Abstract
Bi-allelic pathogenic variants in MERTK cause retinitis pigmentosa (RP). Since deletions of more than one exon have been reported repeatedly for MERTK, CNV (copy number variation) analysis of next-generation sequencing (NGS) data has proven important in molecular genetic diagnostics of MERTK. CNV analysis was performed on NGS data of 677 individuals with inherited retinal diseases (IRD) and confirmed by quantitative RT-PCR analysis. Clinical evaluation was based on retrospective records. Clinical re-examination included visual field examination, dark adaption, scotopic and photopic full-field electroretinograms (ffERG), multifocal ERG (mfERG) and optic coherence tomography (OCT). Fourteen variants were detected in MERTK in six individuals, three of which were deletions of more than one exon. Clinical examinations of five out of six individuals revealed a severe phenotype with early-onset generalized retinal dystrophy with night blindness and progressive visual field loss; however, one individual had a milder phenotype. Three individuals had hearing impairments. We show that deletions represent a substantial part of the causative variants in MERTK and emphasize that CNV analysis should be included in the molecular genetic diagnostics of IRDs.
1. Introduction
Inherited retinal diseases (IRD) encompass a broad range of clinical diagnoses characterized by the progressive loss of photoreceptors, with variable ages of onset and variable clinical representation. IRD is highly genetically heterogeneous with more than 250 associated genes (https://sph.uth.edu/retnet/), which show all modes of Mendelian inheritance (autosomal dominant, autosomal recessive, X-linked), mitochondrial inheritance, and even digenic inheritance [1]. The majority of the genes encode proteins exerting their function in either the photoreceptor or in the retinal pigment epithelium (RPE) cells. They are involved in various cellular processes such as RNA splicing, visual cycle, primary cilia function and phototransduction [2]. Retinitis pigmentosa (RP) is the most frequent of the IRD specific diagnostic subgroups with a worldwide prevalence of 1:3500 [3]. RP is characterized by dysfunctional rod photoreceptors in the early stages of disease, causing night blindness; but eventually, the cone photoreceptors may also be affected, leading to visual field constriction and central vision loss [2].
More than 120 genes are known to be associated with RP [2] and MERTK (MER tyrosinase kinase protooncogene) is associated with autosomal recessive RP type 38 (OMIM 613862) [4]. A recent review shows that the MERTK associated RP comprises around 2% of RP [5]. MERTK is located at 2q14.1 and consists of 19 exons encoding a receptor tyrosine kinase of 999 amino acids belonging to the TAM (TYRO3/AXL/MER) receptor family [6]. It is highly expressed on the apical membrane of the RPE cells as well as in the monocytes/macrophages, ovary, prostate, testis, lung and kidney [7]. MERTK is involved in the internalization of the photoreceptor outer segment (POS) prior to phagocytosis in RPE [8]. MERTK ligands GAS6 (growth arrest-specific protein 6) and PROS1 (protein S) is secreted by the RPE cells and binds to phosphatidylserines (PS) on the surface of POS and to MERTK on the surface of RPE cells; this leads to MERTK phosphorylation and activation of phagocytosis of POS of RPE cells [9,10]. Pathogenic variants in MERTK cause a severe RP phenotype with early age of onset and early involvement of the macular region, very often leading to blindness. The disease is characterized by night blindness in the childhood or teens, abnormal color vision, reduced visual acuity and visual field constriction [4].
More than 90 disease-causing or probably disease-causing variants are reported in HGMD® Professional (2020.3), including nine gross deletions, 29 missense variants, 15 nonsense variants, 16 splicing variants, 16 small deletions, three small insertions and three small indels [11]. A founder variant of MERTK (deletion of exons 1 to 7) was found in the Faroe Islands and is responsible for approximately 30% of all RP cases within the Faroese population [12].
Copy number variations (CNV) has become an important contributor to the cause of retinal dystrophies [13,14,15].
We report the molecular genetic findings in MERTK and the clinical representation in six individuals as part of a large screening study of 677 individuals with a clinical diagnosis of IRD [16].
