Comparative Transcriptome Analysis Identifying the Different Molecular Genetic Markers Related to Production Performance and Meat Quality in Longissimus Dorsi Tissues of MG × STH and STH Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation, Collection, and Measures of Production Performance
2.2. RNA Isolation, Complementary DNA Library Construction, and Transcriptome Sequencing
2.3. RNA-Sequencing Data Analysis
2.4. Quantitative Real-Time PCR Assays for Target Genes
2.5. Statistical Analysis of Production Performance in the MG × STH and STH Sheep
3. Results
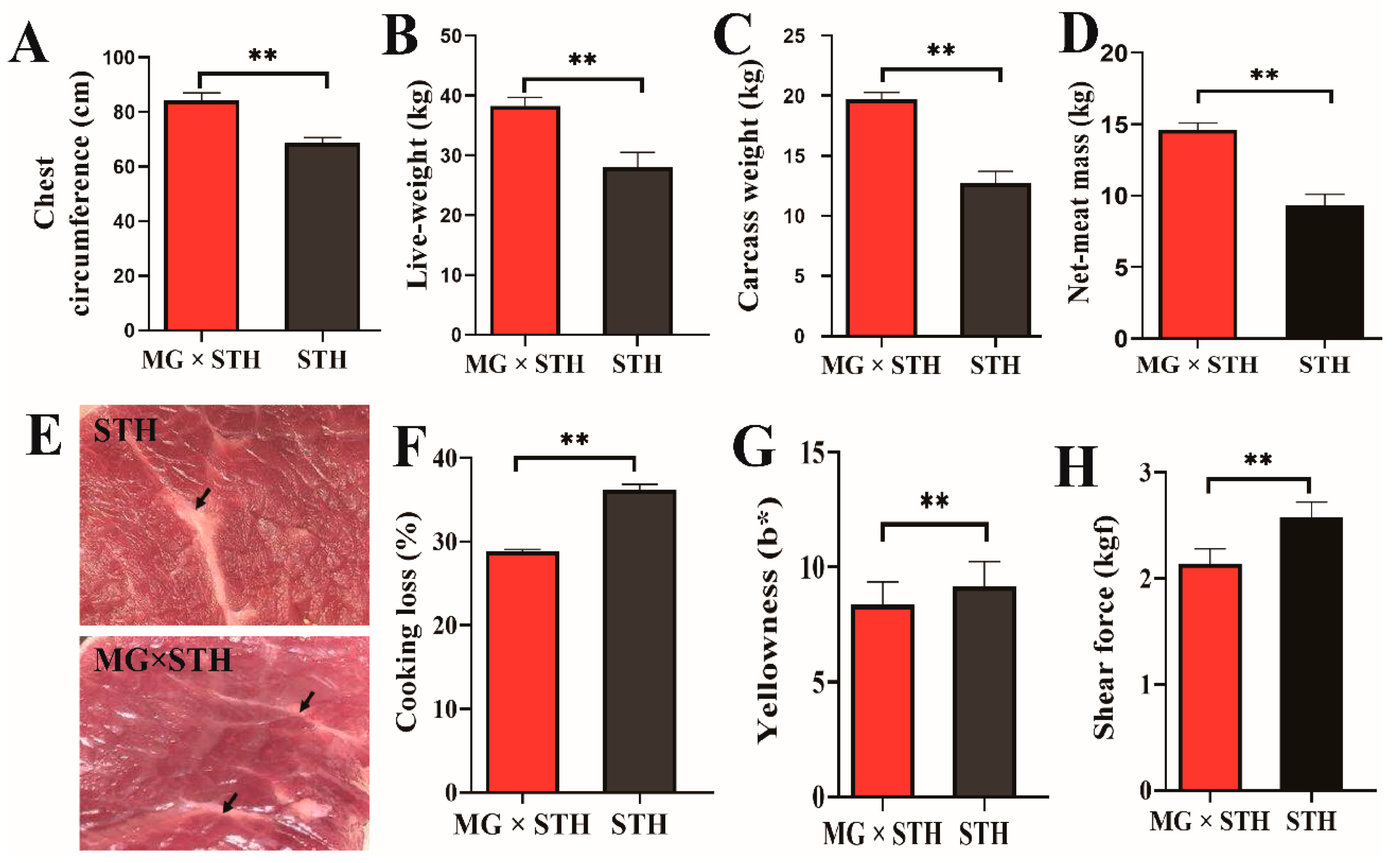
3.1. Differences in Production Performance and Meat Quality between the MG × STH and STH Sheep
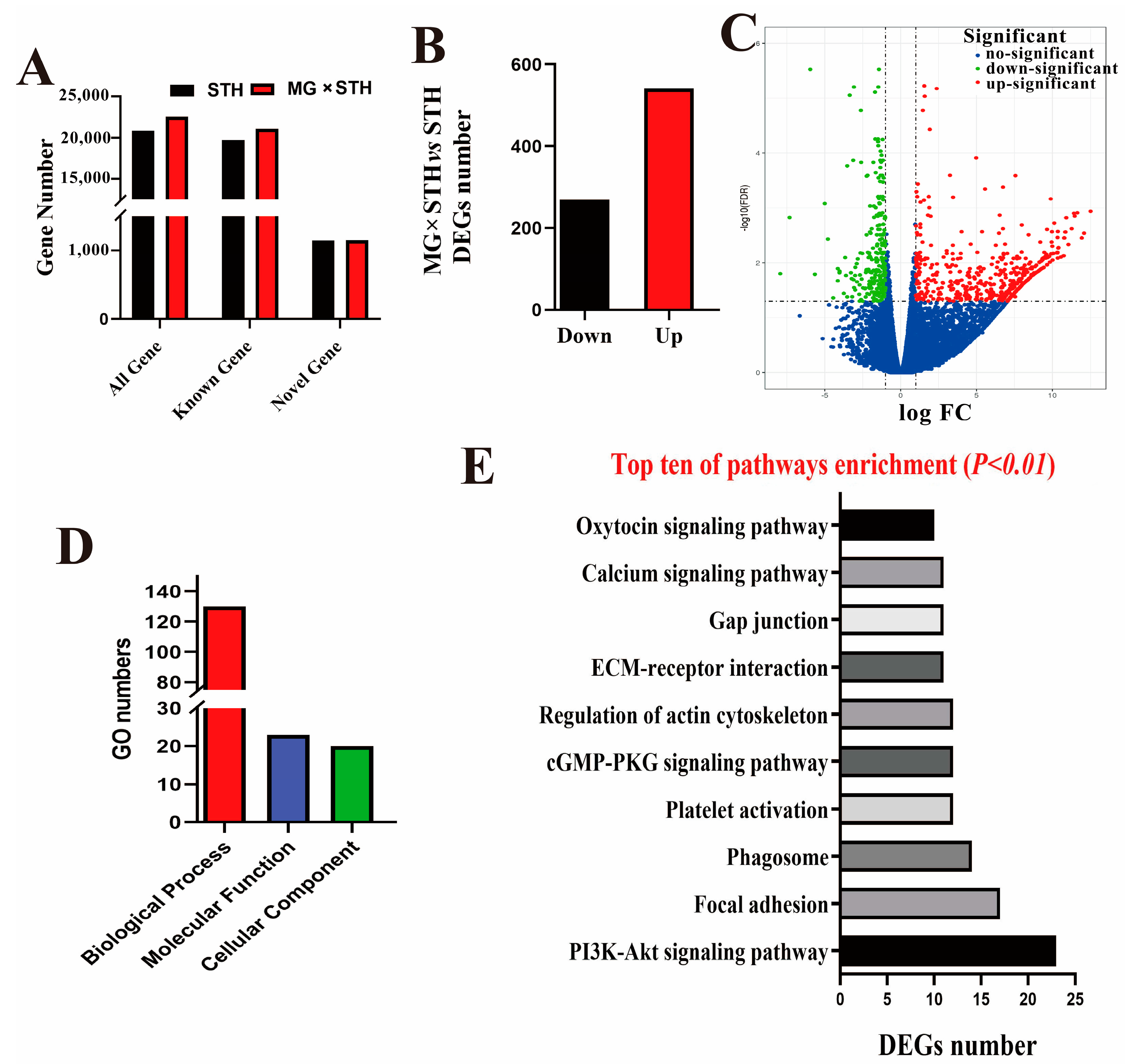
3.2. Transcriptome Analysis of Longissimus Dorsi Tissues from the MG × STH and STH Sheep
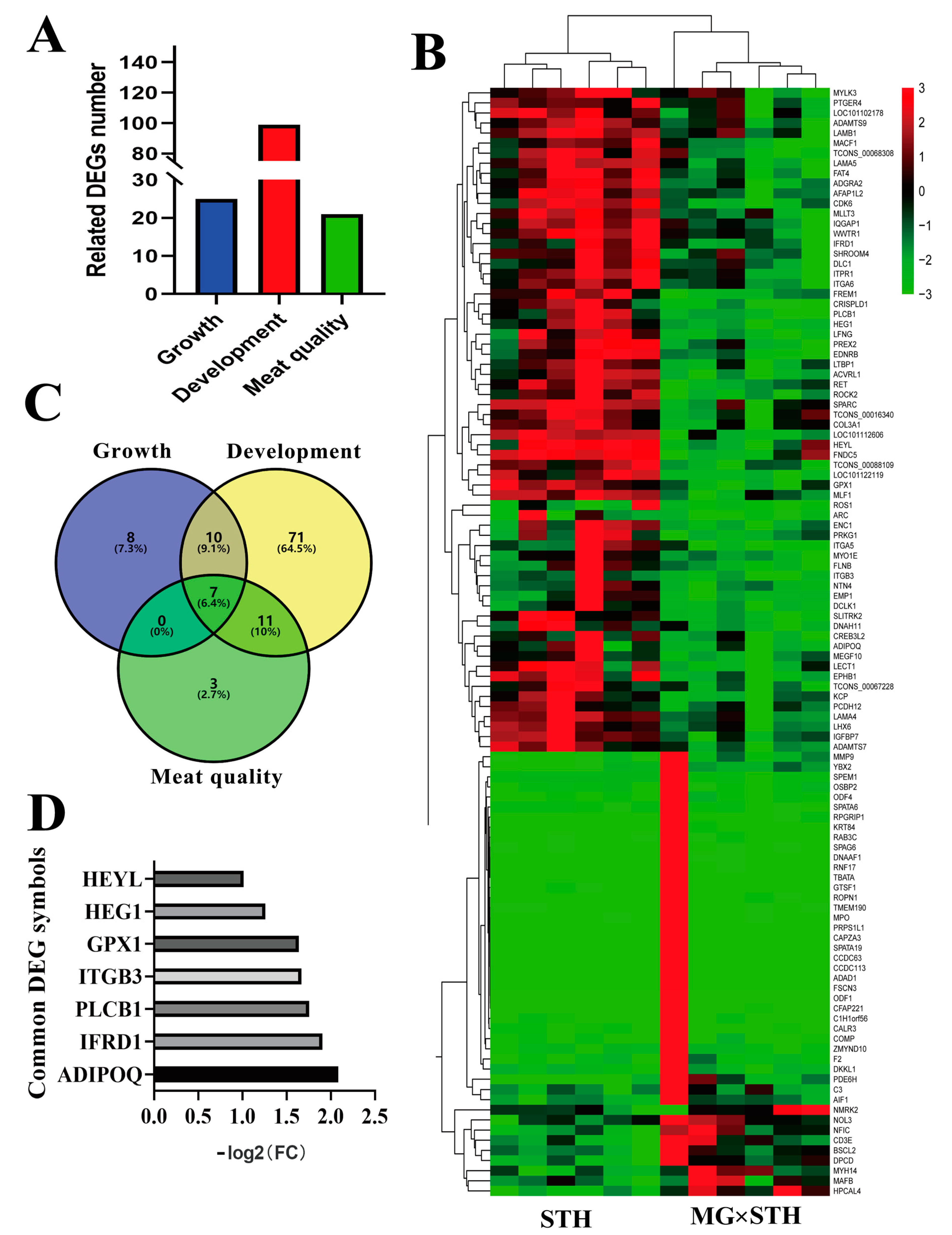
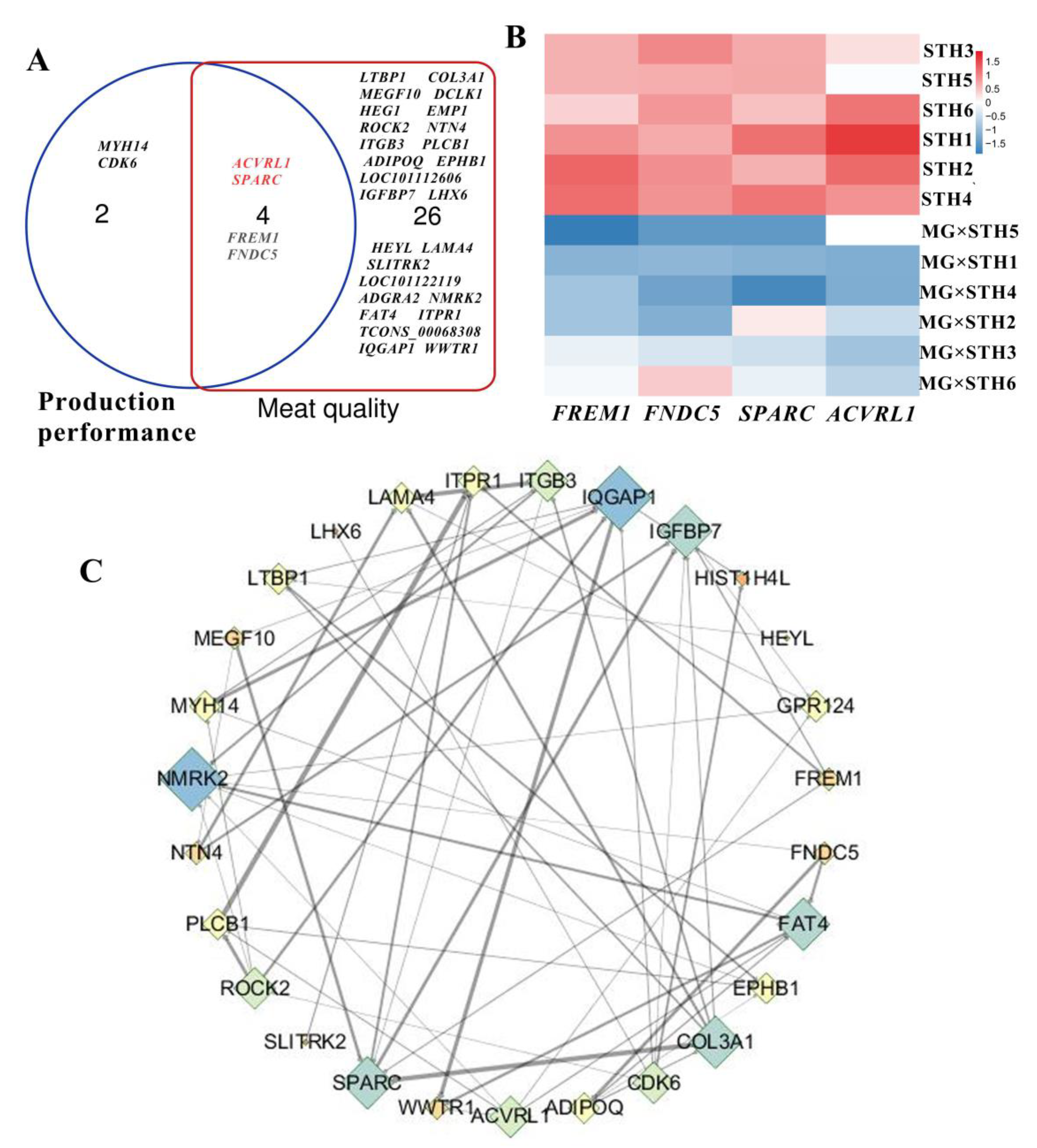
3.3. Identification of the Candidate DEGs Related to the Production Performance and Meat Quality in MG × STH and STH Sheep
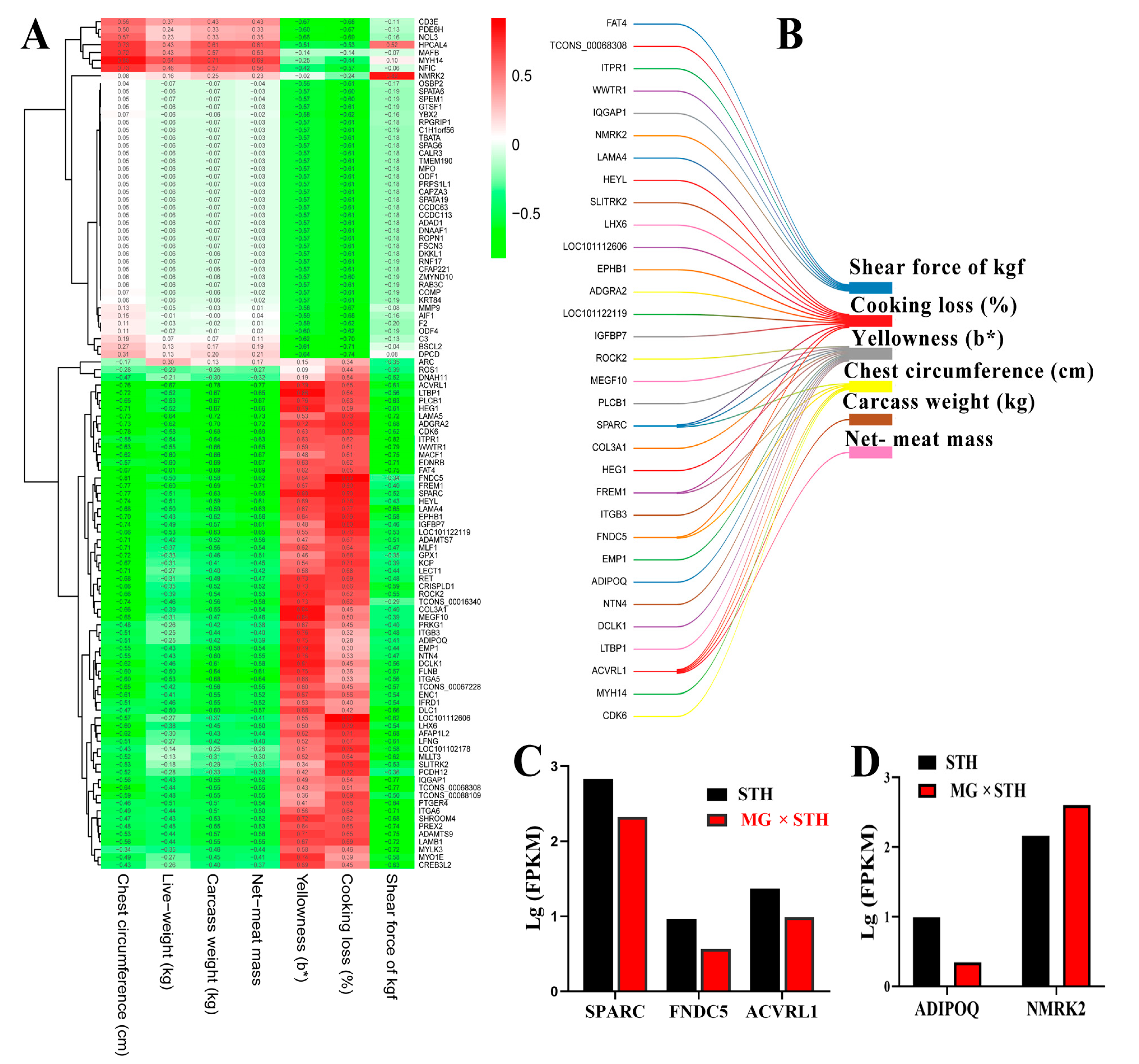
3.4. Correlation Analysis of Multiple Traits and Trait-Associated DEGs in MG × STH and STH Sheep
3.5. Gene Coexpression Analyses of Targeted DEGs Related to Production Performance and Meat Quality in MG × STH and STH Sheep
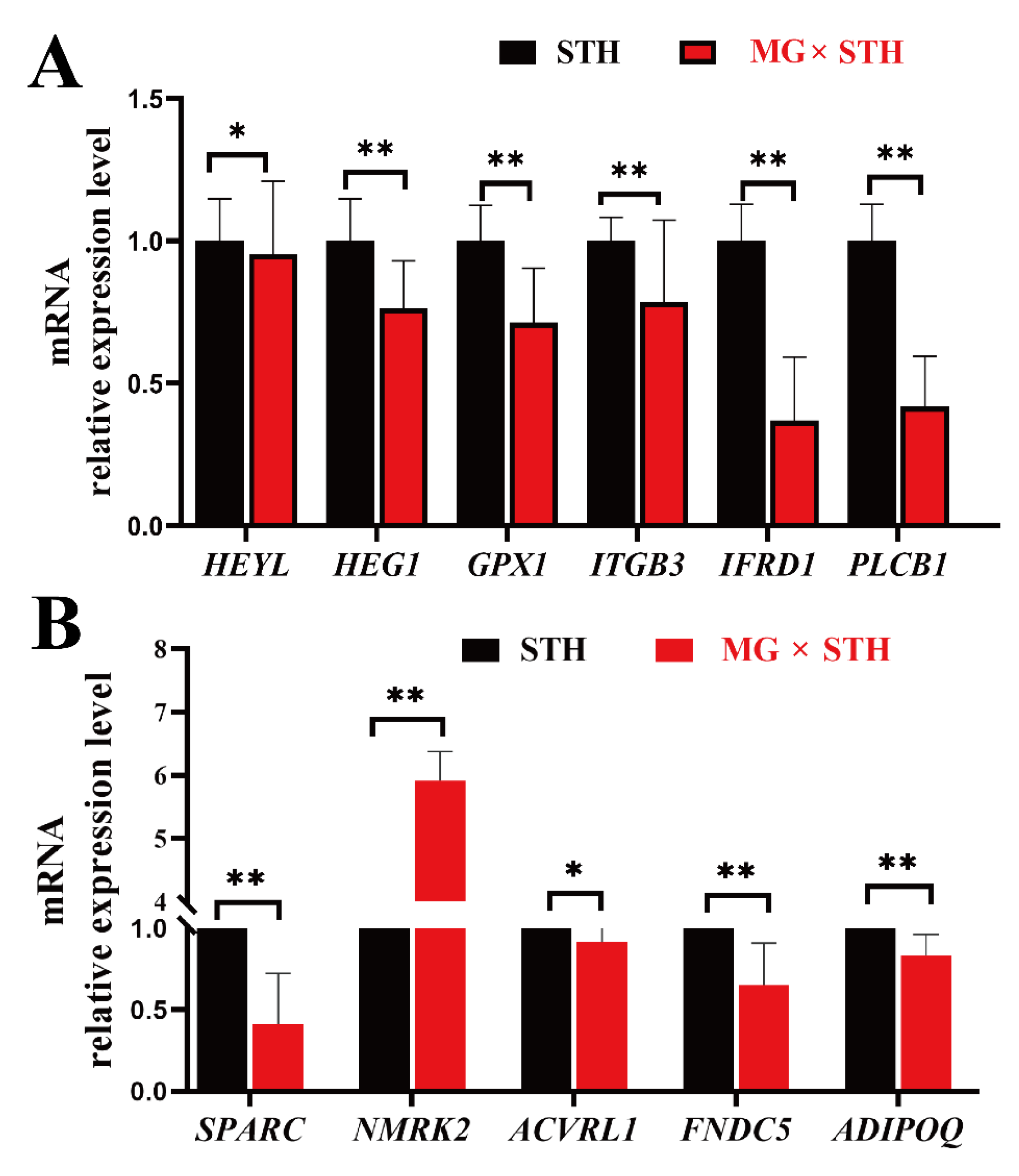
