Maintenance of Yeast Genome Integrity by RecQ Family DNA Helicases
Abstract
1. The RecQ Helicase Family Is Conserved from Bacteria to Humans
2. Domain Structure of the Sgs1 and Rqh1 Helicases
3. Post-Translational Modification of Sgs1 and Rqh1
4. Substrate Preferences of Sgs1 and Rqh1
5. Physical Interactions of Sgs1 and Rqh1
6. Homologous Recombination-Mediated DNA Double-Strand Break Repair
6.1. Genetic Interactions of SGS1 and RQH1 with HR Genes
6.2. Roles of Sgs1 in DSB Repair
6.3. Roles of Sgs1 at the Replication Fork
7. Meiosis
8. Possible Functions in Excision Repair
9. Telomere Length Maintenance
10. Aging and Transcription
11. Other RecQ-Like DNA Helicases in Yeast
12. Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Nakayama, H.; Nakayama, K.; Nakayama, R.; Irino, N.; Nakayama, Y.; Hanawalt, P.C. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: Identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Gen. Genet. MGG 1984, 195, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Willis, N.; Rhind, N. Mus81, Rhp51 (Rad51), and Rqh1 form an epistatic pathway required for the S-phase DNA damage checkpoint. Mol. Biol. Cell 2009, 20, 819–833. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Nakayama, K.; Nakayama, R.; Nakayama, Y. Recombination-deficient mutations and thymineless death in Escherichia coli K12: Reciprocal effects of recBC and recF and indifference of recA mutations. Can. J. Microbiol. 1982, 28, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.I.; Kirk, S.H.; Eisenstark, A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 1998, 52, 591–625. [Google Scholar] [CrossRef]
- Fonville, N.C.; Bates, D.; Hastings, P.J.; Hanawalt, P.C.; Rosenberg, S.M. Role of RecA and the SOS response in thymineless death in Escherichia coli. PLoS Genet. 2010, 6, e1000865. [Google Scholar] [CrossRef]
- Nakayama, K.; Irino, N.; Nakayama, H. The recQ gene of Escherichia coli K12: Molecular cloning and isolation of insertion mutants. Mol. Gen. Genet. MGG 1985, 200, 266–271. [Google Scholar] [CrossRef]
- Bernstein, K.A.; Gangloff, S.; Rothstein, R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010, 44, 393–417. [Google Scholar] [CrossRef]
- Hartung, F.; Puchta, H. The RecQ gene family in plants. J. Plant Physiol. 2006, 163, 287–296. [Google Scholar] [CrossRef]
- Ellis, N.A.; Groden, J.; Ye, T.-Z.; Straughen, J.; Lennon, D.J.; Ciocci, S.; Proytcheva, M.; German, J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 1995, 83, 655–666. [Google Scholar] [CrossRef]
- Yu, C.-E.; Oshima, J.; Fu, Y.-H.; Wijsman, E.M.; Hisama, F.; Alisch, R.; Matthews, S.; Nakura, J.; Miki, T.; Ouais, S. Positional cloning of the Werner’s syndrome gene. Science 1996, 272, 258–262. [Google Scholar] [CrossRef]
- Kitao, S.; Lindor, N.M.; Shiratori, M.; Furuichi, Y.; Shimamoto, A. Rothmund–Thomson syndrome responsible gene, RECQL4: Genomic structure and products. Genomics 1999, 61, 268–276. [Google Scholar] [CrossRef]
- Kitao, S.; Shimamoto, A.; Goto, M.; Miller, R.W.; Smithson, W.A.; Lindor, N.M.; Furuichi, Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999, 22, 82. [Google Scholar] [CrossRef] [PubMed]
- Yankiwski, V.; Marciniak, R.A.; Guarente, L.; Neff, N.F. Nuclear structure in normal and Bloom syndrome cells. Proc. Natl. Acad. Sci. USA 2000, 97, 5214–5219. [Google Scholar] [CrossRef] [PubMed]
- Bachrati, C.Z.; Hickson, I.D. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem. J. 2003, 374, 577–606. [Google Scholar] [CrossRef] [PubMed]
- Kusano, K.; Berres, M.E.; Engels, W.R. Evolution of the RECQ family of helicases: A drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics 1999, 151, 1027–1039. [Google Scholar]
- Yamagata, K.; Kato, J.-I.; Shimamoto, A.; Goto, M.; Furuichi, Y.; Ikeda, H. Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: Implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA 1998, 95, 8733–8738. [Google Scholar] [CrossRef]
- Myung, K.; Datta, A.; Chen, C.; Kolodner, R.D. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 2001, 27, 113. [Google Scholar] [CrossRef]
- Sinclair, D.A.; Mills, K.; Guarente, L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 1997, 277, 1313–1316. [Google Scholar] [CrossRef]
- Gangloff, S.; McDonald, J.P.; Bendixen, C.; Arthur, L.; Rothstein, R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994, 14, 8391–8398. [Google Scholar] [CrossRef]
- Watt, P.M.; Louis, E.J.; Borts, R.H.; Hickson, I.D. Sgs1: A eukaryotic homolog of E. coil RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 1995, 81, 253–260. [Google Scholar] [CrossRef]
- Stewart, E.; Chapman, C.R.; Al-Khodairy, F.; Carr, A.M.; Enoch, T. rqh1+, a fission yeast gene related to the Bloom‘s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997, 16, 2682–2692. [Google Scholar] [CrossRef] [PubMed]
- Davey, S.; Han, C.S.; Ramer, S.A.; Klassen, J.C.; Jacobson, A.; Eisenberger, A.; Hopkins, K.M.; Lieberman, H.B.; Freyer, G.A. Fission yeast rad12+ regulates cell cycle checkpoint control and is homologous to the Bloom’s syndrome disease gene. Mol. Cell. Biol. 1998, 18, 2721–2728. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.M.; Lindsay, H.D.; Munday, C.A.; Carr, A.M. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 1997, 17, 6868–6875. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.B.; Hickson, I.D. RecQ helicases: Conserved guardians of genomic integrity. In DNA Helicases and DNA Motor Proteins; Springer: Berlin/Heidelberg, Germany, 2013; pp. 161–184. [Google Scholar]
- Bernstein, D.A.; Keck, J.L. Domain mapping of Escherichia coli RecQ defines the roles of conserved N-and C-terminal regions in the RecQ family. Nucleic Acids Res. 2003, 31, 2778–2785. [Google Scholar] [CrossRef]
- Sharma, S.; Doherty, K.M.; Brosh, R.M. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 2006, 398, 319–337. [Google Scholar] [CrossRef]
- Bansal, R.; Arya, V.; Sethy, R.; Rakesh, R.; Muthuswami, R. RecA-like domain 2 of DNA-dependent ATPase A domain, a SWI2/SNF2 protein, mediates conformational integrity and ATP hydrolysis. Biosci. Rep. 2018, 38, BSR20180568. [Google Scholar] [CrossRef]
- Tanner, N.K.; Cordin, O.; Banroques, J.; Doere, M.; Linder, P. The Q motif: A newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 2003, 11, 127–138. [Google Scholar] [CrossRef]
- Killoran, M.P.; Keck, J.L. Sit down, relax and unwind: Structural insights into RecQ helicase mechanisms. Nucleic Acids Res. 2006, 34, 4098–4105. [Google Scholar] [CrossRef]
- Bernstein, D.A.; Zittel, M.C.; Keck, J.L. High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J. 2003, 22, 4910–4921. [Google Scholar] [CrossRef]
- Newman, J.A.; Savitsky, P.; Allerston, C.K.; Pike, A.C.W.; Pardon, E.; Steyaert, J.; Arrowsmith, C.H.; Von Delft, F.; Bountra, C.; Edwards, A.; et al. Crystal structure of the Bloom’s syndrome helicase indicates a role for the HRDC domain in conformational changes. Nucleic Acids Res 2013, 10, 5221–5535. [Google Scholar] [CrossRef]