2. Materials and Methods
2.1. Editorial Policies and Ethical Considerations
The project was approved by The National Committee on Health Research Ethics, Denmark (project ID 1301394, 1418960 and 1809595). The project was performed according to the Declaration of Helsinki and approved by the Regional Ethics Committee. Written informed consent was obtained before the molecular genetic testing.
2.2. Clinical Evaluation
Clinical examinations were performed by the Eye clinic at the Kennedy Center, Rigshospitalet (former National Eye Clinic for the Visually Impaired). The clinical evaluation was based on available retrospective clinical records with information on clinical diagnosis, clinical history, fundus changes, OCT imaging, slit lamp examination, ERG, visual acuity and Goldmann visual fields. Furthermore, P152, P155, P156 and affected brother of individual P152 were re-examined after the genetic diagnosis was made.
2.3. Molecular Genetic Analysis
Genomic DNA of 677 individuals with IRD who was sequenced with targeted NGS of 125 genes as described previously [16] (Table S1). In brief, the enriched DNA libraries were sequenced using the Illumina HiSeq 2000 (San Diego, CA, USA). Raw sequencing image files and base-calling were processed with the Illumina Pipeline and raw paired-end low quality reads and adapter sequences were removed using the SOAPnuke software (https://bio.tools/soapdenovo). The remaining high-quality reads were aligned to the human reference genome (GRCh37/hg19) using Burrows–Wheeler Algorithm (BWA) version 0.7.1023, with the MEM algorithm. The SAMtools (version 0.1.19) [17] was used to sort and index SAM/BAM files and the Picard (version 1.117, http://broadinstitute.github.io/picard/) was used to mark PCR-duplicates. Local realignment and base recalibration were performed using GATK (version 3.3-0) [18] and single nucleotide variants (SNVs) and insertions/deletions (InDels) were called with GATK HaplotypeCaller.
CNV analysis for the data was carried out by using the ExomeDepth algorithm based on the coverage data [19]. For each tested individual, the ExomeDepth algorithm builds the most appropriate reference set from the BAM files of a group of samples and ranks the CNV calls by their confidence level. A subset of samples was analyzed using the CNV algorithm in Varseq (Golden Helix, Bozeman, MT, USA). The Varseq software generates a set of matched reference controls and the sample is compared to this set. A ratio and z-score are computed for each region defined in the BED file defining the targeted region. The z-score indicate the number of standard deviations that a sample’s coverage is from the mean coverage of the reference set. CNVs were verified with quantitative RT-PCR (qPCR) using SYBR green and analysis on a 7500 ABI SDS system (Applied Biosystems, Foster City, CA, USA). Breakpoint mapping of the deletion including exon 1 to exon 7 was performed using a standard PCR method for amplification of fragment spanning the breakpoint followed by Sanger sequencing. Primers were designed assuming the breakpoint was in the same region as the deletion found by Ostergaard et al. [12]. Primer sequences for qPCR and breakpoint mapping are listed in Table S2.
Variants were classified according to ACMG guidelines [20] and ACGS guidelines [21]. The CADD score was calculated for missense variants [22]. Variants in splice sites were evaluated using SpliceSiteFinder-like [23,24], MaxEntScan [25], NNSPLICE [26], GeneSplicer [27].
3. Results
3.1. Clinical Characteristics
Clinical data are presented in Table 1 and Figure 1. Pedigrees for three families are shown in Figure 2.

Table 1.
Clinical symptoms.
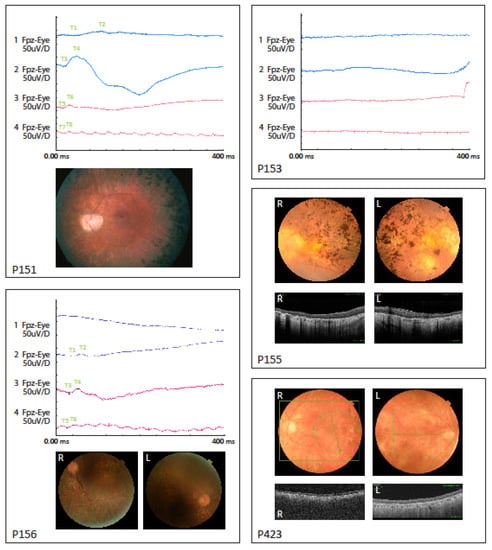
Figure 1.
