Novel Mutations Found in Individuals with Adult-Onset Pompe Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approvals
2.2. Cell Culture
2.3. Genomic DNA and RNA Extraction
2.4. PCR, RT-PCR and qPCR
2.5. Western Blotting
2.6. GAA Enzyme Activity Assay
2.7. Measurement of Urinary Tetrasaccharide
2.8. In Silico Predictions
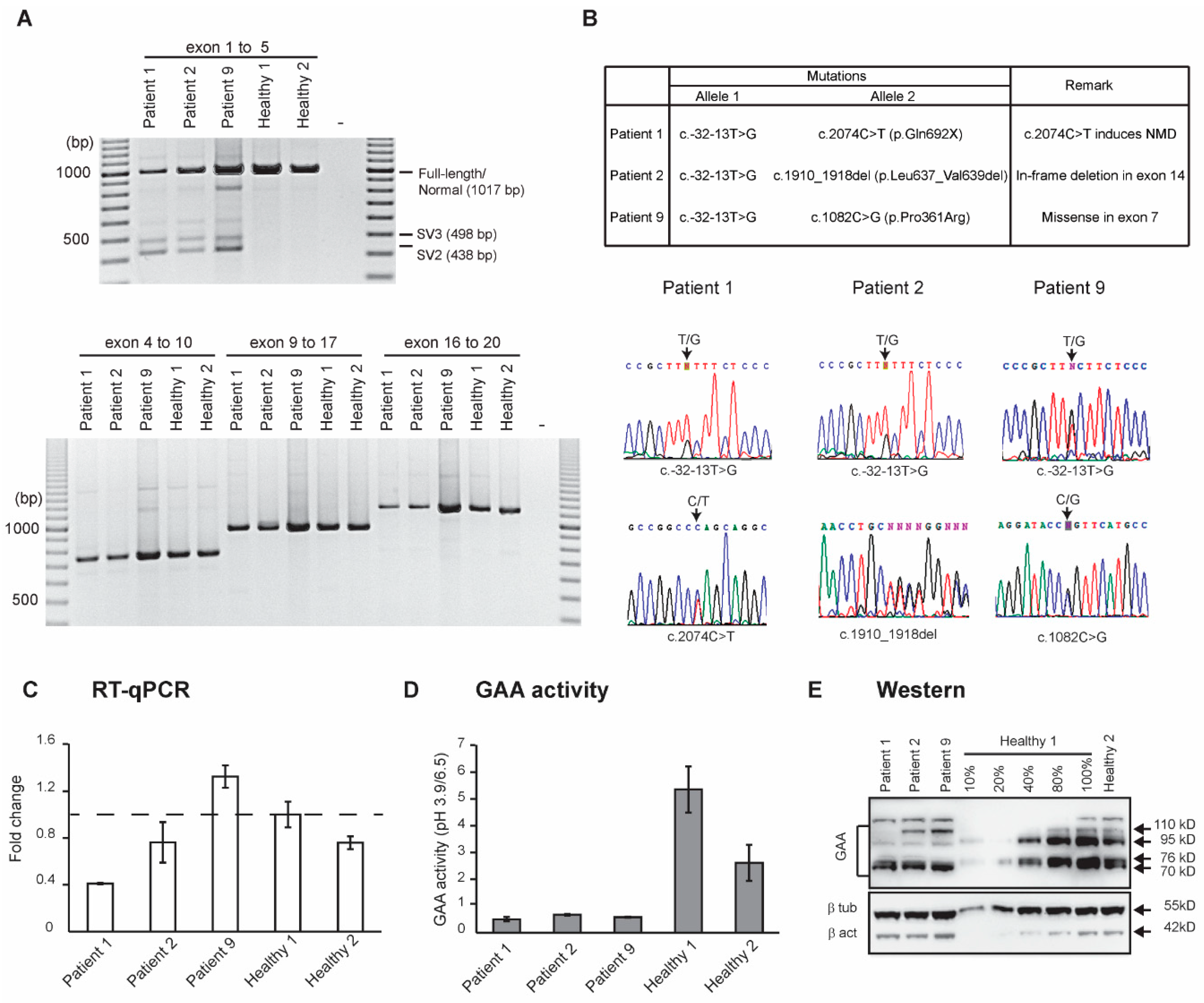
3. Results
3.1. Clinical Assessment
3.2. Molecular Analysis
3.3. In Silico Analysis
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Pompe, J.C. Over idiopatsche hypertrophie van het hart. Ned. Tijdschr. Geenskd 1932, 76, 304–311. [Google Scholar]
- Hers, H.G. Alpha-Glucosidase deficiency in generalized glycogen storage disease (Pompe’s disease). Biochem. J. 1963, 86, 11–16. [Google Scholar] [PubMed]
- Kuo, W.L.; Hirschhorn, R.; Huie, M.L.; Hirschhorn, K. Localization and ordering of acid alpha-glucosidase (GAA) and thymidine kinase (TK1) by fluorescence in situ hybridization. Hum. Genet. 1996, 97, 404–406. [Google Scholar] [CrossRef] [PubMed]
- Ausems, M.G.; Verbiest, J.; Hermans, M.P.; Kroos, M.A.; Beemer, F.A.; Wokke, J.H.; Sandkuijl, L.A.; Reuser, A.J.; van der Ploeg, A.T. Frequency of glycogen storage disease type II in The Netherlands: Implications for diagnosis and genetic counselling. Eur. J. Hum. Genet. 1999, 7, 713–716. [Google Scholar] [CrossRef] [PubMed]
- Matsuishi, T.; Yoshino, M.; Terasawa, K.; Nonaka, I. Childhood acid maltase deficiency. A clinical, biochemical, and morphologic study of three patients. Arch. Neurol. 1984, 41, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Leslie, N.; Bailey, L. Pompe Disease. In GeneReviews®(Internet); Pagon, R.A., Adam, M.P., Ardinger, H.H., Wallace, S.E., Amemiya, A., Bean, L.J.H., Bird, T.D., Ledbetter, N., Mefford, H.C., Smith, R.J.H., et al., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Mehler, M.; DiMauro, S. Residual acid maltase activity in late-onset acid maltase deficiency. Neurology 1977, 27, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Umapathysivam, K.; Hopwood, J.J.; Meikle, P.J. Correlation of acid alpha-glucosidase and glycogen content in skin fibroblasts with age of onset in Pompe disease. Clin. Chim. Acta 2005, 361, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Van der Ploeg, A.T.; Reuser, A.J. Pompe’s disease. Lancet 2008, 372, 1342–1353. [Google Scholar] [CrossRef]
