Horizontally Acquired Homologs of Xenogeneic Silencers: Modulators of Gene Expression Encoded by Plasmids, Phages and Genomic Islands
Abstract
1. Introduction
2. Xenogeneic Silencers
2.1. The H-NS, MvaT, Lsr2 and Rok Families of Xenogeneic Silencers
2.2. Binding to AT-Rich DNA and Oligomerization are Key Features for XS Function
3. Horizontally Acquired Homologs of Xenogeneic Silencers
3.1. Xenogeneic Silencer Homologs Encoded by Plasmids
3.2. Xenogeneic Silencer Homologs Encoded by Bacteriophages
3.3. Xenogeneic Silencer Homologs Encoded by Genomic Islands
3.3.1. Genomic Islands
3.3.2. H-NST, Ler and Hfp (H-NSB)
3.3.3. The Enterobacteriaceae-Associated ROD21-like Genomic Islands
3.3.4. Genomic Islands Encode XS Homologs from Different Families
3.4. Xenogeneic Silencer Homologs, the Growth Phase and the Environmental Conditions
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Jeong, H.; Sung, S.; Kwon, T.; Seo, M.; Caetano-Anollés, K.; Choi, S.H.; Cho, S.; Nasir, A.; Kim, H. HGTree: Database of horizontally transferred genes determined by tree reconciliation. Nucleic Acids Res. 2016, 44, D610–D619. [Google Scholar] [CrossRef] [PubMed]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Paquola, A.C.M.; Asif, H.; de Braganca Pereira, C.A.; Feltes, B.C.; Bonatto, D.; Lima, W.C.; Menck, C.F.M. Horizontal Gene Transfer Building Prokaryote Genomes: Genes Related to Exchange Between Cell and Environment are Frequently Transferred. J. Mol. Evol. 2018, 86, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Darmon, E.; Leach, D.R.F. Bacterial genome instability. Microbiol. Mol. Biol. Rev. 2014, 78, 1–39. [Google Scholar] [CrossRef]
- Platt, T.G.; Bever, J.D.; Fuqua, C. A cooperative virulence plasmid imposes a high fitness cost under conditions that induce pathogenesis. Proc. R. Soc. B Biol. Sci. 2012, 279, 1691–1699. [Google Scholar] [CrossRef]
- San Millan, A.; Toll-Riera, M.; Qi, Q.; MacLean, R.C. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun. 2015, 6, 6845. [Google Scholar] [CrossRef]
- Lamberte, L.E.; Baniulyte, G.; Singh, S.S.; Stringer, A.M.; Bonocora, R.P.; Stracy, M.; Kapanidis, A.N.; Wade, J.T.; Grainger, D.C. Horizontally acquired AT-rich genes in Escherichia coli cause toxicity by sequestering RNA polymerase. Nat. Microbiol. 2017, 2, 16249. [Google Scholar] [CrossRef]
- Navarre, W.W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective Silencing of Foreign DNA with Low GC Content by the H-NS Protein in Salmonella. Science 2006, 313, 236–238. [Google Scholar] [CrossRef]
- Lucchini, S.; Rowley, G.; Goldberg, M.D.; Hurd, D.; Harrison, M.; Hinton, J.C.D. H-NS Mediates the Silencing of Laterally Acquired Genes in Bacteria. PLoS Pathog. 2006, 2, e81. [Google Scholar] [CrossRef]
- Singh, K.; Milstein, J.N.; Navarre, W.W. Xenogeneic Silencing and Its Impact on Bacterial Genomes. Annu. Rev. Microbiol. 2016, 70, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W.; McClelland, M.; Libby, S.J.; Fang, F.C. Silencing of xenogenic DNA by H-NS—facilitation of lateral gene transfer in bacteria by a defence system that recognizes foreign DNA. Genes Dev. 2007, 21, 1456–1471. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.C.; Dorman, C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010, 8, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Will, W.R.; Bale, D.H.; Reid, P.J.; Libby, S.J.; Fang, F.C. Evolutionary expansion of a regulatory network by counter-silencing. Nat. Commun. 2014, 5, 5270. [Google Scholar] [CrossRef] [PubMed]
- Gordon, B.R.G.; Li, Y.; Wang, L.; Sintsova, A.; van Bakel, H.; Tian, S.; Navarre, W.W.; Xia, B.; Liu, J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 5154–5159. [Google Scholar] [CrossRef]
- Yun, C.S.; Takahashi, Y.; Shintani, M.; Takeda, T.; Suzuki-Minakuchi, C.; Okada, K.; Yamane, H.; Nojiri, H. MvaT family proteins encoded on IncP-7 plasmid pCAR1 and the host chromosome regulate the host transcriptome cooperatively but differently. Appl. Environ. Microbiol. 2015, 82, 832–842. [Google Scholar] [CrossRef]