Electroretinograms (ERG) data shown for P151, P153 and P156. Fundus pictures shown for P151, P155, P156 and P423. OCT pictures for P155 and P423. Descriptions of data are listed in Table 1.
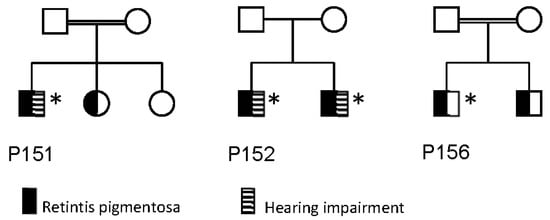
Figure 2.
Pedigrees of families of P151, P152 and P156. Individuals marked with an asterix (*) has been genotyped, and has the genotype listed in Table 2.
In general, the fundoscopic changes were characterized by a pale optic nerve, attenuated retinal vessels, peripheral retinal degeneration and bone spicule hyperpigmentation. Five out of the six individuals showed fundus and/or OCT signs of macular atrophy. The OCT analysis showed severe retinal degeneration and abnormal structure. In two individuals (P152 and P153) the retinal dystrophy was already generalized and severe when full-field ERG was performed, resulting in undetectable responses. This was also seen in the brother of P152. In P151, P156 and P423, earlier full-field ERG measured at disease onset were available and showed reduced rod and cone responses with primary loss of rod function. Additionally, all individuals had night blindness as the initial symptom consistent with the clinical classification of rod-cone dystrophy or RP.
Three individuals (P151, P152 and P155) were diagnosed with hearing impairment. P152 and his brother were diagnosed with a progressive bilateral hearing loss in the higher frequencies (2–4 KHz) diagnosed in their mid-thirties. P151 and P155 had anamnestically unspecific hearing loss in older age, with no hearing curves available.
3.2. Genetic Analysis
Fourteen variants (seven unique variants) were found in six individuals (Table 2). Three variants were the deletion of exons 1 to 7 (P151 and P155). Breakpoint mapping showed that the breakpoints were identical in P151 and P155 and to the breakpoint mapped by Ostergaard et al. [12] (Figure 3). The deletion includes chr2:112,648,150-112,739,208 (GRCh37/hg19). Results from qPCR analysis are shown in Figure S1.

Table 2.
Molecular genetic findings in MERTK.
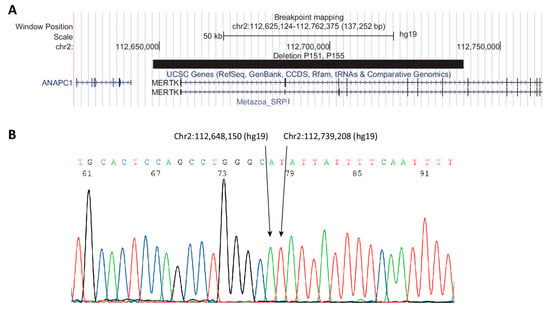
Figure 3.
Breakpoint mapping of deletion in P151 and P155. (A) UCSC (University of California Santa Cruz) browser track showing deletion as a black bar. (B) Sanger sequencing showing the sequence surrounding the breakpoint.
Of the seven unique variants, five had not previously been reported in individuals with retinal dystrophy (c.960+1G>A, c.757+1G>A, c.2060G>T and c.2305A>G). Two of the variants, c.2060G>T; p.(Arg687Leu) and c.2305A>G; p.(Ile769Val)), were found in the same individual (P423). Both variants were apparently homozygous and both were classified as variants of unknown significance (VUS). The variants c.960+1G>A and c.757+1G>A are both splice variants predicted to alter RNA splicing.