- Kroos, M.; Hoogeveen-Westerveld, M.; van der Ploeg, A.; Reuser, A.J. The genotype-phenotype correlation in Pompe disease. Am. J. Med. Genet. C Semin. Med. Genet. 2012, 160C, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Sterne-Weiler, T.; Howard, J.; Mort, M.; Cooper, D.N.; Sanford, J.R. Loss of exon identity is a common mechanism of human inherited disease. Genome Res. 2011, 21, 1563–1571. [Google Scholar] [CrossRef] [PubMed]
- Noensie, E.N.; Dietz, H.C. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol. 2001, 19, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Oba-Shinjo, S.M.; da Silva, R.; Andrade, F.G.; Palmer, R.E.; Pomponio, R.J.; Ciociola, K.M.; M, S.C.; Gutierrez, P.S.; Porta, G.; Marrone, C.D.; et al. Pompe disease in a Brazilian series: Clinical and molecular analyses with identification of nine new mutations. J. Neurol. 2009, 256, 1881–1890. [Google Scholar] [CrossRef]
- Vorgerd, M.; Burwinkel, B.; Reichmann, H.; Malin, J.P.; Kilimann, M.W. Adult-onset glycogen storage disease type II: Phenotypic and allelic heterogeneity in German patients. Neurogenetics 1998, 1, 205–211. [Google Scholar] [CrossRef]
- Pittis, M.G.; Donnarumma, M.; Montalvo, A.L.; Dominissini, S.; Kroos, M.; Rosano, C.; Stroppiano, M.; Bianco, M.G.; Donati, M.A.; Parenti, G.; et al. Molecular and functional characterization of eight novel GAA mutations in Italian infants with Pompe disease. Hum. Mutat. 2008, 29, E27–E36. [Google Scholar] [CrossRef] [PubMed]
- Van den Hout, J.M.; Kamphoven, J.H.; Winkel, L.P.; Arts, W.F.; De Klerk, J.B.; Loonen, M.C.; Vulto, A.G.; Cromme-Dijkhuis, A.; Weisglas-Kuperus, N.; Hop, W.; et al. Long-term intravenous treatment of Pompe disease with recombinant human alpha-glucosidase from milk. Pediatrics 2004, 113, e448–e457. [Google Scholar] [CrossRef]
- Kroos, M.; Hoogeveen-Westerveld, M.; Michelakakis, H.; Pomponio, R.; Van der Ploeg, A.; Halley, D.; Reuser, A.; Consortium, G.A.A.D. Update of the pompe disease mutation database with 60 novel GAA sequence variants and additional studies on the functional effect of 34 previously reported variants. Hum. Mutat. 2012, 33, 1161–1165. [Google Scholar] [CrossRef]
| Primer Names | Sequences (5´ to 3´) | Purpose | Cycling Conditions |
| PCR | Genomic DNA amplification data | 95 °C for 5 min, 35 cycles of 95 °C 30 s, 60 °C 30 s and 72 °C 1 to 4 min | |
| intron 1F | cagtctagacagcagggcaa | ||
| exon 2R | agtaggatgtgccccaggag | ||
| exon 1F2 | cggcctctcagttgggaaa | ||
| exon 2R2 | ggttgccaaggacacga | ||
| exon 2F | tgtaggagctgtccaggcc | ||
| exon 5R2 | ggcattgctgtttagcag | ||
| intron 4F | gatctcggtcttgaaagc | ||
| exon 10R2 | actcagccaccatgtcctcc | ||
| exon 10F | actgccttccccgacttca | ||