3.6. Validation of the DEGs Identified from Transcriptome and Associated Trait Correlation Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, C.; Wang, G.; Wang, J.; Ji, Z.; Dong, F.; Chao, T. Analysis of differential gene expression and novel transcript units of ovine muscle transcriptomes. PLoS ONE 2014, 9, e89817. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; You, S.; Bao, J.; Jia, Y.; Cai, Y.M. Evaluation of the growth performance and meat quality of Mongolian lamb fed grass, hay or pellets of Inner Mongolian native grass. Small Rumin. Res. 2019, 181, 34–38. [Google Scholar] [CrossRef]
- Sun, L.; Bai, M.; Xiang, L.; Zhang, G.; Ma, W.; Jiang, H. Comparative transcriptome profiling of longissimus muscle tissues from Qianhua Mutton Merino and Small Tail Han sheep. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ilisiu, E.; Daraban, S.; Neascu, G.; Ilisiu, V.; Rahman, G. Improvement of lamb production in Romania by crossbreeding of local Tsigai breed with high performance breeds. Landbauf vTI Agric. For. Res. 2010, 60, 259–266. [Google Scholar]
- Sun, L.; Lu, S.; Bai, M.; Xiang, L.; Li, J.; Jia, C.; Jiang, H. Integrative microRNA–mRNA analysis of muscle tissues in Qianhua Mutton Merino and Small Tail Han sheep reveals key roles for oar-miR-655-3p and oar-miR-381-5p. DNA Cell Biol. 2019, 38, 423–435. [Google Scholar] [CrossRef]
- Wei, C.; Wang, H.; Liu, G.; Wu, M.; Cao, J.; Liu, Z.; Liu, R.; Zhao, F.; Zhang, L.; Lu, J.; et al. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genom. 2015, 16, 194–205. [Google Scholar] [CrossRef]