4. Discussion
In this study, we analyzed MERTK as a part of a targeted sequencing panel in individuals with IRD. Variants in MERTK were found in six individuals of which six had a potential molecular genetic diagnosis caused by variants in MERTK. These individuals were part of a cohort of 677, of which 421 had a clinical diagnosis of RP, corresponding to 1.7% of individuals with RP being caused by variants in MERTK, which is comparable to the findings of Audo et al. [5]. A total of 24 CNVs were found in the cohort of 677 individuals (3.5%) and of these four were in MERTK; only EYS with seven CNVs had more deletions/duplications than MERTK [16]. Others have performed CNV analysis using NGS data and found substantially higher yields of causal CNVs [13,15]. However, their numbers are based on unsolved cases. This underscores the importance of CNV analysis in IRDs and reflects that missing genetic explanations of IRDs can be found in already known IRD genes, as structural variants and deep intron variants affecting splicing. Thus, robust bioinformatic pipelines analyzing data from NGS targeted panels (and soon also data from whole-genome sequencing) are necessary, as are methods to detect deep intronic variants (for example RNA seq).
Most individuals had a severe phenotype with an onset of night blindness in the first decade of life and progressive visual field loss during childhood. Individual P423 however, presented with a milder phenotype. P423 had a late-onset (age 53 years) of night blindness and a slow progression of visual acuity and subsequent visual field loss. None of the individuals in our cohort had nystagmus. P423 belongs to a large family with several members affected with RP, however, the closest affected relative was a sister of her paternal grandmother and the affected descendants span three generations resembling autosomal dominant inheritance. Testing of two affected family members showed that these did not have the missense variants in MERTK found in P423. Either the missense variants (both classified as VUS) are not the genetic cause of IRD in P423, or there are two different IRDs in the family, one type explained by MERTK variants and another with a yet unknown genetic cause.
The deletion of exon 1–7 (Chr2:112,648,150-112,739,208 (hg19)) of MERTK was first reported by Ostergaard et al. [12] as a common founder variant in The Faroe Islands. The deletion has since been found in individuals outside The Faroe Islands by Ellingford et al. [32] in a study from Manchester Royal Eye Hospital, and in two individuals in this study (P152 and P155); it is unclear whether the deletion is a hotspot or a founder variant.
The ophthalmological findings in the individuals in this study are consistent with previous descriptions of individuals with MERTK-related retinal dystrophy [5]. Individuals with MERTK-related retinal dystrophy have previously been described both with rod-cone dystrophy and cone-rod dystrophy [5]. In our cohort, the phenotype of all individuals was clinically classified as rod-cone dystrophy with night blindness as the first symptom and verified by full-field ERG with a primary rod dysfunction in four individuals. One individual in our cohort, however, presented with a milder phenotype with later onset and milder disease, but as discussed above the molecular genetic diagnosis in this individual is questionable, although a broadened clinical spectrum of MERTK-related retinal dystrophy cannot be excluded. Clinical variety in terms of disease severity is a known phenomenon in individuals with RP with many other genetic causes [33]. The clinical variety may in part be explained by the type of variant involved, but there may also be additional factors both genetic, epigenetic and environmental factors that influence the phenotype.
To our knowledge hearing impairment has not been reported as part of the MERTK-related phenotype. Individuals P151 and P155 had late-onset hearing loss while the hearing loss in P152 and his brother was of another type. This points to the hearing loss in these individuals might as well be coincidental, as a result of other genetic or non-genetic causes.
In conclusion, this study adds to the understanding of the genetic and clinical characteristics of individuals with MERTK-related retinal dystrophies and underlines that CNV analysis should be added to the genetic evaluation of individuals suspected of having retinal dystrophy. New treatments such as gene therapy for retinal dystrophies are evolving and the understanding of the clinical characteristics of genotype-specific disease entities is important.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/12/1517/s1, Figure S1: qPCR analysis and BMA alignment. Table S1: Genes in gene panel. Table S2: Primers used for qPCR.
Author Contributions
Conceptualization, K.G., K.B.-N., Z.T. and L.B.M.; methodology, K.G. and C.J.; software, C.J. and M.F.; investigation, C.J., M.F., M.B., F.A., H.J., T.R., H.G.G.-K.; writing—original draft preparation, K.G.; writing—review and editing, K.B.-N., Z.T., L.B.M. and M.B.; visualization, K.G.; supervision, K.G.; project administration, K.G.; funding acquisition, K.G., K.B.-N., L.B.M., Z.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Velux Foundation grant number 32700.
