The co-culture condition shows that PsJN (red arrow in f) has no visible effect on the mycelium’s growth on PDA (+/−) medium. The pictures (a,c,d,g) were obtained with a 3D Keyence VHX-2000F microscope.
On BOA (+), the color of the mycelium becomes black resulting from sporulation (Figure 1b,c). When exposed to wavelengths between 355 and 495 nm, followed by a dark period on rich medium, the fungus produces a great number of spores [41,42]. Spores of D. teres are cylindrical in shape with round ends, having a length from 25 to 300 µm and thickness from 7–11 µm [4,5]. Due to their septa, these spores are recognizable from other fungi (Figure 1d) [43]. The spores presenting less than two septa will not germinate and cannot penetrate plant tissues [44]. D. teres infects the plants via the spores, the reproductive structures, which are dispersed largely by the wind or rain and often over long distances.
On PDA (+/−), D. teres covers the entire surface of the culture medium within seven days, denoting a rapid growth (Figure 1e). On this medium, D. teres produces a very small number of spores (Figure 1g) as compared to the BOA (+) medium (Figure 1b) [45]. Since the PDA (+/−) medium is suitable for the development of bacteria and fungi, a co-culture of D. teres with PsJN, was performed. When grown alone on the PDA (+/−) medium (Figure 1e), D. teres has a fluffy mycelium with some feather dusters characteristic of this fungus [46]. Under co-cultivation with PsJN, D. teres has a different phenotype than when growing alone on PDA (+/−) (Figure 1f). Indeed, the mycelium of D. teres is less fluffy and fruiting bodies are present at the periphery, probably to provide a protective barrier against PsJN. PsJN protects indeed several crops against damages caused by different abiotic or biotic stresses and promotes plant growth [17,18,19,47,48,49,50]. According to our results, this strain seems to have no antifungal effect and is, therefore, unable to prevent the development of D. teres.
The results suggest that the variability of the phenotypes observed on the different media would be accompanied by changes in the expression of cell-wall-related genes, since the fungal cell wall is a dynamic structure accommodating the different growth stages and morphologies.
3.2. Identification of CHS Genes in D. teres and Phylogenetic Analysis
The fungal cell wall is a complex and dynamic structure that protects the cell from environmental stresses [10,51]. Given the important role played in fungal physiology, the cell wall is considered as a suitable target for antifungal drugs [6].
Chitin is the most important structural component of the fungal cell wall [9,10,11,52,53]. The enzymes catalyzing the synthesis of chitin are CHSs, which are members of glycosyltransferases from family 2 (GT2), like cellulose synthases. CHSs are able to transfer N-acetyl-D-glucosamine from an activated sugar donor (UDP-N-acetyl-D-glucosamine) to an elongating chitin chain [6,54].
In previous work, seven chitin synthase genes chsA, chsB, chsC, chsD, chsE, csmA and csmB were identified in the model organism A. nidulans encoding CHSs of Classes I, II, III, IV, V VI and VII [22]. The presence of multiple CHSs in many fungi suggests that several CHSs can be used for chitin production at different stages of the fungal life-cycle [10].
According to the classification proposed by Chigira et al. [55] and Choquer et al. [56], in our study, DtCHS1, DtCHS2, DtCHS3, DtCHS4, DtCHS5 and DtCHS7 encode CHSs from Class I, Class II, Class III, Class IV, Class V and Class VII, respectively [55,56]. These enzymes are localized in the plasma membrane [9].
BLASTp analysis carried out using the CHS protein sequences of A. nidulans identified six CHSs in D. teres (hereafter referred to as DtCHS1, 2, 3, 4, 5 and 7 for the protein sequences and DtCHS1, 2, 3, 4, 5 and 7 for the genes) (Table 1). The maximum likelihood phylogenetic analysis carried out using CHS full-length protein sequences from several classes of fungi, notably Dothideomycetes, Sordariomycetes and Leotiomycetes, allowed assigning a phylogenetic relatedness for the D. teres CHS with known orthologs from other species (Figure 2).