- Nagalakshmi, U.; Waern, K.; Snyder, M.J. RNA-Seq: A method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 2010, 89, 4.11.11–4.11.13. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q.J.R. Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing. Reproduction 2013, 145, 587–596. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, G.; Wang, J.; Ji, Z.; Liu, Z.; Pi, X.; Chen, C. Characterization and comparative analyses of muscle transcriptomes in Dorper and small-tailed Han sheep using RNA-Seq technique. PLoS ONE 2013, 8, e72686. [Google Scholar] [CrossRef]
- Miao, X.; Qin, Q.L.X. Genome-wide transcriptome analysis of mRNAs and microRNAs in Dorset and Small Tail Han sheep to explore the regulation of fecundity. Mol. Cell Endocrinol. 2015, 402, 32–42. [Google Scholar] [CrossRef]
- Cheng, X.; Zhao, S.; Yue, Y.; Liu, Z.; Li, H.; Wu, J.R. Comparative analysis of the liver tissue transcriptomes of Mongolian and Lanzhou fat-tailed sheep. Genet. Mol. Res. 2016, 15, e15028572. [Google Scholar] [CrossRef] [PubMed]
- Knapik, J.; Ropka-Molik, K.; Pieszka, M. Genetic and nutritional factors determining the production and quality of sheep meat–A review. Ann. Anim. Sci. 2017, 17, 23–40. [Google Scholar] [CrossRef]
- Kang, X.; Liu, G.; Liu, Y.; Xu, Q.; Zhang, M.; Fang, M. Transcriptome profile at different physiological stages reveals potential mode for curly fleece in Chinese tan sheep. PLoS ONE 2013, 8, e71763. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; He, J.N.; Yu, W.M.; Liu, K.D.; Cheng, M.; Liu, J.F.; He, Y.H.; Zhao, J.S.; Qu, X.X. Transcriptome analysis of skeletal muscle at prenatal stages in Polled Dorset versus Small-tailed Han sheep. Genet. Mol. Res. 2015, 14, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jin, H.