- Mirzaei, H.; Schmidt, K.H. Non-Bloom syndrome-associated partial and total loss-of-function variants of BLM helicase. Proc. Natl. Acad. Sci. USA 2012, 109, 19357–19362. [Google Scholar] [CrossRef] [PubMed]
- Shastri, V.M.; Schmidt, K.H. Cellular defects caused by hypomorphic variants of the Bloom syndrome helicase gene BLM. Mol. Genet. Genom. Med. 2016, 4, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Killoran, M.P.; Keck, J.L. Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. J. Biol. Chem. 2006, 281, 12849–12857. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.-B.; Rigolet, P.; Zargarian, L.; Fermandjian, S.; Xi, X.G. Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom’s syndrome helicase. Nucleic Acids Res. 2005, 33, 3109–3124. [Google Scholar] [CrossRef][Green Version]
- Liu, J.L.; Rigolet, P.; Dou, S.-X.; Wang, P.-Y.; Xi, X.G. The zinc finger motif of Escherichia coli RecQ is implicated in both DNA binding and protein folding. J. Biol. Chem. 2004, 279, 42794–42802. [Google Scholar] [CrossRef]
- Gajiwala, K.S.; Burley, S.K. Winged helix proteins. Curr. Opin. Struct. Biol. 2000, 10, 110–116. [Google Scholar] [CrossRef]
- Rodríguez, A.C.; Stock, D. Crystal structure of reverse gyrase: Insights into the positive supercoiling of DNA. EMBO J. 2002, 21, 418–426. [Google Scholar] [CrossRef]
- Lima, C.D.; Wang, J.C.; Mondragón, A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature 1994, 367, 138. [Google Scholar] [CrossRef]
- Schultz, S.C.; Shields, G.C.; Steitz, T.A. Crystal structure of a CAP-DNA complex: The DNA is bent by 90 degrees. Science 1991, 253, 1001–1007. [Google Scholar] [CrossRef]
- Kitano, K.; Kim, S.-Y.; Hakoshima, T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure 2010, 18, 177–187. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Keck, J.L. Werner helicase wings DNA binding. Structure 2010, 18, 149–151. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, L.; Chan, K.L.; Ralf, C.; Bernstein, D.A.; Garcia, P.L.; Bohr, V.A.; Vindigni, A.; Janscak, P.; Keck, J.L.; Hickson, I.D. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005, 24, 2679–2687. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Macias, M.J.; Bottomley, M.J.; Stier, G.; Linge, J.; Nilges, M.; Bork, P.; Sattler, M. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure 1999, 7, 1557–1566. [Google Scholar] [CrossRef]
- Lillard-Wetherell, K.; Machwe, A.; Langland, G.T.; Combs, K.A.; Behbehani, G.K.; Schonberg, S.A.; German, J.; Turchi, J.J.; Orren, D.K.; Groden, J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004, 13, 1919–1932. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, B.; Gray, M.D.; Oshima, J.; Mian, I.S.; Campisi, J. The premature ageing syndrome protein, WRN, is a 3’->5’ exonuclease. Nat. Genet. 1998, 20, 114–116. [Google Scholar] [CrossRef]
- Shen, J.C.; Gray, M.D.; Oshima, J.; Kamath-Loeb, A.S.; Fry, M.; Loeb, L.A. Werner syndrome protein. I. DNA helicase and DNA exonuclease reside on the same polypeptide. J. Biol. Chem. 1998, 273, 34139–34144. [Google Scholar] [CrossRef]
- Choudhary, S.; Sommers, J.A.; Brosh, R.M., Jr. Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of werner syndrome protein. J. Biol. Chem. 2004, 279, 34603–34613. [Google Scholar] [CrossRef]
- Liberi, G.; Maffioletti, G.; Lucca, C.; Chiolo, I.; Baryshnikova, A.; Cotta-Ramusino, C.; Lopes, M.; Pellicioli, A.; Haber, J.E.; Foiani, M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005, 19, 339–350. [Google Scholar] [CrossRef]
- Cejka, P.; Cannavo, E.; Polaczek, P.; Masuda-Sasa, T.; Pokharel, S.; Campbell, J.L.; Kowalczykowski, S.C. DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2. Nature 2010, 467, 112. [Google Scholar] [CrossRef]
- Pedrazzi, G.; Bachrati, C.Z.; Selak, N.; Studer, I.; Petkovic, M.; Hickson, I.D.; Jiricny, J.; Stagljar, I. The Bloom’s syndrome helicase interacts directly with the human DNA mismatch repair protein hMSH6. Biol. Chem. 2003, 384, 1155–1164. [Google Scholar] [CrossRef]
- Wang, T.-F.; Kung, W.-M. Supercomplex formation between Mlh1–Mlh3 and Sgs1–Top3 heterocomplexes in meiotic yeast cells. Biochem. Biophys. Res. Commun. 2002, 296, 949–953. [Google Scholar] [CrossRef]
- Bermúdez-López, M.; Pociño-Merino, I.; Sánchez, H.; Bueno, A.; Guasch, C.; Almedawar, S.; Bru-Virgili, S.; Garí, E.; Wyman, C.; Reverter, D. ATPase-dependent control of the Mms21 SUMO ligase during DNA repair. PLoS Biol. 2015, 13, e1002089. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez-López, M.; Villoria, M.T.; Esteras, M.; Jarmuz, A.; Torres-Rosell, J.; Clemente-Blanco, A.; Aragon, L. Sgs1’s roles in DNA end resection, HJ dissolution, and crossover suppression require a two-step SUMO regulation dependent on Smc5/6. Genes Dev. 2016, 30, 1339–1356. [Google Scholar] [CrossRef]
- Branzei, D.; Sollier, J.; Liberi, G.; Zhao, X.; Maeda, D.; Seki, M.; Enomoto, T.; Ohta, K.; Foiani, M. Ubc9-and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 2006, 127, 509–522. [Google Scholar] [CrossRef]
- Chakraborty, U.; George, C.M.; Lyndaker, A.M.; Alani, E. A delicate balance between repair and replication factors regulates recombination between divergent DNA sequences in Saccharomyces cerevisiae. Genetics 2016, 202, 525–540. [Google Scholar] [CrossRef]
- Campos-Doerfler, L.; Syed, S.; Schmidt, K.H. Sgs1 Binding to Rad51 Stimulates Homology-Directed DNA Repair in Saccharomyces cerevisiae. Genetics 2018, 208, 125–138. [Google Scholar] [CrossRef]
- Hegnauer, A.M.; Hustedt, N.; Shimada, K.; Pike, B.L.; Vogel, M.; Amsler, P.; Rubin, S.M.; van Leeuwen, F.; Guenole, A.; van Attikum, H.; et al. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 2012, 31, 3768–3783. [Google Scholar] [CrossRef]
- Bachrati, C.Z.; Hickson, I.D. RecQ helicases: Guardian angels of the DNA replication fork. Chromosoma 2008, 117, 219–233. [Google Scholar] [CrossRef]
- Machwe, A.; Lozada, E.M.; Xiao, L.; Orren, D.K. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol. Biol. 2006, 7, 1. [Google Scholar] [CrossRef]
- Niu, H.; Chung, W.H.; Zhu, Z.; Kwon, Y.; Zhao, W.; Chi, P.; Prakash, R.; Seong, C.; Liu, D.; Lu, L.; et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 2010, 467, 108–111. [Google Scholar] [CrossRef]
- Anderson, D.G.; Kowalczykowski, S.C. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell 1997, 90, 77–86. [Google Scholar] [CrossRef]
- Chen, C.F.; Brill, S.J. An essential DNA strand-exchange activity is conserved in the divergent N-termini of BLM orthologs. EMBO J. 2010, 29, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Machwe, A.; Xiao, L.; Groden, J.; Matson, S.W.; Orren, D.K. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 2005, 280, 23397–23407. [Google Scholar] [CrossRef] [PubMed]
- Cheok, C.F.; Wu, L.; Garcia, P.L.; Janscak, P.; Hickson, I.D. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res 2005, 33, 3932–3941. [Google Scholar] [CrossRef]