| exon 15R | tggaacagtgtgtagaggtg | ||
| exon 15F | cgtacagcttcagcgag | ||
| exon 16R | tgcaggtcgtaccatgtg | ||
| exon 16F | caaggactctagcacctgg | ||
| exon 20R | gaatctcccaagtcctgtgadata | ||
| RT-PCR | GAA transcript amplification | 95 °C for 5 min 35 cycles of 95 °C 30 s, 60 °C 30 s and 72 °C 1 min | |
| exon 1F | ggaaactgaggcacggagcg | ||
| exon 5R | ggaccacatccatggcattgc | ||
| exon 4F | gtatatcacaggcctcgccg | ||
| exon 10R | ctggtcatggaactcagcca | ||
| exon 9F | gggggttttcatcaccaacga | ||
| exon 17R | ctgccaagggcctctactgg | ||
| exon 16F | caaggactctagcacctgg | ||
| exon 20R | gaatctcccaagtcctgtga | ||
| RT-qPCR | Full-length GAA transcript (exon 1-2), amplification | 95 °C for 1 min, 40 cycles of 95 °C 3 s, 60 °C 15 s and 72 °C 30 s | |
| exon 1F(q) | tgggaaagctgaggttgtcg | ||
| exon 1-2R(q) | tcctacaggcccgctcc | ||
| TBP transcript amplification | |||
| exon 1-2F(q) | tctttgcagtgacccagcatcac | ||
| exon 2R(q) | cctagagcatctccagcacactct | ||
| Antibodies | Catalogue no. | Source | Dilution |
| rabbit anti-GAA | 137068 | Abcam, Melbourne, Australia | 1:1000 |
| mouse monoclonal anti-β-tubulin | E7 | DSHB, Iowa City, Iowa | 1: 5000 |
| mouse monoclonal anti-β-actin | A5316 | Sigma-Aldrich, Castle Hill NSW | 1: 500,000 |
| Age at Diagnosis | Creatine Kinase (IU/L) (Normal 0–250) | Respiratory Function | Dried Blood Spot GAA Activity (µmol/h/L) (normal 0.3–3.0) | Urine Tetrasaccharides (mmol/mol Creatinine) (Normal <20) | |
|---|---|---|---|---|---|
| Patient 1 | 54 | 320 | Type 2 respiratory failure (pCO2 55 mmHg) | <0.1 | 61 |
| Patient 2 | 44 | Intact | <0.2 | 150 | |
| Patient 9 | 30 | 950 | Intact | 0.8 | 40 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aung-Htut, M.T.; Ham, K.A.; Tchan, M.C.; Fletcher, S.; Wilton, S.D. Novel Mutations Found in Individuals with Adult-Onset Pompe Disease. Genes 2020, 11, 135. https://doi.org/10.3390/genes11020135
Aung-Htut MT, Ham KA, Tchan MC, Fletcher S, Wilton SD. Novel Mutations Found in Individuals with Adult-Onset Pompe Disease. Genes. 2020; 11(2):135. https://doi.org/10.3390/genes11020135
Chicago/Turabian StyleAung-Htut, May T., Kristin A. Ham, Michel C. Tchan, Sue Fletcher, and Steve D. Wilton. 2020. "Novel Mutations Found in Individuals with Adult-Onset Pompe Disease" Genes 11, no. 2: 135. https://doi.org/10.3390/genes11020135
APA StyleAung-Htut, M. T., Ham, K. A., Tchan, M. C., Fletcher, S., & Wilton, S. D. (2020). Novel Mutations Found in Individuals with Adult-Onset Pompe Disease. Genes, 11(2), 135. https://doi.org/10.3390/genes11020135