-G.; Ma, H.-H.; Zhao, Z.H. Comparative analysis on genome-wide DNA methylation in longissimus dorsi muscle between Small Tailed Han and Dorper × Small Tailed Han crossbred sheep. Asian Australas. J. Anim. 2017, 30, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Chulayo, A.Y.; Muchenje, V.; Muchenje, V. Effect of pre-slaughter conditions on physico-chemical characteristics of mutton from three sheep breeds slaughtered at a smallholder rural abattoir. S. Afr. J. Anim. Sci. 2013, 43, 64–68. [Google Scholar] [CrossRef]
- Gabryszuk, M.; Kuźnicka, E.; Horbańczuk, K.; Oprządek, J. Effects of housing systems and the diet supplements on the slaughter value and concentration of mineral elements in the loin muscle of lambs. Asian Australas. J. Anim. 2014, 27, 726–732. [Google Scholar] [CrossRef]
- Przysucha, T.; Grodzki, H.; Gołebiewski, M.; Slósarz, J.; Piotrowski, T. Evaluation of the performance of Scottish Highland beef cattle in Poland. Med. Weter. 2013, 69, 252–254. [Google Scholar]
- Brito, L.F.; McEwan, J.C.; Miller, S.; Bain, W.; Lee, M.; Dodds, K.; Newman, S.-A.; Pickering, N.; Schenkel, F.S.; Clarke, S.J. Genetic parameters for various growth, carcass and meat quality traits in a New Zealand sheep population. Small Rumin. Res. 2017, 154, 81–91. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, J.Y.; Chen, Y.N.; Yuan, F.; Zhang, H.; Yan, F.H.; Wang, M.J.; Wang, G.; Su, M.; Lu, G. Whole genome and transcriptome sequencing of matched primary and peritoneal metastatic gastric carcinoma. Sci. Rep. 2015, 5, 13750. [Google Scholar] [CrossRef]