- Beltrao, P.; Albanèse, V.; Kenner, L.R.; Swaney, D.L.; Burlingame, A.; Villén, J.; Lim, W.A.; Fraser, J.S.; Frydman, J.; Krogan, N.J. Systematic functional prioritization of protein posttranslational modifications. Cell 2012, 150, 413–425. [Google Scholar] [CrossRef]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.; Rush, J.; Fields, S.; Krogan, N.J.; Villén, J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 2013, 10, 676. [Google Scholar] [CrossRef]
- Holt, L.J.; Tuch, B.B.; Villén, J.; Johnson, A.D.; Gygi, S.P.; Morgan, D.O. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 2009, 325, 1682–1686. [Google Scholar] [CrossRef]
- Fricke, W.M.; Kaliraman, V.; Brill, S.J. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 2001, 276, 8848–8855. [Google Scholar] [CrossRef]
- Bodenmiller, B.; Campbell, D.; Gerrits, B.; Lam, H.; Jovanovic, M.; Picotti, P.; Schlapbach, R.; Aebersold, R. PhosphoPep-a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 2008, 26, 1339–1340. [Google Scholar] [CrossRef]
- Bodenmiller, B.; Wanka, S.; Kraft, C.; Urban, J.; Campbell, D.; Pedrioli, P.G.; Gerrits, B.; Picotti, P.; Lam, H.; Vitek, O.; et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 2010, 3, rs4. [Google Scholar] [CrossRef]
- Zhou, C.; Elia, A.E.; Naylor, M.L.; Dephoure, N.; Ballif, B.A.; Goel, G.; Xu, Q.; Ng, A.; Chou, D.M.; Xavier, R.J. Profiling DNA damage-induced phosphorylation in budding yeast reveals diverse signaling networks. Proc. Natl. Acad. Sci. USA 2016, 113, E3667–E3675. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.J.; Sharp, J.A.; Wang, J.C. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 9644–9650. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-Y.; Tsai, C.-H.; Brill, S.J.; Teng, S.-C. Sumoylation of the BLM ortholog, Sgs1, promotes telomere–telomere recombination in budding yeast. Nucleic Acids Res. 2009, 38, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Rog, O.; Miller, K.M.; Ferreira, M.G.; Cooper, J.P. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol. Cell 2009, 33, 559–569. [Google Scholar] [CrossRef]
- Swaffer, M.P.; Jones, A.W.; Flynn, H.R.; Snijders, A.P.; Nurse, P. Quantitative phosphoproteomics reveals the signaling dynamics of Cell-Cycle kinases in the fission Yeast Schizosaccharomyces pombe. Cell Rep. 2018, 24, 503–514. [Google Scholar] [CrossRef]
- Kettenbach, A.N.; Deng, L.; Wu, Y.; Baldissard, S.; Adamo, M.E.; Gerber, S.A.; Moseley, J.B. Quantitative phosphoproteomics reveals pathways for coordination of cell growth and division by the conserved fission yeast kinase pom1. Mol. Cell. Proteom. 2015, 14, 1275–1287. [Google Scholar] [CrossRef]
- Ahmad, F.; Stewart, E. The N-terminal region of the Schizosaccharomyces pombe RecQ helicase, Rqh1p, physically interacts with Topoisomerase III and is required for Rqh1p function. Mol. Genet. Genom. 2005, 273, 102–114. [Google Scholar] [CrossRef]
- Bennett, R.J.; Keck, J.L.; Wang, J.C. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol. 1999, 289, 235–248. [Google Scholar] [CrossRef]
- Sun, H.; Bennett, R.J.; Maizels, N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds GG paired DNAs. Nucleic Acids Res. 1999, 27, 1978–1984. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 1990, 344, 410. [Google Scholar] [CrossRef]
- Huber, M.D.; Duquette, M.L.; Shiels, J.C.; Maizels, N. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006, 358, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.; Wang, S.-W.; Toda, T.; Norbury, C.; Hickson, I.D. Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res. 1999, 27, 4050–4058. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.J.; Noirot-Gros, M.-F.; Wang, J.C. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 2000, 275, 26898–26905. [Google Scholar] [CrossRef] [PubMed]
- Dunø, M.; Thomsen, B.; Westergaard, O.; Krejci, L.; Bendixen, C. Genetic analysis of the Saccharomyces cerevisiae Sgs1 helicase defines an essential function for the Sgs1-Top3 complex in the absence of SRS2 or TOP1. Mol. Gen. Genet. MGG 2000, 264, 89–97. [Google Scholar] [CrossRef]
- Mullen, J.R.; Nallaseth, F.S.; Lan, Y.Q.; Slagle, C.E.; Brill, S.J. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol. Cell. Biol. 2005, 25, 4476–4487. [Google Scholar] [CrossRef]
- Kennedy, J.A.; Daughdrill, G.W.; Schmidt, K.H. A transient α-helical molecular recognition element in the disordered N-terminus of the Sgs1 helicase is critical for chromosome stability and binding of Top3/Rmi1. Nucleic Acids Res. 2013, 41, 10215–10227. [Google Scholar] [CrossRef]
- Wang, F.; Yang, Y.; Singh, T.R.; Busygina, V.; Guo, R.; Wan, K.; Wang, W.; Sung, P.; Meetei, A.R.; Lei, M. Crystal structures of RMI1 and RMI2, two OB-fold regulatory subunits of the BLM complex. Structure 2010, 18, 1159–1170. [Google Scholar] [CrossRef]
- Bocquet, N.; Bizard, A.H.; Abdulrahman, W.; Larsen, N.B.; Faty, M.; Cavadini, S.; Bunker, R.D.; Kowalczykowski, S.C.; Cejka, P.; Hickson, I.D.; et al. Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIalpha and RMI1. Nat. Struct. Mol. Biol. 2014, 21, 261–268. [Google Scholar] [CrossRef]
- Wu, L.; Davies, S.L.; Levitt, N.C.; Hickson, I.D. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 2001, 276, 19375–19381. [Google Scholar] [CrossRef]
- Chiolo, I.; Carotenuto, W.; Maffioletti, G.; Petrini, J.H.; Foiani, M.; Liberi, G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell Biol. 2005, 25, 5738–5751. [Google Scholar] [CrossRef]
- Saffi, J.; Feldmann, H.; Winnacker, E.-L.; Henriques, J.A. Interaction of the yeast Pso5/Rad16 and Sgs1 proteins: Influences on DNA repair and aging. Mutat. Res. DNA Repair 2001, 486, 195–206. [Google Scholar] [CrossRef]
- Mullen, J.R.; Kaliraman, V.; Brill, S.J. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 2000, 154, 1101–1114. [Google Scholar] [PubMed]
- Pedrazzi, G.; Perrera, C.; Blaser, H.; Kuster, P.; Marra, G.; Davies, S.L.; Ryu, G.-H.; Freire, R.; Hickson, I.D.; Jiricny, J. Direct association of Bloom’s syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 2001, 29, 4378–4386. [Google Scholar] [CrossRef] [PubMed]
- Langland, G.; Kordich, J.; Creaney, J.; Goss, K.H.; Lillard-Wetherell, K.; Bebenek, K.; Kunkel, T.A.; Groden, J. The Bloom’s syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J. Biol. Chem. 2001, 276, 30031–30035. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, N.; Goldfarb, T.; Studamire, B.; Alani, E.; Haber, J.E. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 2004, 101, 9315–9320. [Google Scholar] [CrossRef] [PubMed]
- Tronnersjö, S.; Hanefalk, C.; Balciunas, D.; Hu, G.-Z.; Nordberg, N.; Murén, E.; Ronne, H. The jmjN and jmjC domains of the yeast zinc finger protein Gis1 interact with 19 proteins involved in transcription, sumoylation and DNA repair. Mol. Genet. Genom. 2007, 277, 57–70. [Google Scholar] [CrossRef]