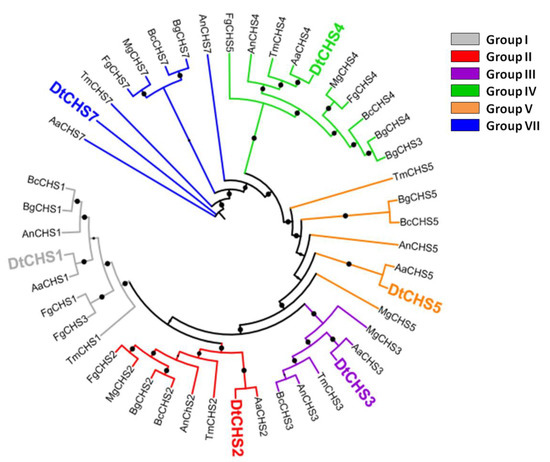
Figure 2.
Maximum likelihood phylogenetic tree (bootstraps: 100) of chitin synthases (CHS) from D. teres (Dt), A. nidulans (An), A. alternata (Aa), B. cinerea (Bc), B. graminis (Bg), F. graminearum (Fg), T. melanosporum (Tm) and M. grisea (Mg). The protein accession numbers are indicated in Table 2. Bootstraps > 80% are represented as black circles on the branches. The bootstrap % value increases with the circle size.
The phylogenetic analysis demonstrates the existence of six CHS classes (Figure 2). Class I, II and IV are present in all fungi, while Classes III, V and VII are particular to filamentous fungi [57]. The number of CHS genes changes according to the species. Most fungal species contain between three and six CHS genes [58]. For example, three CHS are present in S. cerevisiae, four in Candida albicans and eight in A. nidulans [59,60].
CHS1 and CHS2 have overlapping functions in septum formation in A. nidulans [61]. In the same way, CHS1 is crucial for infection-related morphogenesis, since 90% of the chs1 M. oryzae mutants have no septum and, therefore, display severe defects in conidium morphology [9]. CHS1 is also essential for cell wall integrity in Candida albicans [59]. Class IV enzymes contribute to the synthesis of the bulk cell wall chitin [57]. In M. oryzae, CHS1 is important for virulence and plays specific roles during conidiogenesis and appressorium formation. CHS2, CHS3, CHS4 and CHS5 are essential for plant infection and CHS6 is dispensable for pathogenesis [9]. Therefore, individual CHS genes play several roles in hyphal growth, pathogenesis, conidiogenesis and appressorium development.
BLASTp analyses and motif searches reveal similarities between the six identified CHSs from D. teres and other fungal CHSs (Table 3 and Figure 3 and Figure 4). Structurally, CHS1, CHS2 and CHS3 are more similar to each other than to other CHSs in D. teres. As in A. nidulans and other filamentous fungi, Class V CHSs in D. teres have myosin motor-like domains at the N-terminus [11,62]. Myosins are actin-dependent molecular motors and play roles in several cellular processes. More specifically, the head myosin domain binds to actin in an ATP-dependent manner and generates force by ATP hydrolysis [63].

Table 3.
D. teres CHS with predicted transmembrane domains (TMDs), deduced protein lengths and predicted domains.
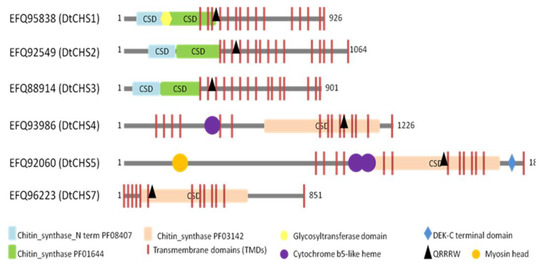
Figure 3.
Schematic representation showing the domain organization of the six putative CHSs from D. teres. Highlighted motifs were identified with Motifscanner and predicted transmembrane domains were identified with Phobius and TMHMM programs. CSD: Chitin Synthase Domain.


Figure 4.
Partial amino acid sequence alignment of the six D. teres CHSs showing the conserved chitin synthase domain (highlighted) and “QRRRW”/“EDR” motifs (underlined). Amino acids indicated with bold characters are those conserved in all CHSs listed here. I-III represent subdomains where conserved amino acids appear at high frequencies.