- Xu, C.; Chen, Y.; Zhang, H.; Chen, Y.; Shen, X.; Shi, C.; Liu, Y.; Yuan, W. Integrated microRNA-mRNA analyses reveal OPLL specific microRNA regulatory network using high-throughput sequencing. Sci. Rep. 2016, 6, 21580. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, B.; Geng, J.; Zhou, J.; Zheng, R.; Chai, J.; Li, F.; Peng, J.; Jiang, S. Integrated analysis of miRNA/mRNA network in placenta identifies key factors associated with labor onset of Large White and Qingping sows. Sci. Rep. 2015, 5, 13074. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome. Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, X.; Zhu, J.; Gu, Y.; Zhao, W.; Zou, J.; Guo, Z. GO-function: Deriving biologically relevant functions from statistically significant functions. Brief. Bioinform. 2012, 13, 216–227. [Google Scholar] [CrossRef]
- Minoru, K.; Michihiro, A.; Susumu, G.; Masahiro, H.; Mika, H.; Masumi, I.; Toshiaki, K.; Shuichi, K.; Shujiro, O.; Toshiaki, T. KEGG for linking genomes to life and the environment. Nucleic. Acids. Res. 2007, 36, 480–484. [Google Scholar]
- Mortimer, S.I.; Fogarty, N.M.; van der Werf, J.H.; Brown, D.J.; Swan, A.A.; Jacob, R.H.; Geesink, G.H.; Hopkins, D.L.; Hocking Edwards, J.E.; Ponnampalam, E.N.J. Genetic correlations between meat quality traits and growth and carcass traits in Merino sheep. J. Anim. Sci. 2018, 96, 3582–3598. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Q.; Zhang, Y.; Cheng, S.; Hu, J.; Ma, Y.; Zhao, X. Comprehensive analysis of microRNA–messenger RNA from White Yak testis reveals the differentially expressed molecules involved in development and reproduction. Int. J. Mol. Sci. 2018, 19, 3083. [Google Scholar] [CrossRef]
- Morandin, C.; Tin, M.M.; Abril, S.; Gómez, C.; Pontieri, L.; Schiøtt, M.; Sundström, L.; Tsuji, K.; Pedersen, J.S.; Helanterä, H.J. Comparative transcriptomics reveals the conserved building blocks involved in parallel evolution of diverse phenotypic traits in ants. Genome. Biol. 2016, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Beccavin, C.; Chevalier, B.; Cogburn, L.A.; Simon, J.; Duclos, M.J. Insulin-like growth factors and body growth in chickens divergently selected for high or low growth rate. J. Endocrinol. 2001, 168, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for visualization and analysis of biological networks. In Data Mining in Proteomics; Humana Press: Totowa, NJ, USA, 2011; pp. 291–303. [Google Scholar]
- Zhang, Q.; Gong, J.; Wang, X.; Wu, X.; Li, Y.; Ma, Y.; Yong, Z.; Zhao, X. Molecular cloning, bioinformatics analysis and expression of insulin-like growth factor 2 from Tianzhu white yak, Bos grunniens. Int. J. Mol.sci. 2014, 15, 504–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, Q.; Gong, J.; Du, J.; Zhang, Y.; Zhao, X.J.G. Yak IGF2 promotes fibroblast proliferation via suppression of IGF1R and PI3KCG expression. Genes 2018, 9, 169. [Google Scholar] [CrossRef]