- Dorman, C.J. H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria. Plasmid 2014, 75, 1–11. [Google Scholar] [CrossRef]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef]
- Helgesen, E.; Fossum-Raunehaug, S.; Skarstad, K. Lack of the H-NS protein results in extended and aberrantly positioned DNA during chromosome replication and segregation in Escherichia coli. J. Bacteriol. 2016, 198, 1305–1316. [Google Scholar] [CrossRef][Green Version]
- Scolari, V.F.; Sclavi, B.; Lagomarsino, M.C. The nucleoid as a smart polymer. Front. Microbiol. 2015, 6, 424. [Google Scholar] [CrossRef][Green Version]
- Dorman, C.J. H-NS: A universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004, 2, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J. Function of nucleoid-associated proteins in chromosome structuring and transcriptional regulation. J. Mol. Microbiol. Biotechnol. 2014, 24, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Krogh, T.J.; Møller-Jensen, J.; Kaleta, C. Impact of chromosomal architecture on the function and evolution of bacterial genomes. Front. Microbiol. 2018, 9, 2019. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W. The Impact of Gene Silencing on Horizontal Gene Transfer and Bacterial Evolution. Adv. Microb. Physiol. 2016, 69, 157–186. [Google Scholar]
- Smits, W.K.; Grossman, A.D. The Transcriptional Regulator Rok Binds A+T-Rich DNA and Is Involved in Repression of a Mobile Genetic Element in Bacillus subtilis. PLoS Genet. 2010, 6. [Google Scholar] [CrossRef]
- Duan, B.; Ding, P.; Hughes, T.R.; Navarre, W.W.; Liu, J.; Xia, B. How bacterial xenogeneic silencer rok distinguishes foreign from self DNA in its resident genome. Nucleic Acids Res. 2018, 46, 10514–10529. [Google Scholar] [CrossRef]
- Bartek, I.L.; Woolhiser, L.K.; Baughn, A.D.; Basaraba, R.J.; Jacobs, W.R.; Lenaerts, A.J.; Voskuil, M.I. Mycobacterium tuberculosis Lsr2 Is a Global Transcriptional Regulator Required for Adaptation to Changing Oxygen Levels and Virulence. mBio 2014, 5, e01106–e01114. [Google Scholar] [CrossRef]
- Hoa, T.T.; Tortosa, P.; Albano, M.; Dubnau, D. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 2002, 43, 15–26. [Google Scholar] [CrossRef]
- Castang, S.; Dove, S.L. High-order oligomerization is required for the function of the H-NS family member MvaT in Pseudomonas aeruginosa. Mol. Microbiol. 2010, 78, 916–931. [Google Scholar] [CrossRef]
- Suzuki-Minakuchi, C.; Navarre, W.W. Xenogeneic Silencing and Horizontal Gene Transfer. In DNA Traffic in the Environment; Nishida, H., Oshima, T., Eds.; Springer: Singapore, 2019; pp. 1–27. ISBN 9789811334115. [Google Scholar]
- Marcus, S.L.; Brumell, J.H.; Pfeifer, C.G.; Finlay, B.B. Salmonella pathogenicity islands: Big virulence in small packages. Microbes Infect. 2000, 2, 145–156. [Google Scholar] [CrossRef]
- Maeda, K.; Nojiri, H.; Shintani, M.; Yoshida, T.; Habe, H.; Omori, T. Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J. Mol. Biol. 2003, 326, 21–33. [Google Scholar] [CrossRef]
- Marcoleta, A.E.; Berríos-Pastén, C.; Nuñez, G.; Monasterio, O.; Lagos, R. Klebsiella pneumoniae Asparagine tDNAs Are Integration Hotspots for Different Genomic Islands Encoding Microcin E492 Production Determinants and Other Putative Virulence Factors Present in Hypervirulent Strains. Front. Microbiol. 2016, 7, 849. [Google Scholar] [CrossRef] [PubMed]
- Sherburne, C.K.; Lawley, T.D.; Gilmour, M.W.; Blattner, F.R.; Burland, V.; Grotbeck, E.; Rose, D.J.; Taylor, D.E. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 2000, 28, 2177–2186. [Google Scholar] [CrossRef] [PubMed]
- Gordon, B.R.G.; Li, Y.; Cote, A.; Weirauch, M.T.; Ding, P.; Hughes, T.R.; Navarre, W.W.; Xia, B.; Liu, J. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 10690–10695. [Google Scholar] [CrossRef] [PubMed]
- Lang, B.; Blot, N.; Bouffartigues, E.; Buckle, M.; Geertz, M.; Gualerzi, C.O.; Mavathur, R.; Muskhelishvili, G.; Pon, C.L.; Rimsky, S.; et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007, 35, 6330–6337. [Google Scholar] [CrossRef]