CHSs have several conserved domains which are considered as signature sequences since they are found in all CHSs [64]. Most of the amino acids of these signature motifs were found to be essential for activity [65]. CHSs contain the conserved EDR motif and the pentapeptide QRRRW (Figure 4) which was reported also in CHSs from the chordate Branchiostoma floridae [66]. The importance of these motifs has been studied in yeast. Thereby, in Saccharomyces cerevisiae, mutations of the QRRRW motif lead to a significant decrease in CHSs activity [58]. By comparing the amino acid sequences of the six D. teres CHSs in NCBI, DtCHS1/DtCHS2, DtCHS2/DtCHS3 and DtCHS4/DtCHS5 have similarities, with 43%, 44% and 45% identity, respectively (Figure 3 and Figure 4). DtCHS4 and DtCHS5 are closer with respect to the similarity of amino acid sequences. This sequence similarity is also confirmed by the phylogenetic tree (Figure 2).
3.3. Expression Analysis of Cell Wall-Related Genes in D. teres
The software geNormPLUS [67] was used to analyze gene expression stability across different samples of D. teres: spores and mycelium on BOA (+), mycelium on MP (−), and mycelium on PDA (+/−) in the presence or absence of the bacterium. The stability and transcript levels of the eleven candidate reference genes actin, ApsC, Cos4, GlkA, PfkA, PgiA, SarA, IsdA, H2B, GAPDH and RS14 were investigated with geNormPLUS. Cos4 and PgiA were identified as the best two transcripts for use in normalization of the data (Supplementary Figure S1).
The attention was focused on key genes involved in fungal cell wall biosynthesis. Besides CHSs, genes coding for enzymes involved in β-(1,3;1,4)-glucan and β-(1,3)-glucan synthesis were investigated. Indeed, β-(1,3)-glucan is relevant since it accounts for 65–90% of the total β-glucan content [68].
The β-(1,3)-glucan hydrolyzing enzymes can be separated into exo-β-(1,3)-glucanases and endo-β-(1,3)-glucanases according to their activities [68]. Endo-β-(1,3)-glucanases cut within the chain of glucans, whereas exo-β-(1,3)-glucanases cleave the residues of glucose at the end of the chain [69,70]. During fungal growth, β-(1,3)-glucanases are essential for the remodeling of the cell wall [69]. β-(1,3)-glucans are synthesized by a glucan synthase complex, which uses UDP-glucose as a substrate and extrudes β-(1,3)-glucan chains [70]. FksA encodes the catalytic subunits of the glucan synthase complex and is cell-cycle-regulated [6,71].
The cell wall of ascomycetes also contains β-(1,3;1,4)-glucan accounting for 10% of the glucan content in the A. nidulans cell wall [35,72]. celA encodes a putative mixed-linkage glucan synthase in A. nidulans [22]; one ortholog was found in D. teres and was used in this study.
We also considered a gene encoding a protein kinase, PTK1, which was shown to be required for conidiation, appressoria formation and pathogenicity in D. teres [36].
The hierarchical clustering of the heat map (Figure 5) shows that the CHS genes studied group in different clusters. CHS3, CHS4, CHS5 and CHS7 show a higher expression in spores and in the mycelium grown in the presence of the bacterium. This suggests that the bacterium induces the expression of specific CHSs in the phytopathogenic fungus. This phenomenon could be explained by an effect on the cell wall of the fungus. Indeed, PGPB is able to produce different types of cell-wall-lysing enzymes including chitinases, proteases, cellulases and β-(1,3)-glucanases [73,74]. The fungus could respond to PsJN by increasing the expression of CHSs in the attempt to restore the structural integrity of its cell wall.