- Brand, T.; Van der Westhuizen, E.; Van der Merwe, D.; Hoffman, L.C. Effect of days in feedlot on growth performance and carcass characteristics of Merino, South African Mutton Merino and Dorper lambs. S. Afr. J. Anim. Sci. 2017, 47, 26–33. [Google Scholar] [CrossRef]
- Cloete, J.; Hoffman, L.; Cloete, S. A comparison between slaughter traits and meat quality of various sheep breeds: Wool, dual-purpose and mutton. Meat Sci. 2012, 91, 318–324. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q.; Qin, X.; Guo, Y.; Zhao, H. Genome-wide mRNA-seq profiling reveals predominant down-regulation of lipid metabolic processes in adipose tissues of Small Tail Han than Dorset sheep. Biochem. Biophys. Res. Commun. 2015, 467, 413–420. [Google Scholar] [CrossRef]
- Larzul, C.; Lefaucheur, L.; Ecolan, P.; Gogué, J.; Talmant, A.; Sellier, P.; Le, R.P.; Monin, G. Phenotypic and genetic parameters for longissimus muscle fiber characteristics in relation to growth, carcass, and meat quality traits in large white pigs. J. Anim. Sci. 1997, 75, 3126. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Cao, J.; Wu, M.; Ma, X.; Liu, Z.; Liu, R.; Zhao, F.; Wei, C.; Du, L.; et al. Genome-wide specific selection in three domestic sheep breeds. PLoS ONE 2015, 10, e0128688. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q.; Qin, X.J.G. Genome-wide analysis reveals the differential regulations of mRNAs and miRNAs in Dorset and Small Tail Han sheep muscles. Gene 2015, 562, 188–196. [Google Scholar] [CrossRef]