- Landick, R.; Wade, J.T.; Grainger, D.C. H-NS and RNA polymerase: A love-hate relationship? Curr. Opin. Microbiol. 2015, 24, 53–59. [Google Scholar] [CrossRef]
- Ding, P.; McFarland, K.A.; Jin, S.; Tong, G.; Duan, B.; Yang, A.; Hughes, T.R.; Liu, J.; Dove, S.L.; Navarre, W.W.; et al. A Novel AT-Rich DNA Recognition Mechanism for Bacterial Xenogeneic Silencer MvaT. PLoS Pathog. 2015, 11. [Google Scholar] [CrossRef]
- Summers, E.L.; Meindl, K.; Usón, I.; Mitra, A.K.; Radjainia, M.; Colangeli, R.; Alland, D.; Arcus, V.L. The Structure of the Oligomerization Domain of Lsr2 from Mycobacterium tuberculosis Reveals a Mechanism for Chromosome Organization and Protection. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Arold, S.T.; Leonard, P.G.; Parkinson, G.N.; Ladbury, J.E. H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl. Acad. Sci. USA. 2010, 107, 15728–15732. [Google Scholar] [CrossRef]
- Suzuki-Minakuchi, C.; Kawazuma, K.; Matsuzawa, J.; Vasileva, D.; Fujimoto, Z.; Terada, T.; Okada, K.; Nojiri, H. Structural similarities and differences in H-NS family proteins revealed by the N-terminal structure of TurB in Pseudomonas putida KT2440. FEBS Lett. 2016, 590, 3583–3594. [Google Scholar] [CrossRef]
- Bhat, A.P.; Shin, M.; Choy, H.E. Identification of high-specificity H-NS binding site in LEE5 promoter of enteropathogenic Esherichia coli (EPEC). J. Microbiol. 2014, 52, 626–629. [Google Scholar] [CrossRef] [PubMed]
- Johansson, J.; Eriksson, S.; Sondén, B.; Wai, S.N.; Uhlin, B.E. Heteromeric interactions among nucleoid-associated bacterial proteins: Localization of StpA-stabilizing regions in H-NS of Escherichia coli. J. Bacteriol. 2001, 183, 2343–2347. [Google Scholar] [CrossRef] [PubMed]
- Picker, M.A.; Wing, H.J. H-NS, Its Family Members and Their Regulation of Virulence Genes in Shigella Species. Genes 2016, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Rimsky, S.; Buc, H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 1996, 178, 4335–4343. [Google Scholar] [CrossRef]
- Shahul Hameed, U.F.; Liao, C.; Radhakrishnan, A.K.; Huser, F.; Aljedani, S.S.; Zhao, X.; Momin, A.A.; Melo, F.A.; Guo, X.; Brooks, C.; et al. H-NS uses an autoinhibitory conformational switch for environment-controlled gene silencing. Nucleic Acids Res. 2019, 47, 2666–2680. [Google Scholar] [CrossRef]
- Leh, H.; Khodr, A.; Bouger, M.-C.; Sclavi, B.; Rimsky, S.; Bury-Moné, S. Bacterial-chromatin structural proteins regulate the bimodal expression of the Locus of Enterocyte Effacement (LEE) pathogenicity island in enteropathogenic Escherichia coli. mBio 2017, 8. [Google Scholar] [CrossRef]
- Queiroz, M.H.; Madrid, C.; Paytubi, S.; Balsalobre, C.; Juárez, A. Integration host factor alleviates H-NS silencing of the Salmonella enterica serovar Typhimurium master regulator of SPI1, hilA. Microbiology 2011, 157, 2504–2514. [Google Scholar] [CrossRef]
- Shintani, M.; Suzuki-Minakuchi, C.; Nojiri, H. Nucleoid-associated proteins encoded on plasmids: Occurrence and mode of function. Plasmid 2015, 80, 32–44. [Google Scholar] [CrossRef]
- Piña-Iturbe, A.; Ulloa-Allendes, D.; Pardo-Roa, C.; Coronado-Arrázola, I.; Salazar-Echegarai, F.J.; Sclavi, B.; González, P.A.; Bueno, S.M. Comparative and phylogenetic analysis of a novel family of Enterobacteriaceae-associated genomic islands that share a conserved excision/integration module. Sci. Rep. 2018, 8, 10292. [Google Scholar] [CrossRef]
- Skennerton, C.T.; Angly, F.E.; Breitbart, M.; Bragg, L.; He, S.; McMahon, K.D.; Hugenholtz, P.; Tyson, G.W. Phage encoded H-NS: A potential achilles heel in the bacterial defence system. PLoS ONE 2011, 6, e20095. [Google Scholar] [CrossRef]