Acknowledgments
We thank the participants and families who participated in the study. We thank Kate Nielsen and Luljeta Ramadani for technical assistance. We thank Jette Bune Rasmussen for preparing the figures. This work was funded by the Velux foundation grant number 32700 (C.J., L.B.M., Z.T., K.B.-N., H.J., K.G.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kajiwara, K.; Berson, E.L.; Dryja, T.P. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science 1994, 264, 1604–1608. [Google Scholar] [CrossRef]
- Nash, B.M.; Wright, D.C.; Grigg, J.R.; Bennetts, B.; Jamieson, R.V. Retinal dystrophies, genomic applications in diagnosis and prospects for therapy. Transl. Pediatr. 2015, 4, 139–163. [Google Scholar] [CrossRef]
- Bertelsen, M.; Jensen, H.; Bregnhoj, J.F.; Rosenberg, T. Prevalence of generalized retinal dystrophy in Denmark. Ophthalmic Epidemiol. 2014, 21, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Gal, A.; Li, Y.; Thompson, D.A.; Weir, J.; Orth, U.; Jacobson, S.G.; Apfelstedt-Sylla, E.; Vollrath, D. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat. Genet. 2000, 26, 270–271. [Google Scholar] [CrossRef] [PubMed]
- Audo, I.; Mohand-Said, S.; Boulanger-Scemama, E.; Zanlonghi, X.; Condroyer, C.; Demontant, V.; Boyard, F.; Antonio, A.; Mejecase, C.; El Shamieh, S.; et al. MERTK mutation update in inherited retinal diseases. Hum. Mutat. 2018, 39, 887–913. [Google Scholar] [CrossRef] [PubMed]
- Lemke, G. Biology of the TAM receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a009076. [Google Scholar] [CrossRef]
- Linger, R.M.; Keating, A.K.; Earp, H.S.; Graham, D.K. TAM receptor tyrosine kinases: Biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv. Cancer Res. 2008, 100, 35–83. [Google Scholar] [CrossRef]
- Feng, W.; Yasumura, D.; Matthes, M.T.; LaVail, M.M.; Vollrath, D. Mertk triggers uptake of photoreceptor outer segments during phagocytosis by cultured retinal pigment epithelial cells. J. Biol. Chem. 2002, 277, 17016–17022. [Google Scholar] [CrossRef]
- Hall, M.O.; Prieto, A.L.; Obin, M.S.; Abrams, T.A.; Burgess, B.L.; Heeb, M.J.; Agnew, B.J. Outer segment phagocytosis by cultured retinal pigment epithelial cells requires Gas6. Exp. Eye Res. 2001, 73, 509–520. [Google Scholar] [CrossRef]
- Law, A.L.; Parinot, C.; Chatagnon, J.; Gravez, B.; Sahel, J.A.; Bhattacharya, S.S.; Nandrot, E.F. Cleavage of Mer tyrosine kinase (MerTK) from the cell surface contributes to the regulation of retinal phagocytosis. J. Biol. Chem. 2015, 290, 4941–4952. [Google Scholar] [CrossRef]
- Stenson, P.D.; Ball, E.V.; Mort, M.; Phillips, A.D.; Shaw, K.; Cooper, D.N. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr. Protoc. Bioinform. 2012, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ostergaard, E.; Duno, M.; Batbayli, M.; Vilhelmsen, K.; Rosenberg, T. A novel MERTK deletion is a common founder mutation in the Faroe Islands and is responsible for a high proportion of retinitis pigmentosa cases. Mol. Vis. 2011, 17, 1485–1492. [Google Scholar] [PubMed]
- Zampaglione, E.; Kinde, B.; Place, E.M.; Navarro-Gomez, D.; Maher, M.; Jamshidi, F.; Nassiri, S.; Mazzone, J.A.; Finn, C.; Schlegel, D.; et al. Copy-number variation contributes 9% of pathogenicity in the inherited retinal degenerations. Genet. Med. 2020, 22, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Van Schil, K.; Karlstetter, M.; Aslanidis, A.; Dannhausen, K.; Azam, M.; Qamar, R.; Leroy, B.P.; Depasse, F.; Langmann, T.; De Baere, E. Autosomal recessive retinitis pigmentosa with homozygous rhodopsin mutation E150K and non-coding cis-regulatory variants in CRX-binding regions of SAMD7. Sci. Rep. 2016, 6, 21307. [Google Scholar] [CrossRef]
