Genomic Investigation into the Virulome, Pathogenicity, Stress Response Factors, Clonal Lineages, and Phylogenetic Relationship of Escherichia coli Strains Isolated from Meat Sources in Ghana
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Identification of E. coli
2.1.1. Sample Collection
2.1.2. Isolation of E. coli
2.2. Selection of Isolates for Genome Sequencing and Assembly
2.3. Genome Annotation and Visualisation
2.4. WGS-Based Molecular Typing of Isolates
2.5. Detection of the Stress Response Mechanisms, CRISPR Array, and Restriction-Modification System (R-M System)
2.6. Assessment of Pathogenic Potential
2.7. Virulome Analysis
2.8. Comparative Phylogenomic Analysis and Metadata Insights
3. Results
3.1. E. coli Strains and Data Source for Comparative Genome Analysis
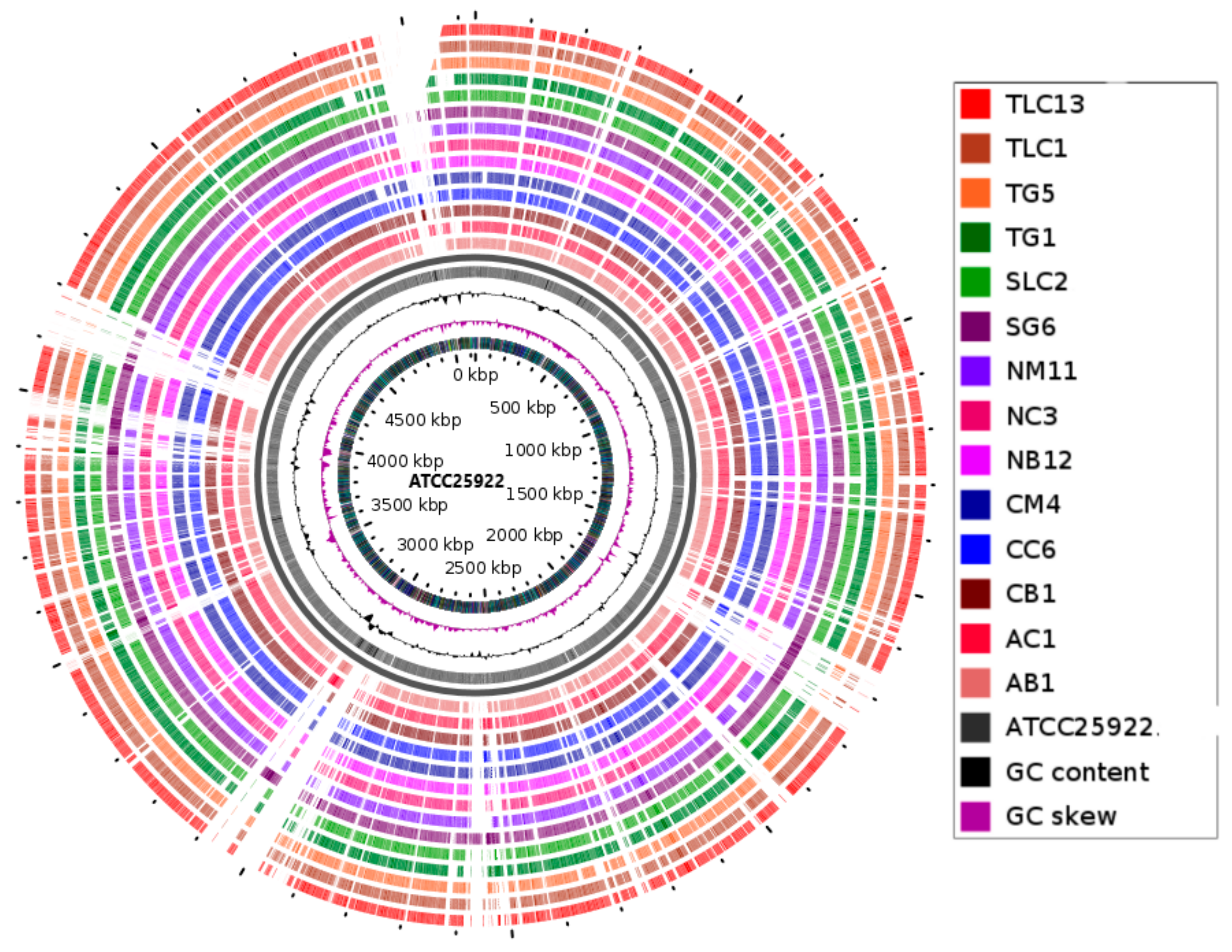