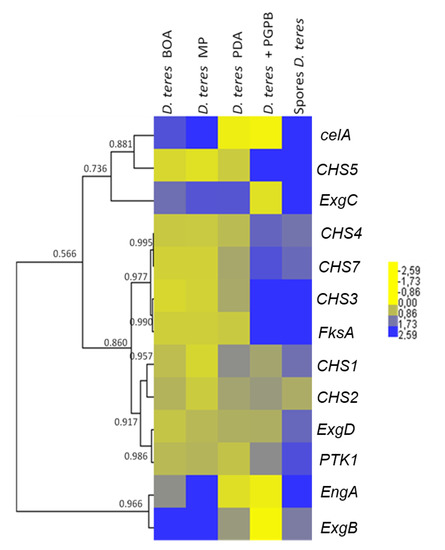
Figure 5.
Heatmap hierarchical clustering with correlation coefficients of genes related to cell wall synthesis in D. teres. The heat map hierarchical clustering was generated using Pearson correlation in complete linkage.
Some CHS genes, such as CHS1 and CHS2, showed a tendency towards decreased expression on the poor medium MP (-) (Figure 5 and Figure 6). This finding may indicate that these genes have a role in vegetative growth and the lower expression reflects the slower mycelium growth rate in a nutrient-poor environment.
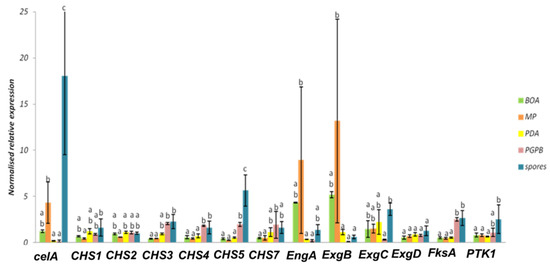
Figure 6.
Normalized relative expression of cell wall genes of D. teres cultivated on different media. Different letters indicate statistically different values (p-value < 0.05) among the groups of the one-way ANOVA with Tukey’s post-hoc test. Error bars represent the standard deviation of four independent biological replicates.
In D. teres, EngA is more expressed in the mycelium cultivated on the poor medium (MP (-) medium) (Figure 5 and Figure 6). Likewise, the exo-β-(1,3)-glucanases ExgB is expressed at higher levels on the poor medium (MP (-) medium) (Figure 5 and Figure 6). However, the differences are not statistically significant and only show a trend.
FksA is more expressed in spores and the mycelium with PsJN (Figure 5 and Figure 6). According to Figure 6, the expression of celA is significantly higher in D. teres spores, as compared to the other conditions.
PTK1 has a slightly larger expression in the spores as compared to the other experimental conditions (Figure 5 and Figure 6). This is in agreement with the reported role of PTK1 in conidiation [36].
Our results show that the expression of CHS4, CHS5 and FksA is higher in the mycelium cultivated on PDA (+/−) medium with PsJN, as compared to the growth on BOA (+), MP (−) and PDA (+/−) (Figure 6). According to Figure 1, PsJN has an effect on the phenotype of D. teres. This could be due to a possible secretion of hydrolytic enzymes by PsJN, acting on the integrity of the fungal cell wall [73,74]. D. teres could also perceive PsJN as a stress and thus strengthen its cell wall.
This hypothesis will have to be confirmed and validated by future experiments.
4. Conclusions
To the best of our knowledge, this is the first study devoted to the cell wall-related genes of D. teres. We identify key genes involved in the biosynthesis/remodeling of D. teres cell wall and differentially expressed in spores and/or in the mycelium depending on the culture media used. We also identify some cell wall biosynthetic genes induced by PsJN, a plant growth-promoting bacterium. Since PsJN seems to disturb fungal growth, it is reasonable to hypothesize that it could produce cell wall degrading enzymes causing a response in D. teres at the gene level. Additionally, we propose a list of potential candidate reference genes for qPCR analysis in D. teres. Our study paves the way to follow-up studies aiming at a functional characterization of cell wall genes of this economically relevant pathogen.
Supplementary Materials
The following is available online at https://www.mdpi.com/2073-4425/11/3/300/s1, Figure S1: Ranking of eleven candidate reference genes in D. teres according to the parameter M computed by geNormPLUS. The increase in stability of the candidate genes is determined by a decrease in the M value.