- Bujakowska, K.M.; Fernandez-Godino, R.; Place, E.; Consugar, M.; Navarro-Gomez, D.; White, J.; Bedoukian, E.C.; Zhu, X.; Xie, H.M.; Gai, X.; et al. Copy-number variation is an important contributor to the genetic causality of inherited retinal degenerations. Genet. Med. 2017, 19, 643–651. [Google Scholar] [CrossRef]
- Jespersgaard, C.; Fang, M.; Bertelsen, M.; Dang, X.; Jensen, H.; Chen, Y.; Bech, N.; Dai, L.; Rosenberg, T.; Zhang, J.; et al. Molecular genetic analysis using targeted NGS analysis of 677 individuals with retinal dystrophy. Sci. Rep. 2019, 9, 1219. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Plagnol, V.; Curtis, J.; Epstein, M.; Mok, K.Y.; Stebbings, E.; Grigoriadou, S.; Wood, N.W.; Hambleton, S.; Burns, S.O.; Thrasher, A.J.; et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 2012, 28, 2747–2754. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Ellard Sian, B.E.L.; Ian, B.; Natalie, F.; Clare, T.; Martina, O.; Diana, M.E.; Stephen, A.; Richard, S.; Deans Zandra, C.D.; Tracy, L.; et al. ACGS Best Practice Guidelines for Variant Classification 2019. Available online: https://www.acgs.uk.com/quality/best-practice-guidelines/#VariantGuidelines (accessed on 4 December 2020).
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Q. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 1998, 7, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, M.B.; Senapathy, P. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987, 15, 7155–7174. [Google Scholar] [CrossRef] [PubMed]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Lin, X.; Salzberg, S.L. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 2001, 29, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Eisenberger, T.; Neuhaus, C.; Khan, A.O.; Decker, C.; Preising, M.N.; Friedburg, C.; Bieg, A.; Gliem, M.; Charbel Issa, P.; Holz, F.G.; et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: The example of retinal dystrophies. PLoS ONE 2013, 8, e78496. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, V.W.; Feng, Y.; Tian, X.; Li, F.Y.; Truong, C.; Wang, G.; Chiang, P.W.; Lewis, R.A.; Wong, L.J. Dependable and efficient clinical utility of target capture-based deep sequencing in molecular diagnosis of retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6213–6223. [Google Scholar] [CrossRef]
- Ellingford, J.M.; Campbell, C.; Barton, S.; Bhaskar, S.; Gupta, S.; Taylor, R.L.; Sergouniotis, P.I.; Horn, B.; Lamb, J.A.; Michaelides, M.; et al. Validation of copy number variation analysis for next-generation sequencing diagnostics. Eur. J. Hum. Genet. 2017, 25, 719–724. [Google Scholar] [CrossRef]
- Oishi, M.; Oishi, A.; Gotoh, N.; Ogino, K.; Higasa, K.; Iida, K.; Makiyama, Y.; Morooka, S.; Matsuda, F.; Yoshimura, N. Comprehensive molecular diagnosis of a large cohort of Japanese retinitis pigmentosa and Usher syndrome patients by next-generation sequencing. Investig. Ophthalmol. Vis. Sci. 2014, 55, 7369–7375. [Google Scholar] [CrossRef]
- Ellingford, J.M.; Horn, B.; Campbell, C.; Arno, G.; Barton, S.; Tate, C.; Bhaskar, S.; Sergouniotis, P.I.; Taylor, R.L.; Carss, K.J.; et al. Assessment of the incorporation of CNV surveillance into gene panel next-generation sequencing testing for inherited retinal diseases. J. Med. Genet. 2018, 55, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Berger, W.; Kloeckener-Gruissem, B.; Neidhardt, J. The molecular basis of human retinal and vitreoretinal diseases. Prog. Retin. Eye Res. 2010, 29, 335–375. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).