- Chaudhury, I.; Koepp, D.M. Degradation of Mrc1 promotes recombination-mediated restart of stalled replication forks. Nucleic Acids Res. 2016, 45, 2558–2570. [Google Scholar] [CrossRef]
- Albers, M.; Diment, A.; Muraru, M.; Russell, C.S.; Beggs, J.D. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA 2003, 9, 138–150. [Google Scholar] [CrossRef]
- Piya, G.; Mueller, E.N.; Haas, H.K.; Ghospurkar, P.L.; Wilson, T.M.; Jensen, J.L.; Colbert, C.L.; Haring, S.J. Characterization of the Interaction between Rfa1 and Rad24 in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0116512. [Google Scholar] [CrossRef]
- Iacovella, M.G.; Golfieri, C.; Massari, L.F.; Busnelli, S.; Pagliuca, C.; Dal Maschio, M.; Infantino, V.; Visintin, R.; Mechtler, K.; Ferreira-Cerca, S. Rio1 promotes rDNA stability and downregulates RNA polymerase I to ensure rDNA segregation. Nat. Commun. 2015, 6, 6643. [Google Scholar] [CrossRef]
- Iacovella, M.G.; Bremang, M.; Basha, O.; Giacò, L.; Carotenuto, W.; Golfieri, C.; Szakal, B.; Dal Maschio, M.; Infantino, V.; Beznoussenko, G.V. Integrating Rio1 activities discloses its nutrient-activated network in Saccharomyces cerevisiae. Nucleic Acids Res. 2018, 46, 7586–7611. [Google Scholar] [CrossRef]
- Chin, J.K.; Bashkirov, V.I.; Heyer, W.-D.; Romesberg, F.E. Esc4/Rtt107 and the control of recombination during replication. DNA Repair 2006, 5, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Driscoll, R.; Castellucci, F.; Foiani, M.; Jackson, S.P.; Branzei, D. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol. Biol. Cell 2009, 20, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Böhm, S.; Mihalevic, M.J.; Casal, M.A.; Bernstein, K.A. Disruption of SUMO-targeted ubiquitin ligases Slx5–Slx8/RNF4 alters RecQ-like helicase Sgs1/BLM localization in yeast and human cells. DNA Repair 2015, 26, 1–14. [Google Scholar] [CrossRef]
- Bonner, J.N.; Choi, K.; Xue, X.; Torres, N.P.; Szakal, B.; Wei, L.; Wan, B.; Arter, M.; Matos, J.; Sung, P. Smc5/6 mediated sumoylation of the Sgs1-Top3-Rmi1 complex promotes removal of recombination intermediates. Cell Rep. 2016, 16, 368–378. [Google Scholar] [CrossRef]
- Wong, J.; Nakajima, Y.; Westermann, S.; Shang, C.; Kang, J.-S.; Goodner, C.; Houshmand, P.; Fields, S.; Chan, C.S.; Drubin, D. A protein interaction map of the mitotic spindle. Mol. Biol. Cell 2007, 18, 3800–3809. [Google Scholar] [CrossRef]
- Pietrobon, V.; Freon, K.; Hardy, J.; Costes, A.; Iraqui, I.; Ochsenbein, F.; Lambert, S.A. The chromatin assembly factor 1 promotes Rad51-dependent template switches at replication forks by counteracting D-loop disassembly by the RecQ-type helicase Rqh1. PLoS Biol. 2014, 12, e1001968. [Google Scholar] [CrossRef]
- Kibe, T.; Ono, Y.; Sato, K.; Ueno, M. Fission yeast Taz1 and RPA are synergistically required to prevent rapid telomere loss. Mol. Biol. Cell 2007, 18, 2378–2387. [Google Scholar] [CrossRef]
- Vo, T.V.; Das, J.; Meyer, M.J.; Cordero, N.A.; Akturk, N.; Wei, X.; Fair, B.J.; Degatano, A.G.; Fragoza, R.; Liu, L.G. A proteome-wide fission yeast interactome reveals network evolution principles from yeasts to human. Cell 2016, 164, 310–323. [Google Scholar] [CrossRef]
- Watts, F.; Skilton, A.; Ho, J.-Y.; Boyd, L.K.; Trickey, M.; Gardner, L.; Ogi, F.-X.; Outwin, E. The role of Schizosaccharomyces pombe SUMO ligases in genome stability. Biochem. Soc. Trans. 2007. [Google Scholar] [CrossRef]
- McDonald, K.R.; Guise, A.J.; Pourbozorgi-Langroudi, P.; Cristea, I.M.; Zakian, V.A.; Capra, J.A.; Sabouri, N. Pfh1 is an accessory replicative helicase that interacts with the replisome to facilitate fork progression and preserve genome integrity. PLoS Genet. 2016, 12, e1006238. [Google Scholar] [CrossRef] [PubMed]
- Barlow, J.H.; Rothstein, R. Timing is everything: Cell cycle control of Rad52. Cell Div. 2010, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Watt, P.M.; Hickson, I.D.; Borts, R.H.; Louis, E.J. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 1996, 144, 935–945. [Google Scholar] [PubMed]
- Ii, M.; Ii, T.; Mironova, L.I.; Brill, S.J. Epistasis analysis between homologous recombination genes in Saccharomyces cerevisiae identifies multiple repair pathways for Sgs1, Mus81-Mms4 and RNase H2. Mutat. Res. 2011, 714, 33–43. [Google Scholar] [CrossRef]
- Shor, E.; Gangloff, S.; Wagner, M.; Weinstein, J.; Price, G.; Rothstein, R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 2002, 162, 647–662. [Google Scholar]
- Bessler, J.B.; Torre, J.Z.; Zakian, V.A. The Pif1p subfamily of helicases: Region-specific DNA helicases? Trends Cell Biol. 2001, 11, 60–65. [Google Scholar] [CrossRef]
- Wagner, M.; Price, G.; Rothstein, R. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics 2006, 174, 555–573. [Google Scholar] [CrossRef]
- Lu, J.; Mullen, J.R.; Brill, S.J.; Kleff, S.; Romeo, A.M.; Sternglanz, R. Human homologues of yeast helicase. Nature 1996, 383, 678. [Google Scholar] [CrossRef]
- Gangloff, S.; Soustelle, C.; Fabre, F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000, 25, 192. [Google Scholar] [CrossRef]
- Maftahi, M.; Hope, J.C.; Delgado-Cruzata, L.; Han, C.S.; Freyer, G.A. The severe slow growth of Δ srs2 Δ rqh1 in Schizosaccharomyces pombe is suppressed by loss of recombination and checkpoint genes. Nucleic Acids Res. 2002, 30, 4781–4792. [Google Scholar] [CrossRef]
- Doerfler, L.; Harris, L.; Viebranz, E.; Schmidt, K.H. Differential genetic interactions between Sgs1, DNA-damage checkpoint components and DNA repair factors in the maintenance of chromosome stability. Genome Integr. 2011, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Doerfler, L.; Schmidt, K.H. Exo1 phosphorylation status controls the hydroxyurea sensitivity of cells lacking the Pol32 subunit of DNA polymerases delta and zeta. DNA Repair 2014, 24, 26–36. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gravel, S.; Chapman, J.R.; Magill, C.; Jackson, S.P. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008, 22, 2767–2772. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Chung, W.-H.; Shim, E.Y.; Lee, S.E.; Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 2008, 134, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Mimitou, E.P.; Symington, L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 2008, 455, 770. [Google Scholar] [CrossRef]
- Spell, R.M.; Jinks-Robertson, S. Examination of the Roles of Sgs1 and Srs2 Helicases in the Enforcement of Recombination Fidelity in Saccharomyces cerevisiae. Genetics 2004, 168, 1855–1865. [Google Scholar] [CrossRef]
- Schmidt, K.H.; Kolodner, R.D. Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc. Natl. Acad. Sci. USA 2006, 103, 18196–18201. [Google Scholar] [CrossRef]
- Lee, S.-K.; Johnson, R.E.; Yu, S.-L.; Prakash, L.; Prakash, S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 1999, 286, 2339–2342. [Google Scholar] [CrossRef]
- Schmidt, K.H.; Kolodner, R.D. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 2004, 24, 3213–3226. [Google Scholar] [CrossRef]