Author Contributions
A.B., C.J. and G.G. conceived and designed the experiments; A.B. performed the experiments; A.B. and G.G. analyzed the data; A.B., G.G., C.J. and E.A.B. wrote the paper. J.-F.H. and J.R. provided feedback and contributed to interpreting and discussing the results. All authors have given approval to the final version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
This work was supported by grand-Reims and Grand-Est Region. The authors gratefully acknowledge BAYER SAS Lyon for providing the D. teres HE 019 strain, and more specifically Marie-Pascale Latorse, Stéphane Brunet and Catherine Wantier for their technical assistance and participation in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kangor, T.; Sooväli, P.; Tamm, Y.; Tamm, I.; Koppel, M. Malting barley diseases, yield and quality—responses to using various agro-technology regimes. Proc. Latv. Acad. Sci. Sect. B Nat. Exact Appl. Sci. 2017, 71, 57–62. [Google Scholar] [CrossRef]
- Campbell, G.F.; Crous, P.W.; Lucas, J.A. Pyrenophora teres f. maculata, the cause of Pyrenophora leaf spot of barley in South Africa. Mycol. Res. 1999, 103, 257–267. [Google Scholar]
- McLean, M.S.; Howlett, B.J.; Hollaway, G.J. Epidemiology and control of spot form of net blotch (Pyrenophora teres f. maculata) of barley: A review. Crop Pasture Sci. 2009, 60, 303–315. [Google Scholar] [CrossRef]
- Lightfoot, D.J.; Able, A.J. Growth of Pyrenophora teres in planta during barley net blotch disease. Australas. Plant Pathol. 2010, 39, 499–507. [Google Scholar] [CrossRef]
- Liu, Z.; Ellwood, S.R.; Oliver, R.P.; Friesen, T.L. Pyrenophora teres: Profile of an increasingly damaging barley pathogen. Mol. Plant Pathol. 2011, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays News Rev. Mol. Cell. Dev. Biol. 2006, 28, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Georgopapadakou, N.H.; Tkacz, J.S. The fungal cell wall as a drug target. Trends Microbiol. 1995, 3, 98–104. [Google Scholar] [CrossRef]
- Yoshimi, A.; Miyazawa, K.; Abe, K. Cell wall structure and biogenesis in Aspergillus species. Biosci. Biotechnol. Biochem. 2016, 80, 1700–1711. [Google Scholar] [CrossRef]
- Kong, L.A.; Yang, J.; Li, G.-T.; Qi, L.L.; Zhang, Y.J.; Wang, C.F.; Zhao, W.S.; Xu, J.-R.; Peng, Y.L. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2012, 8, e1002526. [Google Scholar] [CrossRef]
- Free, S.J. Fungal cell wall organization and biosynthesis. Adv. Genet. 2013, 81, 33–82. [Google Scholar]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 1–25. [Google Scholar]
- Ellwood, S.R.; Liu, Z.; Syme, R.A.; Lai, Z.; Hane, J.K.; Keiper, F.; Moffat, C.S.; Oliver, R.P.; Friesen, T.L. A first genome assembly of the barley fungal pathogen Pyrenophora teres f. teres. Genome Biol. 2010, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, N.A.; Richards, J.K.; Brueggeman, R.S.; Friesen, T.L. Reference assembly and annotation of the Pyrenophora teres f. teres Isolate 0-1. G3 Genes Genomes Genet. 2018, 8, 1–8. [Google Scholar]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef]
- Esmaeel, Q.; Jacquard, C.; Clément, C.; Sanchez, L.; Barka, E.A. Genome sequencing and traits analysis of Burkholderia strains reveal a promising biocontrol effect against grey mould disease in grapevine (Vitis vinifera L.). World J. Microbiol. Biotechnol. 2019, 35, 1–15. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Ryu, C.-M.; Zhang, S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 2004, 94, 1259–1266. [Google Scholar] [CrossRef]
- Barka, E.A.; Gognies, S.; Nowak, J.; Audran, J.-C.; Belarbi, A. Inhibitory effect of endophyte bacteria on Botrytis cinerea and its influence to promote the grapevine growth. Biol. Control 2002, 24, 135–142. [Google Scholar] [CrossRef]