- Williamson, H.S.; Free, A. A truncated H-NS-like protein from enteropathogenic Escherichia coli acts as an H-NS antagonist. Mol. Microbiol. 2005, 55, 808–827. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.M.; Schneider, G.; Dobrindt, U.; Emödy, L.; Hacker, J.; Uhlin, B.E. Differential effects and interactions of endogenous and horizontally acquired H-NS-like proteins in pathogenic Escherichia coli. Mol. Microbiol. 2010, 75, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Mellies, J.L.; Benison, G.; McNitt, W.; Mavor, D.; Boniface, C.; Larabee, F.J. Ler of pathogenic Escherichia coli forms toroidal protein-DNA complexes. Microbiology 2011, 157, 1123–1133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yun, C.-S.; Suzuki, C.; Naito, K.; Takeda, T.; Takahashi, Y.; Sai, F.; Terabayashi, T.; Miyakoshi, M.; Shintani, M.; Nishida, H.; et al. Pmr, a histone-like protein H1 (H-NS) family protein encoded by the IncP-7 plasmid pCAR1, is a key global regulator that alters host function. J. Bacteriol. 2010, 192, 4720–4731. [Google Scholar] [CrossRef]
- Lang, K.S.; Johnson, T.J. Characterization of Acr2, an H-NS-like protein encoded on A/C2-type plasmids. Plasmid 2016, 87–88, 17–27. [Google Scholar] [CrossRef]
- Deighan, P.; Beloin, C.; Dorman, C.J. Three-way interactions among the Sfh, StpA and H-NS nucleoid-structuring proteins of Shigella flexneri 2a strain 2457T. Mol. Microbiol. 2003, 48, 1401–1416. [Google Scholar] [CrossRef]
- Beloin, C.; Deighan, P.; Doyle, M.; Dorman, C.J. Shigella flexneri 2a strain 2457T expresses three members of the H-NS-like protein family: Characterization of the Sfh protein. Mol. Genet. Genom. 2003, 270, 66–77. [Google Scholar] [CrossRef]
- Forns, N.; Baños, R.C.; Balsalobre, C.; Juárez, A.; Madrid, C. Temperature-dependent conjugative transfer of R27: Role of chromosome- and plasmid-encoded Hha and H-NS proteins. J. Bacteriol. 2005, 187, 3950–3959. [Google Scholar] [CrossRef]
- Doyle, M.; Fookes, M.; Ivens, A.; Mangan, M.W.; Wain, J.; Dorman, C.J. An H-NS-like Stealth Protein Aids Horizontal Transmission in Bacteria. Science 2007, 315, 251–252. [Google Scholar] [CrossRef]
- Flowers, J.J.; He, S.; Malfatti, S.; Del Rio, T.G.; Tringe, S.G.; Hugenholtz, P.; McMahon, K.D. Comparative genomics of two “Candidatus Accumulibacter” clades performing biological phosphorus removal. ISME J. 2013, 7, 2301–2314. [Google Scholar] [CrossRef]
- Singh, P.K.; Ramachandran, G.; Durán-Alcalde, L.; Alonso, C.; Wu, L.J.; Meijer, W.J.J. Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ. Microbiol. 2012, 14, 2812–2825. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, E.; Hünnefeld, M.; Popa, O.; Frunzke, J. Impact of Xenogeneic Silencing on Phage–Host Interactions. J. Mol. Biol. 2019, 431, 4670–4683. [Google Scholar] [CrossRef] [PubMed]
- Miyakoshi, M.; Shintani, M.; Terabayashi, T.; Kai, S.; Yamane, H.; Nojiri, H. Transcriptome analysis of Pseudomonas putida KT2440 harboring the completely sequenced IncP-7 plasmid pCAR1. J. Bacteriol. 2007, 189, 6849–6860. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Alarcón, C.; Singer, R.S.; Johnson, T.J. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Carraro, N.; Matteau, D.; Luo, P.; Rodrigue, S.; Burrus, V. The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination. PLoS Genet. 2014, 10, e1004714. [Google Scholar] [CrossRef]
- Dillon, S.C.; Cameron, A.D.S.; Hokamp, K.; Lucchini, S.; Hinton, J.C.D.; Dorman, C.J. Genome-wide analysis of the H-NS and Sfh regulatory networks in Salmonella Typhimurium identifies a plasmid-encoded transcription silencing mechanism. Mol. Microbiol. 2010, 76, 1250–1265. [Google Scholar] [CrossRef]
- Sun, Z.; Vasileva, D.; Suzuki-Minakuchi, C.; Okada, K.; Luo, F.; Igarashi, Y.; Nojiri, H. Differential protein-protein binding affinities of H-NS family proteins encoded on the chromosome of Pseudomonas putida KT2440 and IncP-7 plasmid pCAR1. Biosci. Biotechnol. Biochem. 2018, 82, 1640–1646. [Google Scholar] [CrossRef]
- Navarre, W.W. H-NS as a Defence System. In Bacterial Chromatin; Springer: Dordrecht, The Netherlands, 2010; pp. 251–322. ISBN 9789048134731. [Google Scholar]
- Hatfull, G.F.; Jacobs-Sera, D.; Lawrence, J.G.; Pope, W.H.; Russell, D.A.; Ko, C.-C.; Weber, R.J.; Patel, M.C.; Germane, K.L.; Edgar, R.H.; et al. Comparative Genomic Analysis of 60 Mycobacteriophage Genomes: Genome Clustering, Gene Acquisition, and Gene Size. J. Mol. Biol. 2010, 397, 119–143. [Google Scholar] [CrossRef]
- Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Karthikeyan, T.; Wadsworth, C.; Lewis, J.A.; Jacobs-Sera, D.; Falbo, J.; Gross, J.; Pannunzio, N.R.; et al. Origins of highly mosaic mycobacteriophage genomes. Cell 2003, 113, 171–182. [Google Scholar] [CrossRef]
- Pfeifer, E.; Hünnefeld, M.; Popa, O.; Polen, T.; Kohlheyer, D.; Baumgart, M.; Frunzke, J. Silencing of cryptic prophages in Corynebacterium glutamicum. Nucleic Acids Res. 2016, 44, 10117–10131. [Google Scholar]
- Hacker, J.; Kaper, J.B. Pathogenicity Islands and The Evolution of Microbes. Annu. Rev. Microbiol. 2000, 54, 641–679. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.T.; Ronson, C.W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Genetics 1998, 95, 5145–5149. [Google Scholar] [CrossRef] [PubMed]
- Che, D.; Hasan, M.S.; Chen, B. Identifying Pathogenicity Islands in Bacterial Pathogenomics Using Computational Approaches. Pathogens 2014, 3, 36–56. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.P. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: Sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002, 30, 866–875. [Google Scholar] [CrossRef]
- Wozniak, R.A.F.; Fouts, D.E.; Spagnoletti, M.; Colombo, M.M.; Ceccarelli, D.; Garriss, G.; Déry, C.; Burrus, V.; Waldor, M.K. Comparative ICE genomics: Insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 2009, 5. [Google Scholar] [CrossRef]
- Daccord, A.; Ceccarelli, D.; Rodrigue, S.; Burrus, V. Comparative Analysis of Mobilizable Genomic Islands. J. Bacteriol. 2013, 195, 606–614. [Google Scholar] [CrossRef]
- Coluzzi, C.; Guédon, G.; Devignes, M.D.; Ambroset, C.; Loux, V.; Lacroix, T.; Payot, S.; Leblond-Bourget, N. A Glimpse into the World of Integrative and Mobilizable Elements in Streptococci Reveals an Unexpected Diversity and Novel Families of Mobilization Proteins. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Murphy, R.A.; Boyd, E.F. Three Pathogenicity Islands of Vibrio cholerae Can Excise from the Chromosome and Form Circular Intermediates. J. Bacteriol. 2008, 190, 636–647. [Google Scholar] [CrossRef]
- Quiroz, T.S.; Nieto, P.A.; Tobar, H.E.; Salazar-Echegarai, F.J.; Lizana, R.J.; Quezada, C.P.; Santiviago, C.A.; Araya, D.V.; Riedel, C.A.; Kalergis, A.M.; et al. Excision of an Unstable Pathogenicity Island in Salmonella enterica Serovar Enteritidis Is Induced during Infection of Phagocytic Cells. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Lautner, M.; Schunder, E.; Herrmann, V.; Heuner, K. Regulation, integrase-dependent excision, and horizontal transfer of genomic islands in Legionella pneumophila. J. Bacteriol. 2013, 195, 1583–1597. [Google Scholar] [CrossRef]
- Manson, J.M.; Gilmore, M.S. Pathogenicity island integrase cross-talk: A potential new tool for virulence modulation. Mol. Microbiol. 2006, 61, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, M.R.; Rozovsky, S.; Boyd, E.F. Pathogenicity island cross talk mediated by recombination directionality factors facilitates excision from the chromosome. J. Bacteriol. 2016, 198, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Haskett, T.L.; Terpolilli, J.J.; Ramachandran, V.K.; Verdonk, C.J.; Poole, P.S.; O’Hara, G.W.; Ramsay, J.P. Sequential induction of three recombination directionality factors directs assembly of tripartite integrative and conjugative elements. PLOS Genet. 2018, 14, e1007292. [Google Scholar] [CrossRef] [PubMed]
- Sentchilo, V.; Czechowska, K.; Pradervand, N.; Minoia, M.; Miyazaki, R.; van der Meer, J.R. Intracellular excision and reintegration dynamics of the ICEclc genomic island of Pseudomonas knackmussii sp. strain B13. Mol. Microbiol. 2009, 72, 1293–1306. [Google Scholar] [CrossRef]
- Penadés, J.R.; Christie, G.E. The Phage-Inducible Chromosomal Islands: A Family of Highly Evolved Molecular Parasites. Annu. Rev. Virol. 2015, 2, 181–201. [Google Scholar] [CrossRef]