- Gao, Q.; Mok, H.-P.; Zhuang, J. Secreted modular calcium-binding proteins in pathophysiological processes and embryonic development. Chin. Med. J. 2019, 132, 2476. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, S.; Wang, X.; Zhang, Q.; He, Y.; Zhang, X.; Yang, L.; Shi, J. Comparative Transcriptome Analysis Identifying the Different Molecular Genetic Markers Related to Production Performance and Meat Quality in Longissimus Dorsi Tissues of MG × STH and STH Sheep. Genes 2020, 11, 183. https://doi.org/10.3390/genes11020183
Cheng S, Wang X, Zhang Q, He Y, Zhang X, Yang L, Shi J. Comparative Transcriptome Analysis Identifying the Different Molecular Genetic Markers Related to Production Performance and Meat Quality in Longissimus Dorsi Tissues of MG × STH and STH Sheep. Genes. 2020; 11(2):183. https://doi.org/10.3390/genes11020183
Chicago/Turabian StyleCheng, Shuru, Xueying Wang, Quanwei Zhang, Yuqin He, Xia Zhang, Lei Yang, and Jinping Shi. 2020. "Comparative Transcriptome Analysis Identifying the Different Molecular Genetic Markers Related to Production Performance and Meat Quality in Longissimus Dorsi Tissues of MG × STH and STH Sheep" Genes 11, no. 2: 183. https://doi.org/10.3390/genes11020183
APA StyleCheng, S., Wang, X., Zhang, Q., He, Y., Zhang, X., Yang, L., & Shi, J. (2020). Comparative Transcriptome Analysis Identifying the Different Molecular Genetic Markers Related to Production Performance and Meat Quality in Longissimus Dorsi Tissues of MG × STH and STH Sheep. Genes, 11(2), 183. https://doi.org/10.3390/genes11020183