3.2. Genomic Visualisation and Annotation
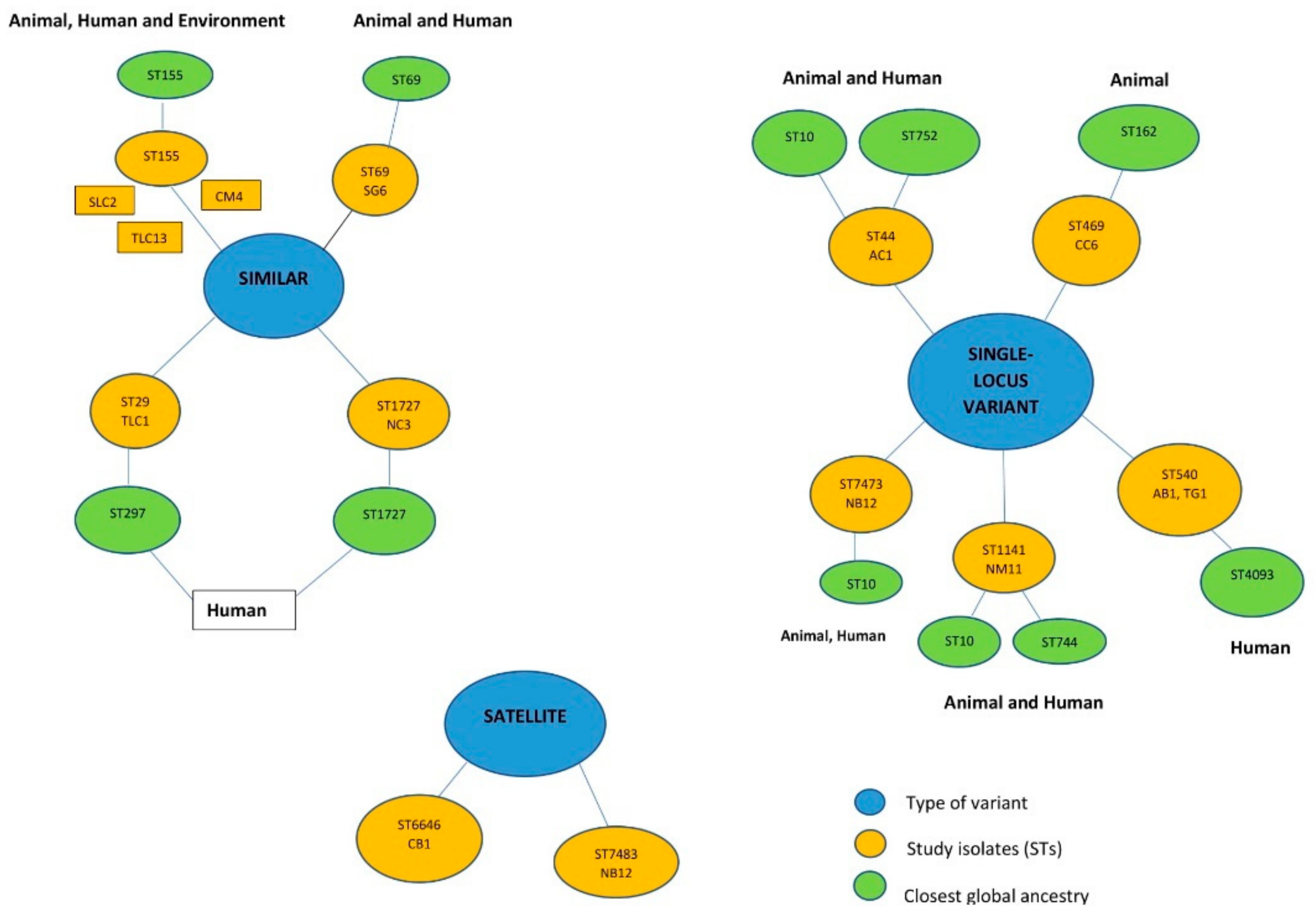
3.3. In Silico Typing of the E. coli Isolates
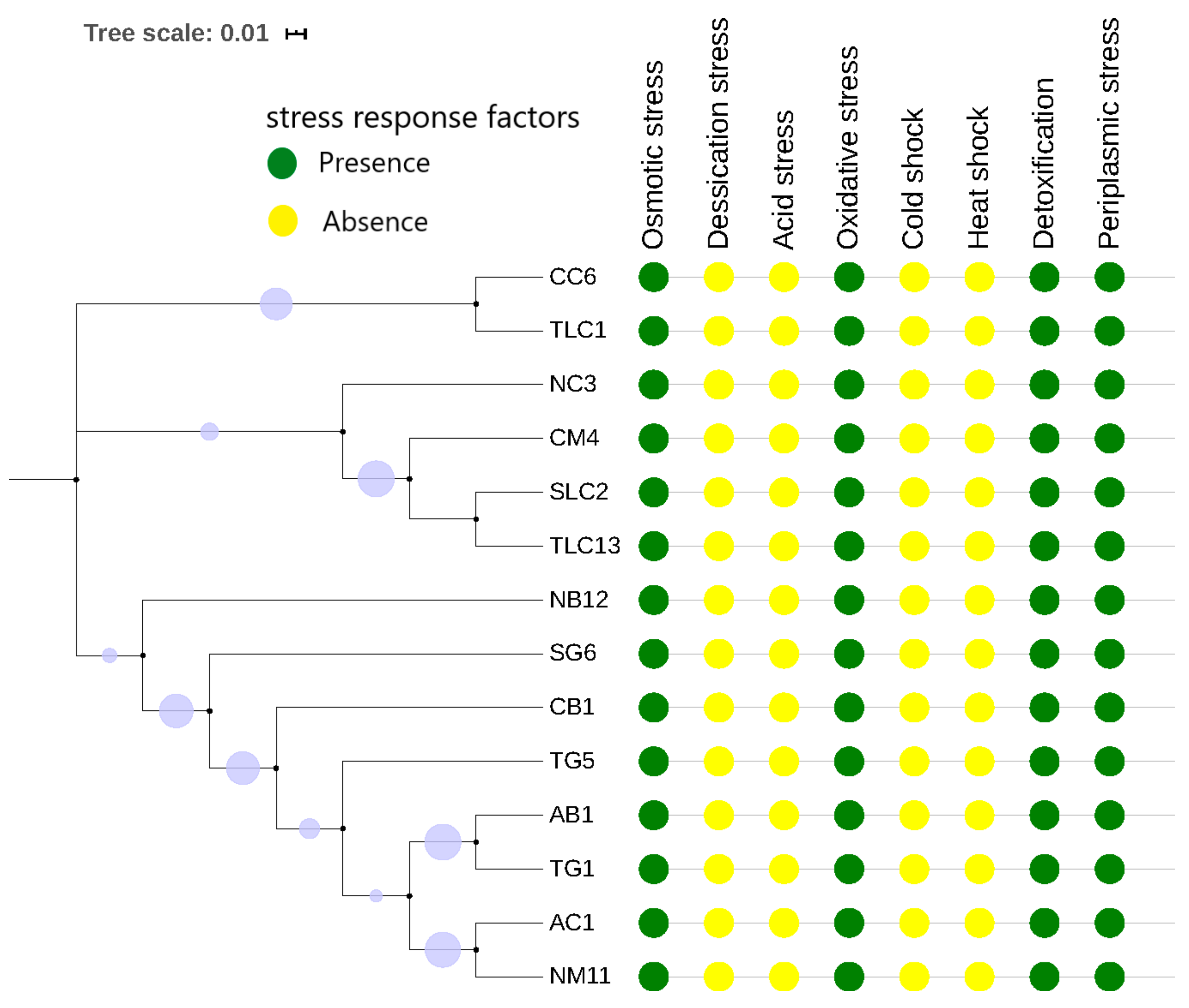
3.4. Genomic Prediction of Defence Systems (CRISPR-Cas Elements and R-M System) and Stress Response Factors
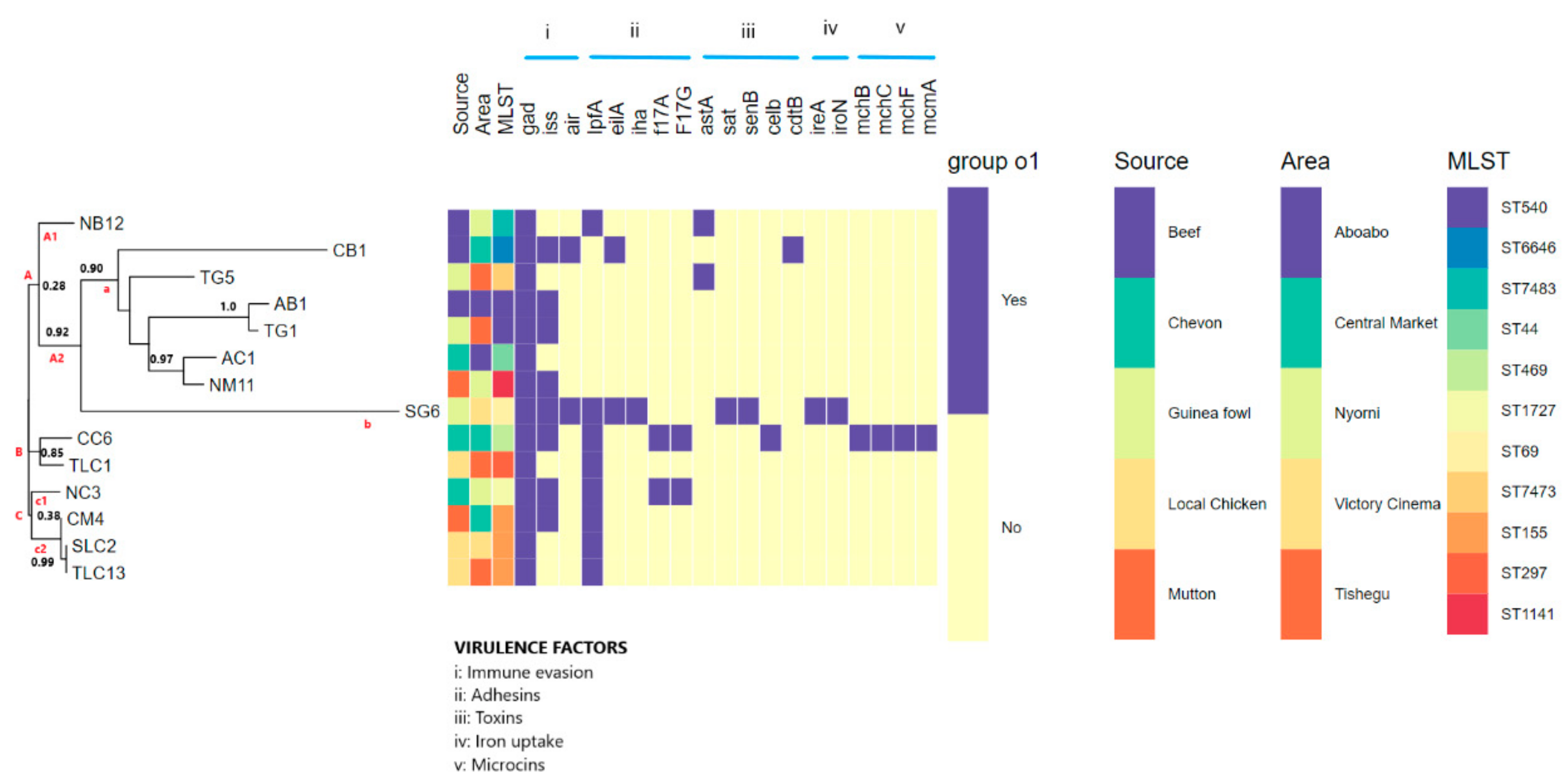
3.5. Pathogenic Potential and Putative Virulence Factor Predictions
3.6. Phylogenomic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Octavia, S.; Lan, R. The Family Enterobacteriaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2014; pp. 225–286. ISBN 9783642389221. [Google Scholar]
- Feng, P.; Weagant, S.D.; Grant, M.A.; Burkhardt, W. Enumeration of Escherichia coli and the Coliform Bacteria. In Bacteriological Analytical Manual; Food & Drug Administration (FDA): Silver Spring, MD, USA, 2018; pp. 21–34. [Google