- Doublet, B.; Boyd, D.; Mulvey, M.R.; Cloeckaert, A. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 2005, 55, 1911–1924. [Google Scholar] [CrossRef]
- Haskett, T.L.; Terpolilli, J.J.; Bekuma, A.; O’Hara, G.W.; Sullivan, J.T.; Wang, P.; Ronson, C.W.; Ramsay, J.P. Assembly and transfer of tripartite integrative and conjugative genetic elements. Proc. Natl. Acad. Sci. USA 2016, 113, 12268–12273. [Google Scholar] [CrossRef]
- Levine, J.A.; Hansen, A.-M.; Michalski, J.M.; Hazen, T.H.; Rasko, D.A.; Kaper, J.B. H-NST induces LEE expression and the formation of attaching and effacing lesions in enterohemorrhagic Escherichia coli. PLoS ONE 2014, 9, e86618. [Google Scholar] [CrossRef]
- Elliott, S.J.; Wainwright, L.A.; McDaniel, T.K.; Jarvis, K.G.; Deng, Y.K.; Lai, L.C.; McNamara, B.P.; Donnenberg, M.S.; Kaper, J.B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 1998, 28, 1–4. [Google Scholar] [CrossRef]
- Ooka, T.; Ogura, Y.; Katsura, K.; Seto, K.; Kobayashi, H.; Kawano, K.; Tokuoka, E.; Furukawa, M.; Harada, S.; Yoshino, S.; et al. Defining the genome features of Escherichia albertii, an emerging enteropathogen closely related to Escherichia coli. Genome Biol. Evol. 2015, 7, 3170–3179. [Google Scholar]
- Deng, W.; Li, Y.; Vallance, B.A.; Finlay, B.B. Locus of Enterocyte Effacement from Citrobacter rodentium: Sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 2001, 69, 6323–6335. [Google Scholar] [CrossRef]
- Mellies, J.L.; Elliott, S.J.; Sperandio, V.; Donnenberg, M.S.; Kaper, J.B. The Per regulon of enteropathogenic Escherichia coli: Identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 1999, 33, 296–306. [Google Scholar] [CrossRef]
- Winardhi, R.S.; Gulvady, R.; Mellies, J.L.; Yan, J. Locus of enterocyte effacement-encoded regulator (Ler) of pathogenic Escherichia coli competes off histone-like nucleoid-structuring protein (H-NS) through noncooperative DNA binding. J. Biol. Chem. 2014, 289, 13739–13750. [Google Scholar] [CrossRef] [PubMed]
- Guignot, J.; Segura, A.; Tran Van Nhieu, G. The Serine Protease EspC from Enteropathogenic Escherichia coli Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System. PLoS Pathog. 2015, 11, e1005013. [Google Scholar] [CrossRef] [PubMed]
- Elliott, S.J.; Sperandio, V.; Girón, J.A.; Mellies, J.L.; Wainwright, L.; Hutcheson, S.W.; McDaniel, T.K.; Kaper, J.B. The Locus of Enterocyte Effacement (LEE)-Encoded Regulator Controls Expression of Both LEE- and Non-LEE-Encoded Virulence Factors in Enteropathogenic and Enterohemorrhagic Escherichia coli. Infect. Immun. 2000, 68, 6115–6126. [Google Scholar] [CrossRef] [PubMed]
- Lathem, W.W.; Grys, T.E.; Witowski, S.E.; Torres, A.G.; Kaper, J.B.; Tarr, P.I.; Welch, R.A. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 2002, 45, 277–288. [Google Scholar] [CrossRef]
- Torres, A.G.; López-Sánchez, G.N.; Milflores-Flores, L.; Patel, S.D.; Rojas-López, M.; Martínez De La Peña, C.F.; Arenas-Hernández, M.M.P.; Martínez-Laguna, Y. Ler and H-NS, regulators controlling expression of the long polar fimbriae of Escherichia coli O157:H7. J. Bacteriol. 2007, 189, 5916–5928. [Google Scholar] [CrossRef]
- Thomson, N.R.; Clayton, D.J.; Windhorst, D.; Vernikos, G.; Davidson, S.; Churcher, C.; Quail, M.A.; Stevens, M.; Jones, M.A.; Watson, M.; et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 2008, 18, 1624–1637. [Google Scholar] [CrossRef]
- Feasey, N.A.; Hadfield, J.; Keddy, K.H.; Dallman, T.J.; Jacobs, J.; Deng, X.; Wigley, P.; Barquist, L.; Langridge, G.C.; Feltwell, T.; et al. Distinct Salmonella Enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat. Genet. 2016, 48, 1211–1217. [Google Scholar] [CrossRef]
- Newman, R.M.; Salunkhe, P.; Godzik, A.; Reed, J.C. Identification and Characterization of a Novel Bacterial Virulence Factor That Shares Homology with Mammalian Toll/Interleukin-1 Receptor Family Proteins. Infect. Immun. 2006, 74, 594–601. [Google Scholar] [CrossRef]