- Kaliraman, V.; Mullen, J.R.; Fricke, W.M.; Bastin-Shanower, S.A.; Brill, S.J. Functional overlap between Sgs1–Top3 and the Mms4–Mus81 endonuclease. Genes Dev. 2001, 15, 2730–2740. [Google Scholar] [CrossRef]
- Ooi, S.L.; Shoemaker, D.D.; Boeke, J.D. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 2003, 35, 277. [Google Scholar] [CrossRef]
- Mimitou, E.P.; Symington, L.S. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010, 29, 3358–3369. [Google Scholar] [CrossRef]
- Torres, J.Z.; Schnakenberg, S.L.; Zakian, V.A. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: Viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 2004, 24, 3198–3212. [Google Scholar] [CrossRef]
- Fabre, F.; Chan, A.; Heyer, W.-D.; Gangloff, S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 2002, 99, 16887–16892. [Google Scholar] [CrossRef]
- Boddy, M.N.; Lopez-Girona, A.; Shanahan, P.; Interthal, H.; Heyer, W.-D.; Russell, P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 2000, 20, 8758–8766. [Google Scholar] [CrossRef]
- Anderson, R.M.; Sinclair, D.A. Yeast RecQ Helicases: Clues to DNA Repair, Genome Stability and Aging. Mol. Mech. Werner’s Syndr. 2004, 78. [Google Scholar]
- Caspari, T.; Murray, J.M.; Carr, A.M. Cdc2—Cyclin B kinase activity links Crb2 and Rqh1—Topoisomerase III. Genes Dev. 2002, 16, 1195–1208. [Google Scholar] [CrossRef]
- Doe, C.L.; Ahn, J.S.; Dixon, J.; Whitby, M.C. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002, 277, 32753–32759. [Google Scholar] [CrossRef]
- Ashton, T.M.; Hickson, I.D. Yeast as a model system to study RecQ helicase function. DNA Repair 2010, 9, 303–314. [Google Scholar] [CrossRef]
- Laursen, L.V.; Ampatzidou, E.; Andersen, A.H.; Murray, J.M. Role for the fission yeast RecQ helicase in DNA repair in G2. Mol. Cell. Biol. 2003, 23, 3692–3705. [Google Scholar] [CrossRef][Green Version]
- Budd, M.E.; Campbell, J.L. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 1995, 92, 7642–7646. [Google Scholar] [CrossRef]
- Gnügge, R.; Symington, L.S. Keeping it real: MRX—Sae2 clipping of natural substrates. Genes Dev. 2017, 31, 2311–2312. [Google Scholar] [CrossRef]
- Xue, C.; Wang, W.; Crickard, J.B.; Moevus, C.J.; Kwon, Y.; Sung, P.; Greene, E.C. Regulatory control of Sgs1 and Dna2 during eukaryotic DNA end resection. Proc. Natl. Acad. Sci. USA 2019, 116, 6091–6100. [Google Scholar] [CrossRef]
- Adkins, N.L.; Niu, H.; Sung, P.; Peterson, C.L. Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 2013, 20, 836. [Google Scholar] [CrossRef]
- Kalocsay, M.; Hiller, N.J.; Jentsch, S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 2009, 33, 335–343. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Krebs, J.E.; Peterson, C.L. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006, 20, 2437–2449. [Google Scholar] [CrossRef]
- Bernstein, K.A.; Reid, R.J.; Sunjevaric, I.; Demuth, K.; Burgess, R.C.; Rothstein, R. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol. Biol. Cell 2011, 22, 1599–1607. [Google Scholar] [CrossRef]
- Godin, S.; Wier, A.; Kabbinavar, F.; Bratton-Palmer, D.S.; Ghodke, H.; Van Houten, B.; VanDemark, A.P.; Bernstein, K.A. The Shu complex interacts with Rad51 through the Rad51 paralogues Rad55-Rad57 to mediate error-free recombination. Nucleic Acids Res. 2013, 41, 4525–4534. [Google Scholar] [CrossRef]
- She, Z.; Gao, Z.Q.; Liu, Y.; Wang, W.J.; Liu, G.F.; Shtykova, E.V.; Xu, J.H.; Dong, Y.H. Structural and SAXS analysis of the budding yeast SHU-complex proteins. FEBS Lett. 2012, 586, 2306–2312. [Google Scholar] [CrossRef]
- Sugiyama, T.; Kowalczykowski, S.C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002, 277, 31663–31672. [Google Scholar] [CrossRef]
- Crickard, J.B.; Xue, C.; Wang, W.; Kwon, Y.; Sung, P.; Greene, E.C. The RecQ helicase Sgs1 drives ATP-dependent disruption of Rad51 filaments. Nucleic Acids Res. 2019, 47, 4694–4706. [Google Scholar] [CrossRef] [PubMed]
- Antony, E.; Tomko, E.J.; Xiao, Q.; Krejci, L.; Lohman, T.M.; Ellenberger, T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell 2009, 35, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Kodadek, T. Inhibition of protein-mediated homologous pairing by a DNA helicase. J. Biol. Chem. 1991, 266, 9712–9718. [Google Scholar] [PubMed]
- Morel, P.; Hejna, J.A.; Ehrlich, S.D.; Cassuto, E. Antipairing and strand transferase activities of E. coli helicase II (UvrD). Nucleic Acids Res. 1993, 21, 3205–3209. [Google Scholar] [CrossRef][Green Version]
- Fasching, C.L.; Cejka, P.; Kowalczykowski, S.C.; Heyer, W.-D. Top3-Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol. Cell 2015, 57, 595–606. [Google Scholar] [CrossRef]
- Cejka, P.; Plank, J.L.; Bachrati, C.Z.; Hickson, I.D.; Kowalczykowski, S.C. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1–Top3. Nat. Struct. Mol. Biol. 2010, 17, 1377–1382. [Google Scholar] [CrossRef]
- Menolfi, D.; Delamarre, A.; Lengronne, A.; Pasero, P.; Branzei, D. Essential roles of the Smc5/6 complex in replication through natural pausing sites and endogenous DNA damage tolerance. Mol. Cell 2015, 60, 835–846. [Google Scholar] [CrossRef]
- Bermúdez-López, M.; Villoria, M.T.; Esteras, M.; Jarmuz, A.; Torres-Rosell, J.; Clemente-Blanco, A.; Aragon, L. Sgs1’s Roles in DNA End Resection, Hj Dissolution, and Crossover Suppression Require a Two-Step Sumo Regulation Dependent on Smc5/6. Genes Dev. 2016, 11, 1339–1356. [Google Scholar]
- Doe, C.L.; Dixon, J.; Osman, F.; Whitby, M.C. Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000, 19, 2751–2762. [Google Scholar] [CrossRef]
- Hope, J.C.; Maftahi, M.; Freyer, G.A. A postsynaptic role for Rhp55/57 that is responsible for cell death in Δrqh1 mutants following replication arrest in Schizosaccharomyces pombe. Genetics 2005, 170, 519–531. [Google Scholar] [CrossRef]
- Langerak, P.; Mejia-Ramirez, E.; Limbo, O.; Russell, P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011, 7, e1002271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-M.; Liu, X.-M.; Ding, Y.-H.; Xiong, L.-Y.; Ren, J.-Y.; Zhou, Z.-X.; Wang, H.-T.; Zhang, M.-J.; Yu, Y.; Dong, M.-Q. Fission yeast Pxd1 promotes proper DNA repair by activating Rad16XPF and inhibiting Dna2. PLoS Biol. 2014, 12, e1001946. [Google Scholar] [CrossRef] [PubMed]
- Nanbu, T.; Nguyễn, L.C.; Habib, A.G.; Hirata, N.; Ukimori, S.; Tanaka, D.; Masuda, K.; Takahashi, K.; Yukawa, M.; Tsuchiya, E. Fission yeast Exo1 and Rqh1-Dna2 redundantly contribute to resection of uncapped telomeres. PLoS ONE 2015, 10, e0140456. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Frei, C.; Gasser, S.M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000, 14, 81–96. [Google Scholar] [PubMed]
- Cobb, J.A.; Bjergbaek, L.; Shimada, K.; Frei, C.; Gasser, S.M. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003, 22, 4325–4336. [Google Scholar] [CrossRef] [PubMed]