- Miotto-Vilanova, L.; Jacquard, C.; Courteaux, B.; Worthman, L.; Michel, J.; Clément, C.; Barka, E.A.; Sanchez, L. Burkholderia phytofirmans PsJN confers grapevine resistance against Botrytis cinerea via a direct antimicrobial effect combined with a better resource mobilization. Front. Plant Sci. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Sharma, V.K.; Nowak, J. Enhancement of verticillium wilt resistance in tomato transplants by in vitro co-culture of seedlings with a plant growth promoting rhizobacterium (Pseudomonas sp. strain PsJN). Can. J. Microbiol. 1998, 44, 528–536. [Google Scholar] [CrossRef]
- Beauvais, A.; Fontaine, T.; Aimanianda, V.; Latgé, J.-P. Aspergillus cell wall and biofilm. Mycopathologia 2014, 178, 371–377. [Google Scholar] [CrossRef]
- Onesirosan, P.T.; Banttari, E.E. Effect of light and temperature upon sporulation of Helminthosporium teres in culture. Phytopathology 1969, 59, 906–909. [Google Scholar]
- de Groot, P.W.J.; Brandt, B.W.; Horiuchi, H.; Ram, A.F.J.; de Koster, C.G.; Klis, F.M. Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet. Biol. 2009, 46, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Causier, B.E.; Milling, R.J.; Foster, S.G.; Adams, D.J. Characterization of chitin synthase from Botrytis cinerea. Microbiology 1994, 140, 2199–2205. [Google Scholar] [CrossRef]
- Zhang, Z.; Hall, A.; Perfect, E.; Gurr, S.J. Differential expression of two Blumeria graminis chitin synthase genes. Mol. Plant Pathol. 2000, 1, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Balestrini, R.; Sillo, F.; Kohler, A.; Schneider, G.; Faccio, A.; Tisserant, E.; Martin, F.; Bonfante, P. Genome-wide analysis of cell wall-related genes in Tuber melanosporum. Curr. Genet. 2012, 58, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information (NCBI). Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 2 March 2020).
- Motif Scan. Available online: https://myhits.isb-sib.ch/cgibin/motif_scan (accessed on 10 January 2020).
- Clustal Omega. Available online: https://www.ebi.ac.uk/Tools/services/web/toolresult.ebi?jobId=clustalo-I20181219-133935-0983-50653640-p1m (accessed on 10 January 2020).
- TMHMM (v. 2.0). Available online: http://www.cbs.dtu.dk/services/TMHMM-2.0/ (accessed on 10 January 2020).
- Phobius. Available online: http://phobius.sbc.su.se/ (accessed on 10 January 2020).
- Mangeot-Peter, L.; Legay, S.; Hausman, J.F.; Esposito, S.; Guerriero, G. Identification of reference genes for RT-qPCR data normalization in Cannabis sativa stem tissues. Int. J. Mol. Sci. 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Bohle, K.; Jungebloud, A.; Göcke, Y.; Dalpiaz, A.; Cordes, C.; Horn, H.; Hempel, D.C. Selection of reference genes for normalisation of specific gene quantification data of Aspergillus niger. J. Biotechnol. 2007, 132, 353–358. [Google Scholar] [CrossRef]
- Dilger, M.; Felsenstein, F.G.; Schwarz, G. Identification and quantitative expression analysis of genes that are differentially expressed during conidial germination in Pyrenophora teres. Mol. Genet. Genom. 2003, 270, 147–155. [Google Scholar] [CrossRef]
- Guerriero, G.; Silvestrini, L.; Legay, S.; Maixner, F.; Sulyok, M.; Hausman, J.F.; Strauss, J. Deletion of the celA gene in Aspergillus nidulans triggers overexpression of secondary metabolite biosynthetic genes. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Ruiz-Roldán, M.C.; Maier, F.J.; Schäfer, W. PTK1, a mitogen-activated-protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity of Pyrenophora teres on barley. Mol. Plant-Microbe Interact. MPMI 2001, 14, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, A.J. Java Treeview- extensible visualization of microarray data. Bioinforma. Oxf. Engl. 2004, 20, 3246–3248. [Google Scholar] [CrossRef]