- Coward, C.; Sait, L.; Williams, L.; Humphrey, T.J.; Cogan, T.; Maskell, D.J. Investigation into the role of five Salmonella enterica serovar Enteritidis genomic islands in colonization of the chicken reproductive tract and other organs following oral challenge. FEMS Microbiol. Lett. 2012, 336, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.A.; Blondel, C.J.; Quezada, C.P.; Porwollik, S.; Andrews-polymenis, H.L.; Toro, C.S.; Mcclelland, M.; Santiviago, C.A. Infection of Mice by Salmonella enterica Serovar Enteritidis Involves Additional Genes That Are Absent in the Genome of Serovar Typhimurium. Infect. Immun. 2012, 80, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Roa, C.; Salazar, G.A.; Noguera, L.; Salazar-Echegarai, F.J.; Vallejos, O.P.; Suazo, I.; Schultz, B.M.; Coronado-Arrazola, I.; Kalergis, A.M.; Bueno, S.M. Pathogenicity island excision during an infection by Salmonella enterica serovar Enteritidis is required for crossing the intestinal epithelial barrier in mice to cause systemic infection. PLOS Pathog. 2019, 15. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Echegarai, F.J.; Tobar, H.E.; Nieto, P.A.; Riedel, C.A.; Bueno, S.M. Conjugal Transfer of the Pathogenicity Island ROD21 in Salmonella enterica serovar Enteritidis Depends on Environmental Conditions. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Tobar, H.E.; Salazar-Echegarai, F.J.; Nieto, P.A.; Palavecino, C.E.; Sebastian, V.P.; Riedel, C.A.; Kalergis, A.M.; Bueno, S.M. Chromosomal Excision of a New Pathogenicity Island Modulates Salmonella Virulence In Vivo. Curr. Gene Ther. 2013, 13, 240–249. [Google Scholar] [CrossRef]
- Bueno, S.M.; Santiviago, C.A.; Murillo, A.A.; Fuentes, J.A.; Trombert, A.N.; Rodas, P.I.; Youderian, P.; Mora, G.C. Precise Excision of the Large Pathogenicity Island, SPI7, in Salmonella enterica Serovar Typhi. J. Bacteriol. 2004, 186, 3202–3213. [Google Scholar] [CrossRef] [PubMed]
- García, J.; Madrid, C.; Juárez, A.; Pons, M. New Roles for Key Residues in Helices H1 and H2 of the Escherichia coli H-NS N-terminal Domain: H-NS Dimer Stabilization and Hha Binding. J. Mol. Biol. 2006, 359, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Madrid, C.; García, J.; Pons, M.; Juárez, A. Molecular evolution of the H-NS protein: Interaction with Hha-like proteins is restricted to Enterobacteriaceae. J. Bacteriol. 2007, 189, 265–268. [Google Scholar] [CrossRef]
- Ali, S.S.; Whitney, J.C.; Stevenson, J.; Robinson, H.; Howell, P.L.; Navarre, W.W. Structural Insights into the Regulation of Foreign Genes in Salmonella by the Hha/H-NS Complex. J. Biol. Chem. 2013, 288, 13356–13369. [Google Scholar] [CrossRef]
- Gao, Y.; Foo, Y.H.; Winardhi, R.S.; Tang, Q.; Yan, J.; Kenney, L.J. Charged residues in the H-NS linker drive DNA binding and gene silencing in single cells. Proc. Natl. Acad. Sci. USA 2017, 114, 12560–12565. [Google Scholar] [CrossRef]
- Desai, P.T.; Porwollik, S.; Long, F.; Cheng, P.; Wollam, A.; Clifton, S.W.; Weinstock, G.M.; McClelland, M. Evolutionary Genomics of Salmonella enterica Subspecies. mBio 2013, 4. [Google Scholar] [CrossRef]
- Lou, L.; Zhang, P.; Piao, R.; Wang, Y. Salmonella Pathogenicity Island 1 (SPI-1) and Its Complex Regulatory Network. Front. Cell. Infect. Microbiol. 2019, 9, 270. [Google Scholar] [CrossRef] [PubMed]
- Ares, M.A.; Fernández-Vázquez, J.L.; Rosales-Reyes, R.; Jarillo-Quijada, M.D.; von Bargen, K.; Torres, J.; González-y-Merchand, J.A.; Alcántar-Curiel, M.D.; De la Cruz, M.A. H-NS Nucleoid Protein Controls Virulence Features of Klebsiella pneumoniae by Regulating the Expression of Type 3 Pili and the Capsule Polysaccharide. Front. Cell. Infect. Microbiol. 2016, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, C.-T. A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I. Bioinformatics 2004, 20, 612–622. [Google Scholar] [CrossRef]
- Zhang, C.-T.; Zhang, R. Genomic islands in Rhodopseudomonas palustris. Nat. Biotechnol. 2004, 22, 1078–1079. [Google Scholar] [CrossRef]