- Saffi, J.; Pereira, V.R.; Henriques, J.A.P. Importance of the Sgs1 helicase activity in DNA repair of Saccharomyces cerevisiae. Curr. Genet. 2000, 37, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Ui, A.; Seki, M.; Ogiwara, H.; Lai, M.S.; Yamamoto, K.; Tada, S.; Enomoto, T. Activation of a novel pathway involving Mms1 and Rad59 in sgs1 cells. Biochem. Biophys. Res. Commun. 2007, 356, 1031–1037. [Google Scholar] [CrossRef]
- Versini, G.; Comet, I.; Wu, M.; Hoopes, L.; Schwob, E.; Pasero, P. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 2003, 22, 1939–1949. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Kumar, S.; Hartsuiker, E.; Maftahi, M.; Carr, A.M.; Freyer, G.A.; Burhans, W.C.; Huberman, J.A. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 7472–7477. [Google Scholar] [CrossRef]
- Mankouri, H.W.; Morgan, A. The DNA helicase activity of yeast Sgs1p is essential for normal lifespan but not for resistance to topoisomerase inhibitors. Mech. Ageing Dev. 2001, 122, 1107–1120. [Google Scholar] [CrossRef]
- Wang, J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996, 65, 635–692. [Google Scholar] [CrossRef] [PubMed]
- Sogo, J.M.; Lopes, M.; Foiani, M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 2002, 297, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, I.; Bentsen, I.B.; Andersen, A.H.; Gasser, S.M.; Bjergbaek, L. A Rad53 independent function of Rad9 becomes crucial for genome maintenance in the absence of the Recq helicase Sgs1. PLoS ONE 2013, 8, e81015. [Google Scholar] [CrossRef] [PubMed]
- Ira, G.; Malkova, A.; Liberi, G.; Foiani, M.; Haber, J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 2003, 115, 401–411. [Google Scholar] [CrossRef]
- Wu, L.; Hickson, I.D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 2003, 426, 870–874. [Google Scholar] [CrossRef]
- Krejci, L.; Van Komen, S.; Li, Y.; Villemain, J.; Reddy, M.S.; Klein, H.; Ellenberger, T.; Sung, P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 2003, 423, 305–309. [Google Scholar] [CrossRef]
- Prakash, R.; Satory, D.; Dray, E.; Papusha, A.; Scheller, J.; Kramer, W.; Krejci, L.; Klein, H.; Haber, J.E.; Sung, P.; et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: Implications for crossover control in mitotic recombination. Genes Dev. 2009, 23, 67–79. [Google Scholar] [CrossRef]
- Mankouri, H.W.; Ngo, H.P.; Hickson, I.D. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell 2007, 18, 4062–4073. [Google Scholar] [CrossRef]
- Panico, E.R.; Ede, C.; Schildmann, M.; Schurer, K.A.; Kramer, W. Genetic evidence for a role of Saccharomyces cerevisiae Mph1 in recombinational DNA repair under replicative stress. Yeast 2010, 27, 11–27. [Google Scholar] [CrossRef]
- Fricke, W.M.; Brill, S.J. Slx1—Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1—Top3. Genes Dev. 2003, 17, 1768–1778. [Google Scholar] [CrossRef]
- Mullen, J.R.; Kaliraman, V.; Ibrahim, S.S.; Brill, S.J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 2001, 157, 103–118. [Google Scholar] [PubMed]
- Cobb, J.A.; Schleker, T.; Rojas, V.; Bjergbaek, L.; Tercero, J.A.; Gasser, S.M. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005, 19, 3055–3069. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Vanoli, F.; Foiani, M. SUMOylation regulates Rad18-mediated template switch. Nature 2008, 456, 915. [Google Scholar] [CrossRef]
- Karras, G.I.; Jentsch, S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 2010, 141, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ball, L.G.; Fan, L.; Hanna, M.; Xiao, W. Sgs1 helicase is required for efficient PCNA monoubiquitination and translesion DNA synthesis in Saccharomyces cerevisiae. Curr. Genet. 2018, 64, 459–468. [Google Scholar] [CrossRef]
- Rockmill, B.; Fung, J.C.; Branda, S.S.; Roeder, G.S. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 2003, 13, 1954–1962. [Google Scholar] [CrossRef]
- Miyajima, A.; Seki, M.; Onoda, F.; Ui, A.; Satoh, Y.; Ohno, Y.; Enomoto, T. Different domains of Sgs1 are required for mitotic and meiotic functions. Genes Genet. Syst. 2000, 75, 319–326. [Google Scholar] [CrossRef]
- Gangloff, S.; de Massy, B.; Arthur, L.; Rothstein, R.; Fabre, F. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 1999, 18, 1701–1711. [Google Scholar] [CrossRef]
- Jessop, L.; Rockmill, B.; Roeder, G.S.; Lichten, M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2006, 2, e155. [Google Scholar] [CrossRef]
- Oh, S.D.; Lao, J.P.; Hwang, P.Y.; Taylor, A.F.; Smith, G.R.; Hunter, N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 2007, 130, 259–272. [Google Scholar] [CrossRef]
- Miyajima, A.; Seki, M.; Onoda, F.; Shiratori, M.; Odagiri, N.; Ohta, K.; Kikuchi, Y.; Ohno, Y.; Enomoto, T. Sgs1 helicase activity is required for mitotic but apparently not for meiotic functions. Mol. Cell. Biol. 2000, 20, 6399–6409. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Agarwal, S.; Roeder, G.S. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 2000, 102, 245–255. [Google Scholar] [CrossRef]
- Oh, S.D.; Lao, J.P.; Taylor, A.F.; Smith, G.R.; Hunter, N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell 2008, 31, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Jessop, L.; Lichten, M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell 2008, 31, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Boddy, M.N.; Gaillard, P.-H.L.; McDonald, W.H.; Shanahan, P.; Yates, J.R., III; Russell, P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 2001, 107, 537–548. [Google Scholar] [CrossRef]
- Cromie, G.A.; Hyppa, R.W.; Taylor, A.F.; Zakharyevich, K.; Hunter, N.; Smith, G.R. Single Holliday junctions are intermediates of meiotic recombination. Cell 2006, 127, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Lynn, A.; Soucek, R.; Börner, G.V. ZMM proteins during meiosis: Crossover artists at work. Chromosome Res. 2007, 15, 591–605. [Google Scholar] [CrossRef]
- Cromie, G.A.; Hyppa, R.W.; Smith, G.R. The fission yeast BLM homolog Rqh1 promotes meiotic recombination. Genetics 2008, 179, 1157–1167. [Google Scholar] [CrossRef][Green Version]
- Cromie, G.A.; Smith, G.R. Branching out: Meiotic recombination and its regulation. Trends Cell Biol. 2007, 17, 448–455. [Google Scholar] [CrossRef]
- De Boer, J.; Hoeijmakers, J.H. Nucleotide excision repair and human syndromes. Carcinogenesis 2000, 21, 453–460. [Google Scholar] [CrossRef]
- Chalissery, J.; Jalal, D.; Al-Natour, Z.; Hassan, A.H. Repair of oxidative DNA damage in Saccharomyces cerevisiae. DNA Repair 2017, 51, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjoras, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M.; von Kobbe, C.; Sommers, J.A.; Karmakar, P.; Opresko, P.L.; Piotrowski, J.; Dianova, I.; Dianov, G.L.; Bohr, V.A. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001, 20, 5791–5801. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sommers, J.A.; Wu, L.; Bohr, V.A.; Hickson, I.D.; Brosh, R.M. Stimulation of flap endonuclease-1 by the Bloom’s syndrome protein. J. Biol. Chem. 2004, 279, 9847–9856. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sommers, J.A.; Gary, R.K.; Friedrich-Heineken, E.; Hübscher, U.; Brosh, R.M., Jr. The interaction site of Flap Endonuclease-1 with WRN helicase suggests a coordination of WRN and PCNA. Nucleic Acids Res. 2005, 33, 6769–6781. [Google Scholar] [CrossRef][Green Version]