- Moya, P.; Pedemonte, D.; Amengual, S.; Franco, M.; Sisterna, M. Antagonism and modes of action of Chaetomium globosum species group, potential biocontrol agent of barley foliar diseases. Boletin Soc. Argent. Bot. 2016, 51, 569–578. [Google Scholar] [CrossRef]
- Osherov, N.; May, G.S. The molecular mechanisms of conidial germination. FEMS Microbiol. Lett. 2001, 199, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Deadman, M.L.; Cooke, B.M. A method of spore production for Drechslera teres using detached barley leaves. Trans. Br. Mycol. Soc. 1985, 85, 489–493. [Google Scholar] [CrossRef]
- Scott, D.B. Identity of Pyrenophora isolates causing net-type and spot-type lesions on barley. Mycopathologia 1991, 116, 29–35. [Google Scholar] [CrossRef]
- Alcorn, J.L. The Taxonomy of “Helminthosporium” Species. Annu. Rev. Phytopathol. 1988, 26, 37–56. [Google Scholar] [CrossRef]
- Mironenko, N.V.; Afanasenko, O.S.; Filatova, O.A.; Kopahnke, D. Genetic control of virulence of Pyrenophora teres drechs, the causative agent of net blotch in barley. Genetika 2005, 41, 1674–1680. [Google Scholar] [CrossRef]
- Pană, M.; Cristea, S.; Manole, M.S.; Cernat, S.; Zala, C.; Berca, L.M. Research on the influence of temperature, light and culture media on growth and development of Pyrenophora teres fungus (in vitro). Agron. Ser. Sci. Res. Lucr. Stiintifice Ser. Agron. 2015, 58, 147–150. [Google Scholar]
- Jayasena, K.W.; George, E.; Loughman, R.; Hardy, G. First record of the teleomorph stage of Drechslera teres f. maculata in Australia. Australas. Plant Pathol. 2004, 33, 455–456. [Google Scholar] [CrossRef]
- Barka, E.A.; Nowak, J.; Clément, C. Enhancement of chilling resistance of inoculated grapevine plantlets with a plant growth-promoting rhizobacterium, Burkholderia phytofirmans strain PsJN. Appl. Environ. Microbiol. 2006, 72, 7246–7252. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Reiter, B.; Sessitsch, A.; Nowak, J.; Clément, C.; Barka, E.A. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 2005, 71, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Pinedo, I.; Ledger, T.; Greve, M.; Poupin, M.J. Burkholderia phytofirmans PsJN induces long-term metabolic and transcriptional changes involved in Arabidopsis thaliana salt tolerance. Front. Plant Sci. 2015, 6, 1–17. [Google Scholar] [CrossRef]
- Su, F.; Jacquard, C.; Villaume, S.; Michel, J.; Rabenoelina, F.; Clément, C.; Barka, E.A.; Dhondt-Cordelier, S.; Vaillant-Gaveau, N. Burkholderia phytofirmans PsJN reduces impact of freezing temperatures on photosynthesis in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1–13. [Google Scholar] [CrossRef]
- Latgé, J.P. 30 years of battling the cell wall. Med. Mycol. 2017, 55, 4–9. [Google Scholar] [CrossRef]
- Lee, J.I.; Choi, J.H.; Park, B.C.; Park, Y.H.; Lee, M.Y.; Park, H.M.; Maeng, P.J. Differential expression of the chitin synthase genes of Aspergillus nidulans, chsA, chsB, and chsC, in response to developmental status and environmental factors. Fungal Genet. Biol. 2004, 41, 635–646. [Google Scholar] [CrossRef]
- Seidl, V. Chitinases of filamentous fungi: A large group of diverse proteins with multiple physiological functions. Fungal Biol. Rev. 2008, 22, 36–42. [Google Scholar] [CrossRef]
- Orlean, P.; Funai, D. Priming and elongation of chitin chains: Implications for chitin synthase mechanism. Cell Surf. 2019, 5, 1–7. [Google Scholar] [CrossRef]
- Chigira, Y.; Abe, K.; Gomi, K.; Nakajima, T. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr. Genet. 2002, 41, 261–267. [Google Scholar] [CrossRef]