- Bowers, J.R.; Kitchel, B.; Driebe, E.M.; MacCannell, D.R.; Roe, C.; Lemmer, D.; de Man, T.; Rasheed, J.K.; Engelthaler, D.M.; Keim, P.; et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Marsh, J.W.; Mustapha, M.M.; Griffith, M.P.; Evans, D.R.; Ezeonwuka, C.; Pasculle, A.W.; Shutt, K.A.; Sundermann, A.; Ayres, A.M.; Shields, R.K.; et al. Evolution of Outbreak-Causing Carbapenem-Resistant Klebsiella pneumoniae ST258 at a Tertiary Care Hospital over 8 Years. mBio 2019, 10. [Google Scholar] [CrossRef]
- Zhang, X.; Wan, W.; Yu, H.; Wang, M.; Zhang, H.; Lv, J.; Tang, Y.-W.; Kreiswirth, B.N.; Du, H.; Chen, L. New Delhi Metallo-β-Lactamase 5–Producing Klebsiella pneumoniae Sequence Type 258, Southwest China, 2017. Emerg. Infect. Dis. 2019, 25, 1209–1213. [Google Scholar] [CrossRef]
- Hudson, C.M.; Lau, B.Y.; Williams, K.P. Islander: A database of precisely mapped genomic islands in tRNA and tmRNA genes. Nucleic Acids Res. 2015, 43, D48–D53. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.; Dorman, C.J. Reciprocal transcriptional and posttranscriptional growth-phase-dependent expression of sfh, a gene that encodes a paralogue of the nucleoid-associated protein H-NS. J. Bacteriol. 2006, 188, 7581–7591. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Vasileva, D.; Suzuki-Minakuchi, C.; Okada, K.; Luo, F.; Igarashi, Y.; Nojiri, H. Growth phase-dependent expression profiles of three vital H-NS family proteins encoded on the chromosome of Pseudomonas putida KT2440 and on the pCAR1 plasmid. BMC Microbiol. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
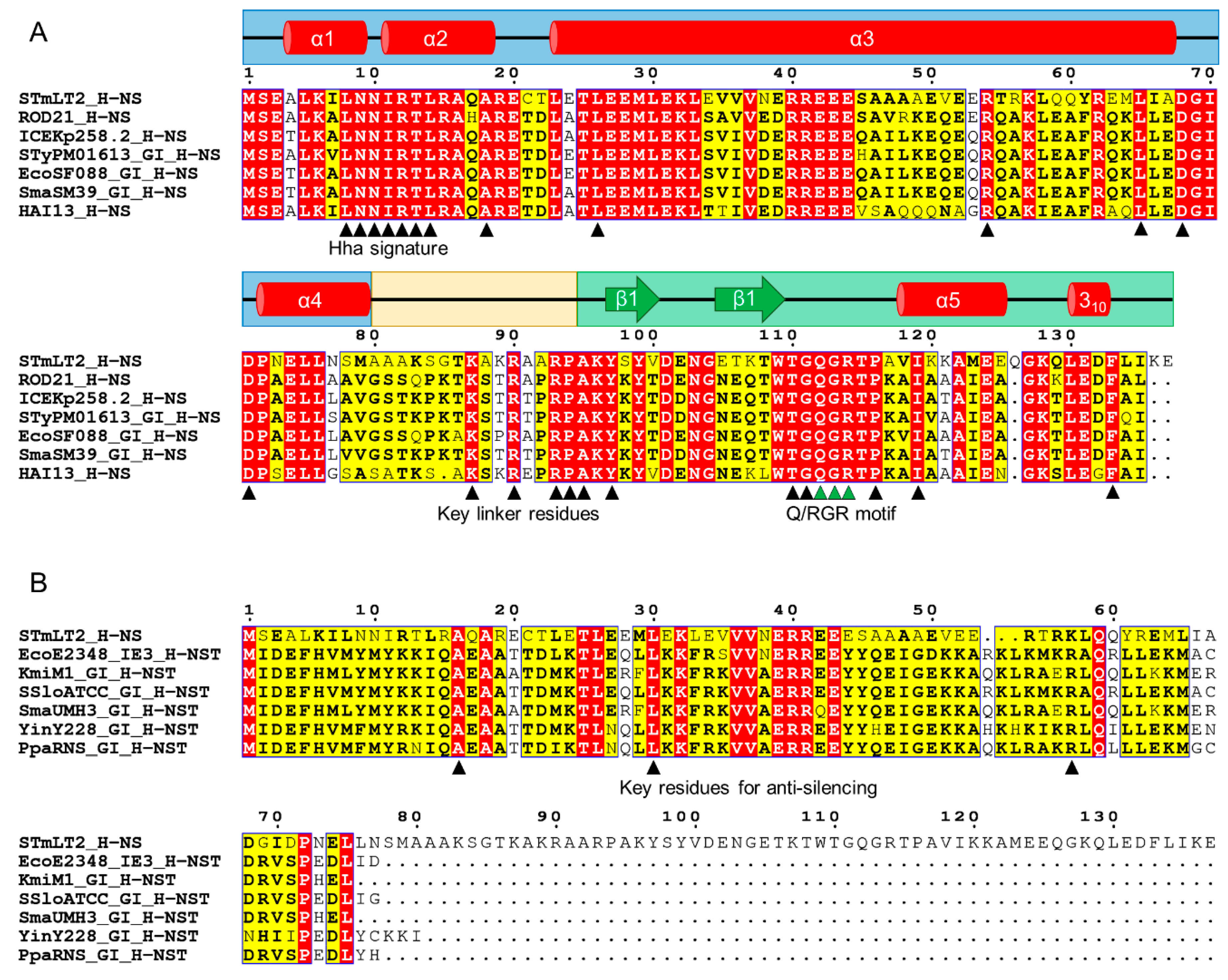
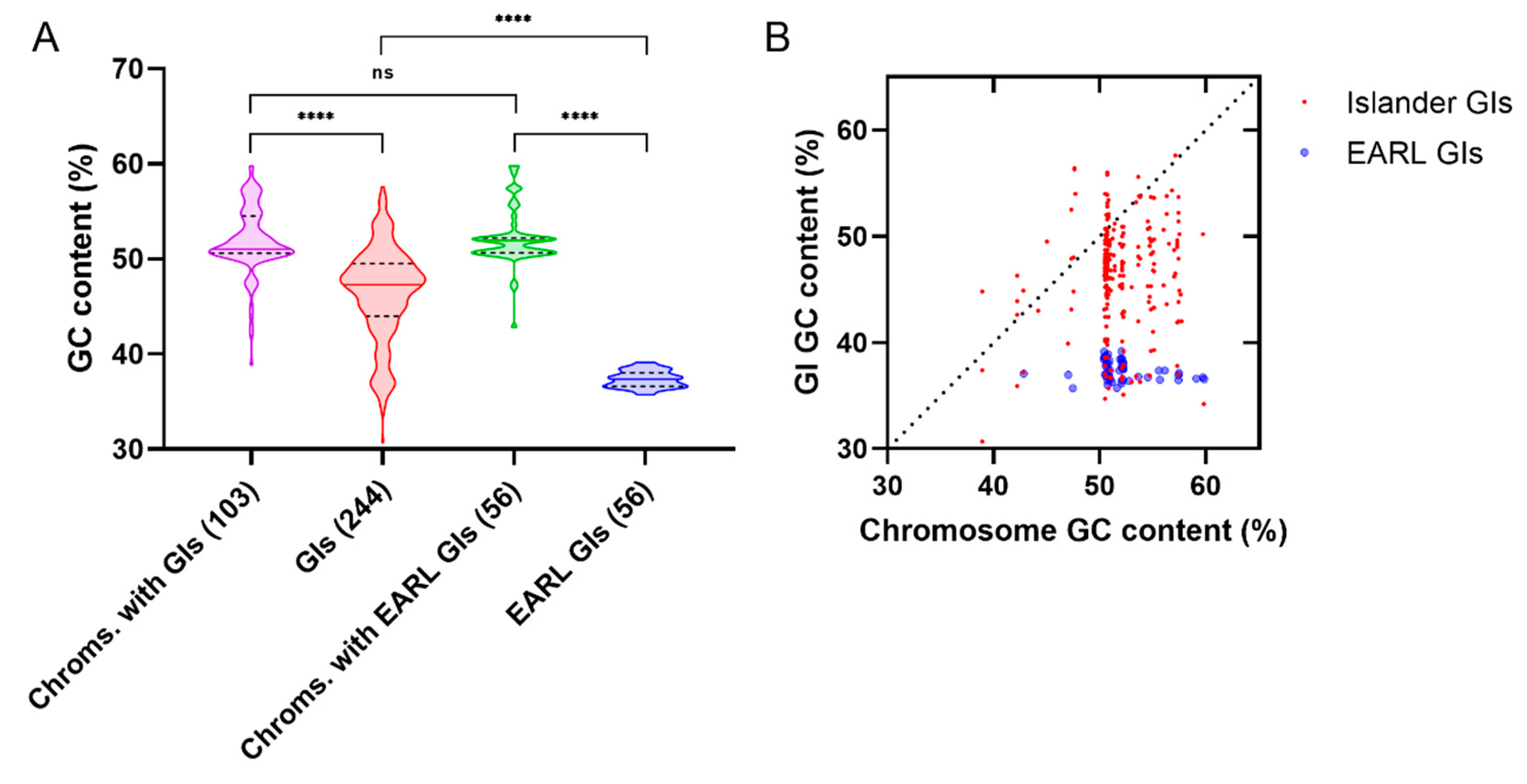
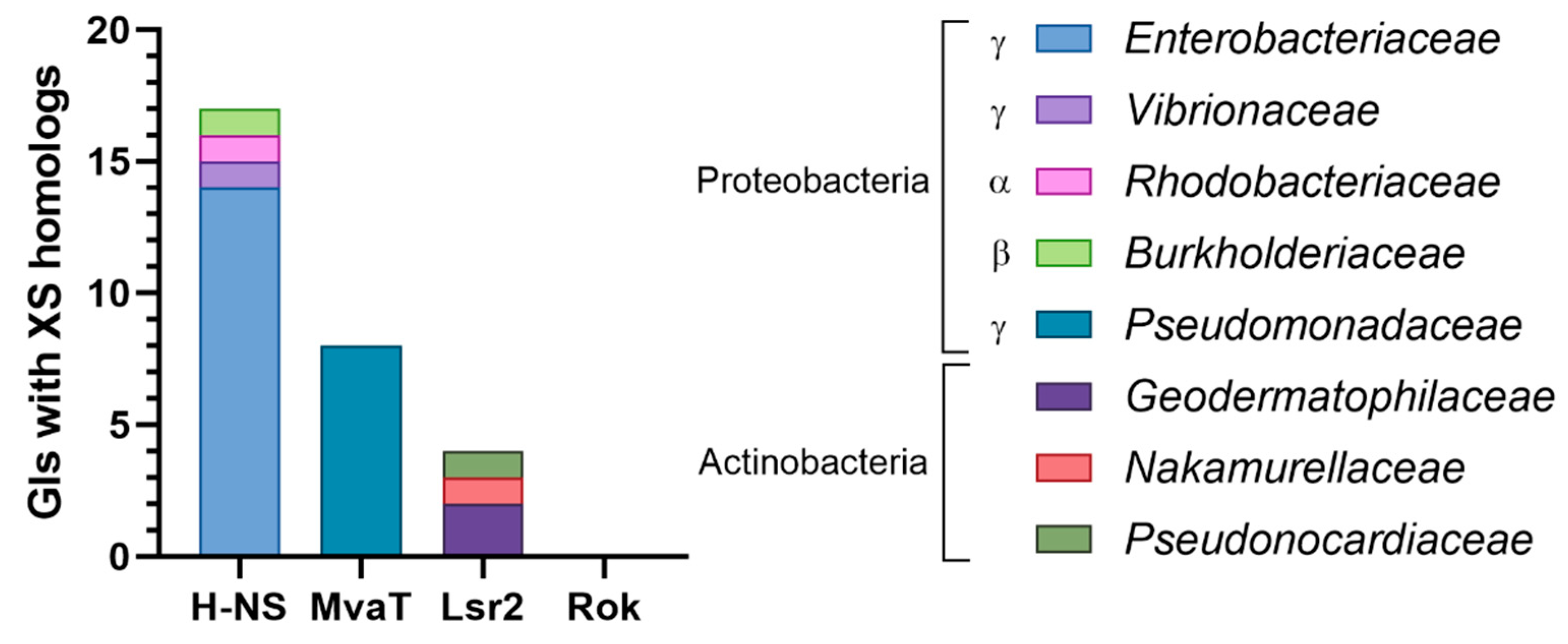
| XS Homolog | Extent of Similarity (with XS) | Host | Location | Protein–Protein Interaction with 1 | References |
|---|---|---|---|---|---|
| Hfp/H-NSB | full-length (H-NS) | Family Enterobacteriaceae | serU island and EARL GIs | H-NS | [50,52,53] |
| H-NST | N-terminal (H-NS) | Family Enterobacteriaceae | serU island and EARL GIs | H-NS | [50,52] |
| Ler | C-terminal (H-NS) | Attaching and effacing pathogens (Enterobacteriaceae) | Locus of Enterocyte Effacement GI | Ler | [54] |
| Pmr | full-length (MvaT) | Pseudomonas putida strain KT2240 | pCAR1 plasmid | Pmr, TurA, TurB, TurE | [55] |
| Acr2 | full-length (H-NS) | Class Gammaproteobacteria | IncA/C plasmids | Unknown | [49,56] |
| Sfh | full-length (H-NS) | Shigella flexneri 2a strain 2457T | pSf-R27 and R27-like plasmids | H-NS, StpA, Sfh | [44,57,58,59,60] |
| H-NSEPV1 | full-length (H-NS) | “Candidatus Accumulibacter phosphatis” | Phage EPV1 | Unknown | [51,61] |
| RokLS20 | C-terminal (Rok) | Bacillus subtilis strain IFO3335 | pLS20 plasmid | Unknown | [62] |
| Lsr2 homologs | full-length (Lsr2) | Phylum Actinobacteria | Plasmids (unclassified) and mycobacteriophages | Unknown | [49,63] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piña-Iturbe, A.; Suazo, I.D.; Hoppe-Elsholz, G.; Ulloa-Allendes, D.; González, P.A.; Kalergis, A.M.; Bueno, S.M. Horizontally Acquired Homologs of Xenogeneic Silencers: Modulators of Gene Expression Encoded by Plasmids, Phages and Genomic Islands. Genes 2020, 11, 142. https://doi.org/10.3390/genes11020142
Piña-Iturbe A, Suazo ID, Hoppe-Elsholz G, Ulloa-Allendes D, González PA, Kalergis AM, Bueno SM. Horizontally Acquired Homologs of Xenogeneic Silencers: Modulators of Gene Expression Encoded by Plasmids, Phages and Genomic Islands. Genes. 2020; 11(2):142. https://doi.org/10.3390/genes11020142
Chicago/Turabian StylePiña-Iturbe, Alejandro, Isidora D. Suazo, Guillermo Hoppe-Elsholz, Diego Ulloa-Allendes, Pablo A. González, Alexis M. Kalergis, and Susan M. Bueno. 2020. "Horizontally Acquired Homologs of Xenogeneic Silencers: Modulators of Gene Expression Encoded by Plasmids, Phages and Genomic Islands" Genes 11, no. 2: 142. https://doi.org/10.3390/genes11020142
APA StylePiña-Iturbe, A., Suazo, I. D., Hoppe-Elsholz, G., Ulloa-Allendes, D., González, P. A., Kalergis, A. M., & Bueno, S. M. (2020). Horizontally Acquired Homologs of Xenogeneic Silencers: Modulators of Gene Expression Encoded by Plasmids, Phages and Genomic Islands. Genes, 11(2), 142. https://doi.org/10.3390/genes11020142