- Trego, K.S.; Chernikova, S.B.; Davalos, A.R.; Perry, J.J.P.; Finger, L.D.; Ng, C.; Tsai, M.-S.; Yannone, S.M.; Tainer, J.A.; Campisi, J. The DNA repair endonuclease XPG interacts directly and functionally with the WRN helicase defective in Werner syndrome. Cell Cycle 2011, 10, 1998–2007. [Google Scholar] [CrossRef]
- Fan, W.; Luo, J. RecQ4 facilitates UV light-induced DNA damage repair through interaction with nucleotide excision repair factor xeroderma pigmentosum group A (XPA). J. Biol. Chem. 2008, 283, 29037–29044. [Google Scholar] [CrossRef]
- Ringvoll, J.; Uldal, L.; Roed, M.A.; Reite, K.; Baynton, K.; Klungland, A.; Eide, L. Mutations in the RAD27 and SGS1 genes differentially affect the chronological and replicative lifespan of yeast cells growing on glucose and glycerol. FEMS Yeast Res. 2007, 7, 848–859. [Google Scholar] [CrossRef]
- Huang, M.-E.; Kolodner, R.D. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol. Cell 2005, 17, 709–720. [Google Scholar] [CrossRef]
- Schulz, V.P.; Zakian, V.A.; Ogburn, C.E.; McKay, J.; Jarzebowicz, A.A.; Martin, G.; Edland, S. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum. Genet. 1996, 97, 750–754. [Google Scholar] [CrossRef]
- Tahara, H.; Tokutake, Y.; Maeda, S.; Kataoka, H.; Watanabe, T.; Satoh, M.; Matsumoto, T.; Sugawara, M.; Ide, T.; Goto, M. Abnormal telomere dynamics of B-lymphoblastoid cell strains from Werner’s syndrome patients transformed by Epstein–Barr virus. Oncogene 1997, 15, 1911. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wyllie, F.S.; Jones, C.J.; Skinner, J.W.; Haughton, M.F.; Wallis, C.; Wynford-Thomas, D.; Faragher, R.G.; Kipling, D. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat. Genet. 2000, 24, 16. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, D.; Martina, M.; Clerici, M.; Lucchini, G.; Longhese, M.P. Multiple pathways regulate 3’ overhang generation at S. cerevisiae telomeres. Mol. Cell 2009, 35, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997, 3, 1271. [Google Scholar] [CrossRef] [PubMed]
- Cesare, A.J.; Reddel, R.R. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 2008, 129, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Johnson, F.B.; Marciniak, R.A.; McVey, M.; Stewart, S.A.; Hahn, W.C.; Guarente, L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001, 20, 905–913. [Google Scholar] [CrossRef]
- Cohen, H.; Sinclair, D.A. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. USA 2001, 98, 3174–3179. [Google Scholar] [CrossRef]
- Huang, P.-H.; Pryde, F.E.; Lester, D.; Maddison, R.L.; Borts, R.H.; Hickson, I.D.; Louis, E.J. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 2001, 11, 125–129. [Google Scholar] [CrossRef]
- Hardy, J.; Churikov, D.; Géli, V.; Simon, M.-N. Sgs1 and Sae2 promote telomere replication by limiting accumulation of ssDNA. Nat. Commun. 2014, 5, 5004. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kozak, M.; Martin, J.D.; Pennock, E.; Johnson, F.B. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol. 2007, 5, e160. [Google Scholar] [CrossRef]
- Luke-Glaser, S.; Luke, B. The Mph1 helicase can promote telomere uncapping and premature senescence in budding yeast. PLoS ONE 2012, 7, e42028. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; O’Rourke, J.J.; Sobinoff, A.P.; Allen, J.A.; Nelson, C.B.; Tomlinson, C.G.; Lee, M.; Reddel, R.R.; Deans, A.J.; Pickett, H.A. The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT). Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Nanbu, T.; Takahashi, K.; Murray, J.M.; Hirata, N.; Ukimori, S.; Kanke, M.; Masukata, H.; Yukawa, M.; Tsuchiya, E.; Ueno, M. Fission yeast RecQ helicase Rqh1 is required for the maintenance of circular chromosomes. Mol. Cell. Biol. 2013, 33, 1175–1187. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McVey, M.; Kaeberlein, M.; Tissenbaum, H.A.; Guarente, L. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics 2001, 157, 1531–1542. [Google Scholar]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef]
- Heo, S.J.; Tatebayashi, K.; Ohsugi, I.; Shimamoto, A.; Furuichi, Y.; Ikeda, H. Bloom’s syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells 1999, 4, 619–625. [Google Scholar] [CrossRef]
- Lee, H.-C.; Wei, Y.-H. Mitochondria and aging. In Advances in Mitochondrial Medicine; Springer: Berlin/Heidelberg, Germany, 2012; pp. 311–327. [Google Scholar]
- Croteau, D.L.; Rossi, M.L.; Canugovi, C.; Tian, J.; Sykora, P.; Ramamoorthy, M.; Wang, Z.; Singh, D.K.; Akbari, M.; Kasiviswanathan, R. RECQL4 localizes to mitochondria and preserves mitochondrial DNA integrity. Aging Cell 2012, 11, 456–466. [Google Scholar] [CrossRef]
- Smeal, T.; Claus, J.; Kennedy, B.; Cole, F.; Guarente, L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 1996, 84, 633–642. [Google Scholar] [CrossRef]
- Barea, F.; Tessaro, S.; Bonatto, D. In silico analyses of a new group of fungal and plant RecQ4-homologous proteins. Comput. Biol. Chem. 2008, 32, 349–358. [Google Scholar] [CrossRef]
- Kwon, S.-H.; Choi, D.-H.; Lee, R.; Bae, S.-H. Saccharomyces cerevisiae Hrq1 requires a long 3′-tailed DNA substrate for helicase activity. Biochem. Biophys. Res. Commun. 2012, 427, 623–628. [Google Scholar] [CrossRef]
- Choi, D.-H.; Lee, R.; Kwon, S.-H.; Bae, S.-H. Hrq1 functions independently of Sgs1 to preserve genome integrity in Saccharomyces cerevisiae. J. Microbiol. 2013, 51, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Groocock, L.M.; Prudden, J.; Perry, J.J.P.; Boddy, M.N. The RecQ4 orthologue Hrq1 is critical for DNA interstrand cross-link repair and genome stability in fission yeast. Mol. Cell. Biol. 2012, 32, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Chan, A.; Zakian, V.A. Hrq1, a homolog of the human RecQ4 helicase, acts catalytically and structurally to promote genome integrity. Cell Rep. 2014, 6, 346–356. [Google Scholar] [CrossRef]
- Choi, D.-H.; Min, M.-H.; Kim, M.-J.; Lee, R.; Kwon, S.-H.; Bae, S.-H. Hrq1 facilitates nucleotide excision repair of DNA damage induced by 4-nitroquinoline-1-oxide and cisplatin in Saccharomyces cerevisiae. J. Microbiol. 2014, 52, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Mayor, T.; Graumann, J.; Bryan, J.; MacCoss, M.J.; Deshaies, R.J. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol. Cell. Proteom. 2007, 6, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Mandell, J.G.; Goodrich, K.J.; Bähler, J.; Cech, T.R. Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase. J. Biol. Chem. 2005, 280, 5249–5257. [Google Scholar] [CrossRef]