- Choquer, M.; Boccara, M.; Gonçalves, I.R.; Soulié, M.-C.; Vidal-Cros, A. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur. J. Biochem. 2004, 271, 2153–2164. [Google Scholar] [CrossRef] [PubMed]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.-L. Characterization of Specific Domains of the Cellulose and Chitin Synthases from Pathogenic Oomycetes; KTH Royal Institute of Technology: Stockholm, Sweden, 2015. [Google Scholar]
- Munro, C.A.; Winter, K.; Buchan, A.; Henry, K.; Becker, J.M.; Brown, A.J.P.; Bulawa, C.E.; Gow, N.A.R. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol. Microbiol. 2001, 39, 1414–1426. [Google Scholar] [CrossRef] [PubMed]
- Rogg, L.E.; Fortwendel, J.R.; Juvvadi, P.R.; Steinbach, W.J. Regulation of expression, activity and localization of fungal chitin synthases. Med. Mycol. 2012, 50, 2–17. [Google Scholar] [CrossRef]
- Ichinomiya, M.; Yamada, E.; Yamashita, S.; Ohta, A.; Horiuchi, H. Class I and class II chitin synthases are involved in septum formation in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 2005, 4, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, H. Functional diversity of chitin synthases of Aspergillus nidulans in hyphal growth, conidiophore development and septum formation. Med. Mycol. 2009, 47, 47–52. [Google Scholar] [CrossRef]
- Lamichhane, A.K.; Garraffo, H.M.; Cai, H.; Walter, P.J.; Kwon-Chung, K.J.; Chang, Y.C. A novel role of fungal type I myosin in regulating membrane properties and its association with d-amino acid utilization in Cryptococcus gattii. MBio 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Nagahashi, S.; Sudoh, M.; Ono, N.; Sawada, R.; Yamaguchi, E.; Uchida, Y.; Mio, T.; Takagi, M.; Arisawa, M.; Yamada-Okabe, H. Characterization of chitin synthase 2 of Saccharomyces cerevisiae. Implication of two highly conserved domains as possible catalytic sites. J. Biol. Chem. 1995, 270, 13961–13967. [Google Scholar] [CrossRef]
- Merzendorfer, H. Insect chitin synthases: A review. J. Comp. Physiol. 2006, 176, 1–15. [Google Scholar] [CrossRef]
- Guerriero, G. Putative chitin synthases from Branchiostoma floridae show extracellular matrix-related domains and mosaic structures. Genom. Proteom. Bioinform. 2012, 10, 197–207. [Google Scholar] [CrossRef]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fesel, P.H.; Zuccaro, A. β-glucan: Crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet. Biol. 2016, 90, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, T.; Hartland, R.P.; Beauvais, A.; Diaquin, M.; Latge, J.-P. Purification and characterization of an endo-1,3-β-glucanase from Aspergillus fumigatus. Eur. J. Biochem. 1997, 243, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Hartl, L.; Latgé, J.-P. β-1,3-glucan modifying enzymes in Aspergillus fumigatus. Front. Microbiol. 2013, 4, 1–9. [Google Scholar] [CrossRef]
- Park, B.-C.; Park, Y.-H.; Yi, S.; Choi, Y.K.; Kang, E.-H.; Park, H.-M. Transcriptional regulation of fksA, a β-1,3-glucan synthase gene, by the APSES protein StuA during Aspergillus nidulans development. J. Microbiol. 2014, 52, 940–947. [Google Scholar] [CrossRef]
- Samar, D.; Kieler, J.B.; Klutts, J.S. Identification and deletion of Tft1, a predicted glycosyltransferase necessary for cell wall β-1,3;1,4-glucan synthesis in Aspergillus fumigatus. PLoS ONE 2015, 10, e0117336. [Google Scholar] [CrossRef]
- Jadhav, H.; Shaikh, S.; Sayyed, R. Role of hydrolytic enzymes of rhizoflora in biocontrol of fungal phytopathogens: An overview. In Rhizotrophs: Plant Growth Promotion to Bioremediation; Springer: Singapore, 2017; Volume 2, pp. 183–203. [Google Scholar]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 1–16. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).