- Legrand, M.; Chan, C.L.; Jauert, P.A.; Kirkpatrick, D.T. The contribution of the S-phase checkpoint genes MEC1 and SGS1 to genome stability maintenance in Candida albicans. Fungal Genet. Biol. 2011, 48, 823–830. [Google Scholar] [CrossRef]
- Rai, M.N.; Balusu, S.; Gorityala, N.; Dandu, L.; Kaur, R. Functional genomic analysis of Candida glabrata-macrophage interaction: Role of chromatin remodeling in virulence. PLoS Pathog. 2012, 8, e1002863. [Google Scholar] [CrossRef]
- Schmidt, K.H.; Viebranz, E.; Doerfler, L.; Lester, C.; Rubenstein, A. Formation of complex and unstable chromosomal translocations in yeast. PLoS ONE 2010, 5, e12007. [Google Scholar] [CrossRef]
- Schmidt, K.H.; Wu, J.; Kolodner, R.D. Control of Translocations between Highly Diverged Genes by Sgs1, the Saccharomyces cerevisiae Homolog of the Bloom’s Syndrome Protein. Mol. Cell. Biol. 2006, 26, 5406–5420. [Google Scholar] [CrossRef]
- Voter, A.F.; Qiu, Y.; Tippana, R.; Myong, S.; Keck, J.L. A guanine-flipping and sequestration mechanism for G-quadruplex unwinding by RecQ helicases. Nat. Commun. 2018, 9, 4201. [Google Scholar] [CrossRef] [PubMed]

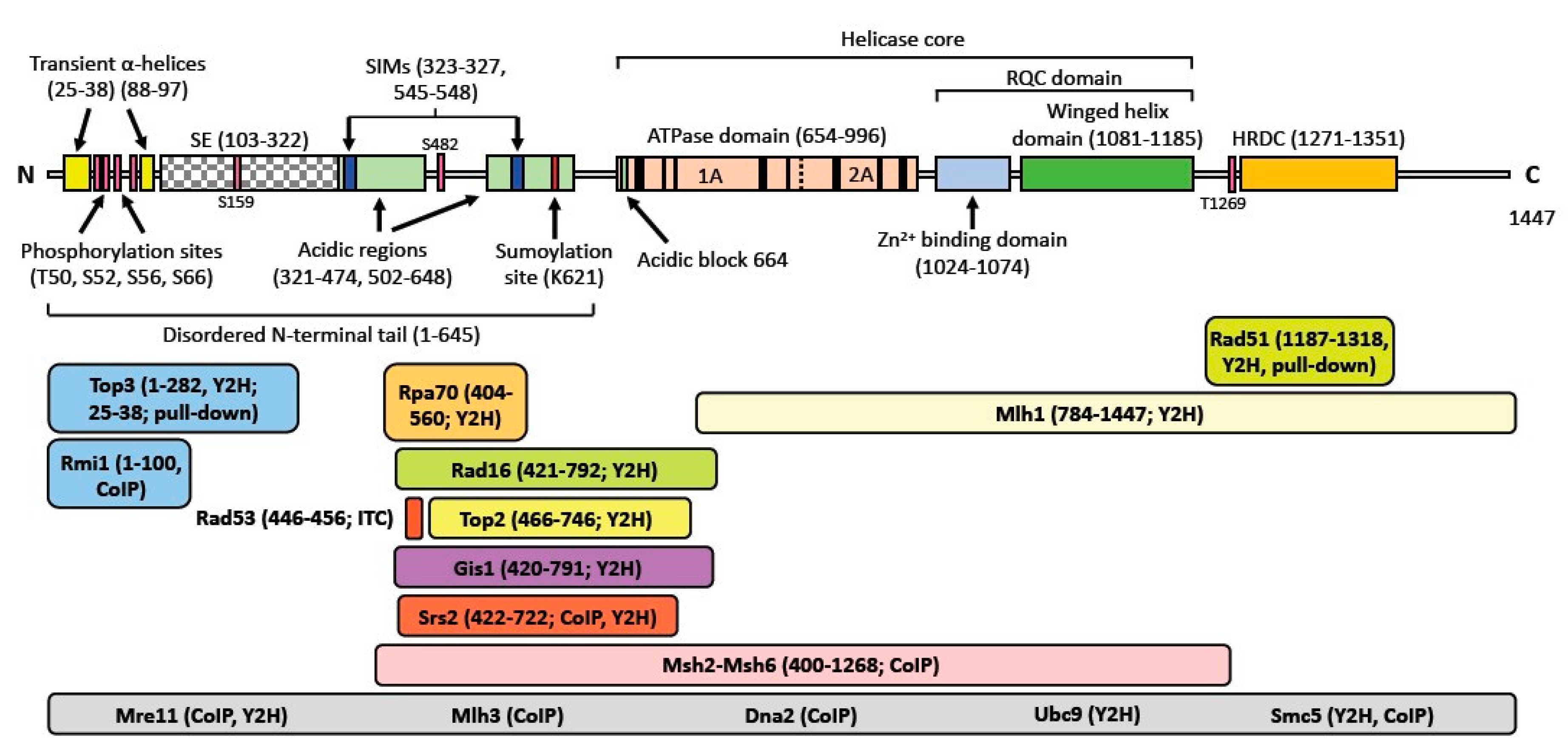
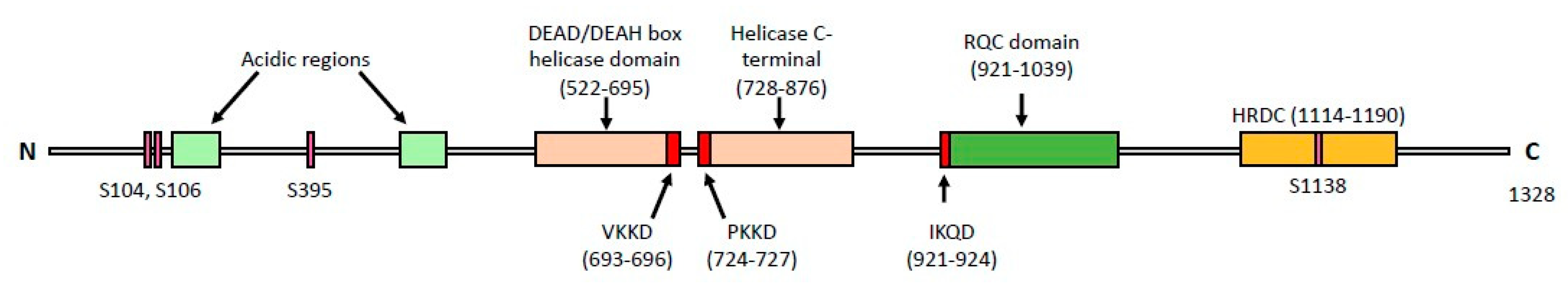
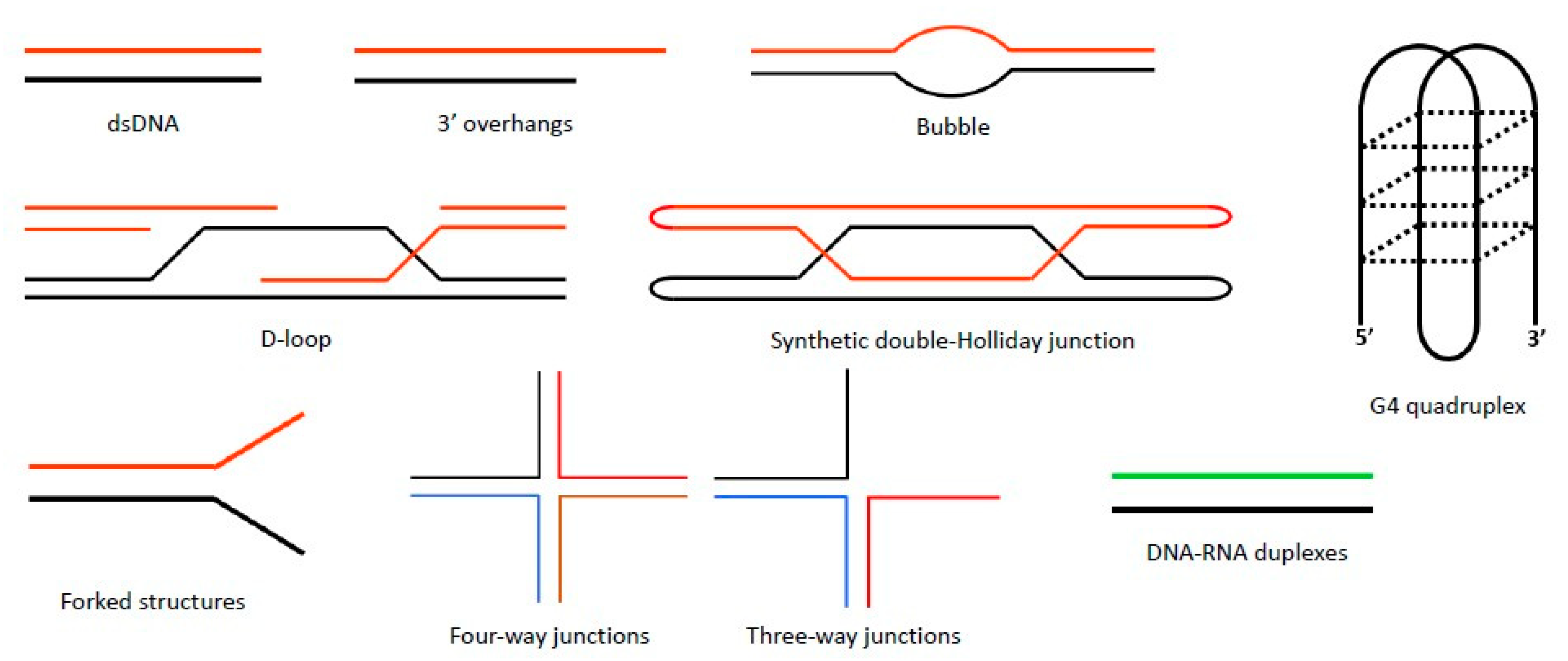

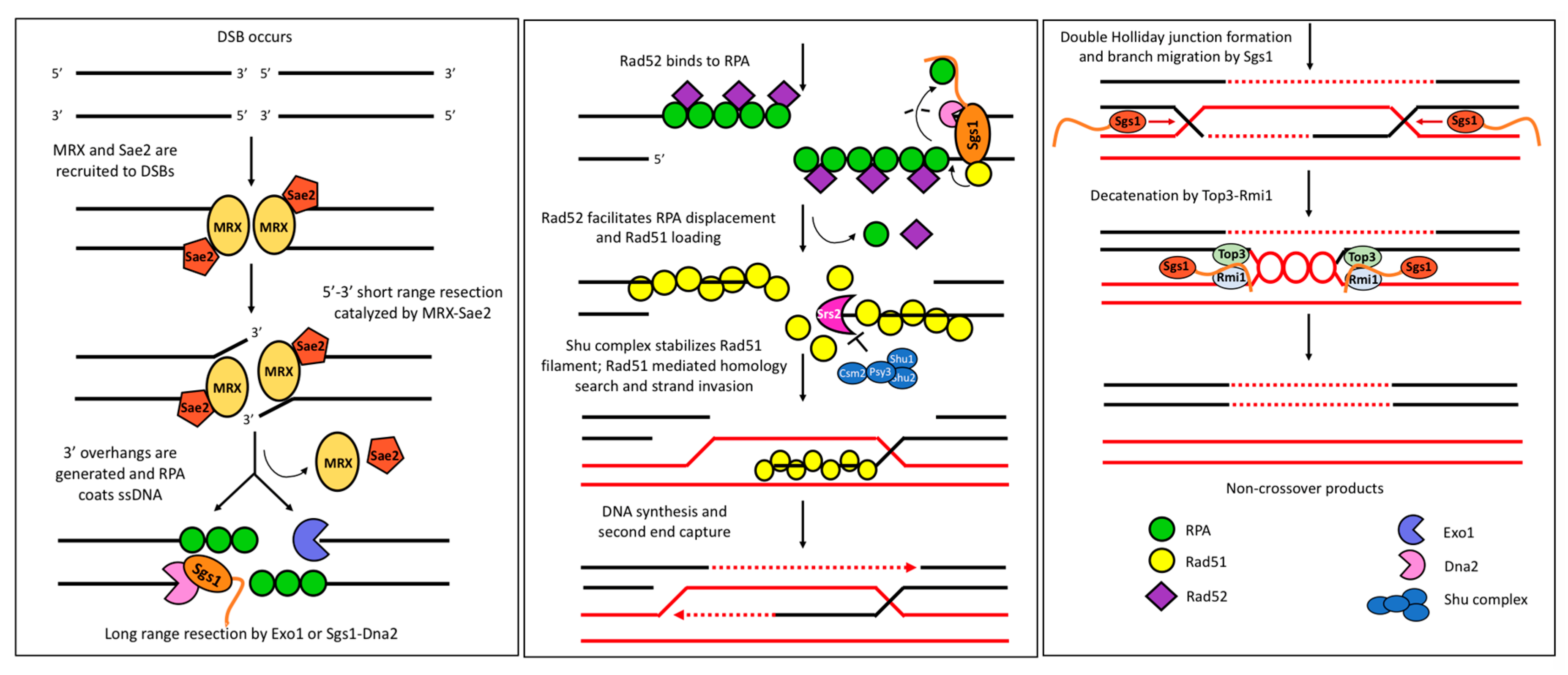
| Binding Partner | Name Description | Assay | Protein Function | Sgs1-Interacting Domain | References |
|---|---|---|---|---|---|
| Bud27 | Bud site selection | Two-hybrid | TOR-dependent gene expression | 421–791 | [97] |
| Dia2 a | Digs Into Agar | Affinity capture-western | Component of SCF E3 ubiquitin ligase complex | Full length | [98] |
| Dna2 | DNA synthesis defective | CoIP | DNA-dependent ATPase, helicase & nuclease | Full length | [51] |
| Gis1 | Glg1-2 Suppressor | Y2H | Histone demethylase & transcription factor | 420–791 | [98] |
| Mlh1 | MutL Homolog | Y2H | DNA mismatch repair | 784–1447 | [95] |
| Mlh3 | MutL Homolog | CoIP | DNA mismatch repair | Full length | [53] |
| Mre11 | Meiotic REcombination | CoIP, Y2H | DSBR; nuclease subunit of MRX | Full length | [50] |
| Prp45 | Pre-mRNA processing | Two-hybrid | Pre-mRNA splicing | 503–739 | [99] |
| Rad16 | RADiation sensitive | Y2H | Nucleotide excision repair | 421–792 | [93] |
| Rad51 | RADiation sensitive | Y2H, pull-down | DSBR; strand exchange protein | 1187–1318 | [58,91] |
| Rad53 | RADiation sensitive | ITC | DNA damage response kinase | 446–456 | [60] |
| Rpa70/Rfa1 | Replication Factor A | Y2H, affinity capture-western, reconstituted complex | Subunit of heterotrimeric RPA (ssDNA binding protein) | 421–792 | [58,100] |
| Rio1 | RIght Open reading frame | Y2H, CoIP | Serine kinase involved in cell cycle regulation and rDNA integrity | Full length | [101,102] |
| Rmi1 | RecQ Mediated genome Instability | CoIP | DSBR; subunit of Sgs1-Top3-Rmi1 complex | 1–100 | [87] |
| Rtt107/Esc4 | Regulator of Ty1 Transposition | Y2H | DNA repair during S phase; recruits Smc5/6 to DSBs | Full length | [103] |
| Smt3 | Suppressor of Mif Two | Y2H | Ubiquitin like protein | Full length | [104,105,106] |
| Srs2 | Suppressor of Rad Six | DNA helicase; antirecombinase | 422–722 | [92] | |
| Stu2 | Suppressor of TUbulin | Y2H | Microtubule associated protein | Full length | [107] |
| Top2 | TOPoisomerase | Y2H | Relaxes both positively & negatively supercoiled DNA | 466–746 | [20,86,94] |
| Top3 | TOPoisomerase | Y2H | Relaxes negatively supercoiled DNA, subunit of Sgs1-Top3-Rmi1 complex | 1–282 | [19,71,85,86] |
| Ubc9 | UBiquitin-Conjugating | Y2H | SUMO-conjugating enzyme | Full length | [56] |
| Binding Partner | Name Description | Assay | Protein Description | Rqh1-Interacting Domain | Reference |
|---|---|---|---|---|---|
| CAF-1 a (subunit Pcf1) | Chromatin assembly factor | CoIP | Histone chaperone promotes chromatin assembly during DNA repair and replication | Full length | [108] |
| Top3 | Topoisomerase III | CoIP | Relaxes negatively supercoiled DNA | 1–322 | [78] |
| RPA (Rad11) | Replication protein A | CoIP | Binds to ssDNA | Full length | [109] |
| Cdc23 | MCM associated protein Mcm10 | Two-hybrid | Efficient phosphorylation of MCM complex and pre-RC activation | Full length | [110] |
| Nse2 b | Non-SMC element SUMO ligase | Biochemical activity | Component of Smc5-6 required for DNA damage response | Full length | [111] |
| Cbh1 | CENP-B homolog | Two-hybrid | Promotes Swi6 association with centromere causing increased silencing | Full length | [110] |
| Rcl1 | rRNA processing protein | Two-hybrid | Nuclease for 18S rRNA production | Full length | [110] |
| Rmi1 | RecQ mediated genome instability protein | Two-hybrid | Holliday junction dissolution | Full length | [110] |
| Usp104 | U1 snRNP-associated protein | Two-hybrid | Splicing factor | Full length | [110] |
| Pfh1 | PiF1 Helicase homolog | Affinity capture-MS | 5′-3′ DNA helicase promotes fork progression | Full length | [112] |
| Spo7 | Sporulation protein | Two-hybrid | meiotic spindle pole body component | Full length | [110] |
| Atg11 | Autophagy associated protein | Two-hybrid | Scaffold protein in mitophagy | Full length | [110] |
| Pli1 b | SUMO E3 ligase | Biochemical activity | Major SUMO ligase, role in telomere maintenance | Full length | [111] |
| Property a | Sgs1 | Rqh1 |
|---|---|---|
| ATPase/Helicase activity | Yes | Yes |
| PTMs b | Phosphorylation, SUMOylation | Phosphorylation, SUMOylation |
| Nucleolar localization | Yes | Yes |
| Localization in nuclear foci | Yes | Yes c(NLS:1294YSRKRKYSTS1303) |
| Homologous Recombination | Yes | Yes |
| Suppression of homeologous recombination | Yes | n.d. d |
| Suppression of GCRs e | Yes | Yes (only between gene duplications) |
| Maintenance of normal lifespan | Yes | Yes |
| Suppression of meiotic defects | Yes | Yes |
| Meiosis | Yes | Yes |
| Meiotic recombination | Yes | No |
| Activation of intra-S phase checkpoint f | Yes | Yes |
| Stabilization of replicative polymerases at stalled forks | Yes | n.d. |
| Telomere maintenance g | Yes | Yes |
| rDNA maintenance | Yes | Yes |
| Resistance to DNA damaging agents | MMS, HU, CPT h, IR, H2O2, Cisplatin, MMC i | MMS, HU, CPT, MMC, IR, UV |
| Substrates | ssDNA, dsDNA, four-way junctions, forked and G4 DNA | dsDNA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, S.V.; Schmidt, K.H. Maintenance of Yeast Genome Integrity by RecQ Family DNA Helicases. Genes 2020, 11, 205. https://doi.org/10.3390/genes11020205
Gupta SV, Schmidt KH. Maintenance of Yeast Genome Integrity by RecQ Family DNA Helicases. Genes. 2020; 11(2):205. https://doi.org/10.3390/genes11020205
Chicago/Turabian StyleGupta, Sonia Vidushi, and Kristina Hildegard Schmidt. 2020. "Maintenance of Yeast Genome Integrity by RecQ Family DNA Helicases" Genes 11, no. 2: 205. https://doi.org/10.3390/genes11020205
APA StyleGupta, S. V., & Schmidt, K. H. (2020). Maintenance of Yeast Genome Integrity by RecQ Family DNA Helicases. Genes, 11(2), 205. https://doi.org/10.3390/genes11020205