Scholar]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent Advances in Understanding Enteric Pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed]
- Luna-Guevara, J.J.; Arenas-Hernandez, M.M.P.; De La Peña, C.M.; Silva, J.L.; Luna-Guevara, M.L. The Role of Pathogenic E. coli in Fresh Vegetables: Behavior, Contamination Factors, and Preventive Measures. Int. J. Microbiol. 2019, 2019, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D. Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance. Front. Microbiol. 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Leimbach, A.; Hacker, J.; Dobrindt, U.E. coli as an All-Rounder: The Thin Line between Commensalism and Pathogenicity. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 3–32. ISBN 9783642365591. [Google Scholar]
- Bisi-Johnson, M.A.; Obi, C.L.; Vasaikar, S.D.; Baba, K.A.; Hattori, T. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Pathog. 2011, 3, 9. [Google Scholar] [CrossRef]
- Fang, F.C.; Frawley, E.R.; Tapscott, T.; Vázquez-Torres, A. Bacterial Stress Responses during Host Infection. Cell Host Microbe 2016, 20, 133–143. [Google Scholar] [CrossRef]
- Marles-Wright, J.; Lewis, R.J. Stress responses of bacteria. Curr. Opin. Struct. Biol. 2007, 17, 755–760. [Google Scholar] [CrossRef]
- Chung, H.J.; Bang, W.; Drake, M.A. Stress Response of Escherichia coli. Compr. Rev. Food Sci. Food Saf. 2006, 5, 52–64. [Google Scholar] [CrossRef]
- Van, T.T.H.; Chin, J.; Chapman, T.; Tran, L.T.; Coloe, P.J. Safety of raw meat and shellfish in Vietnam: An analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 2008, 124, 217–223. [Google Scholar] [CrossRef]
- Lyhs, U.; Ikonen, I.; Pohjanvirta, T.; Raninen, K.; Perko-Mäkelä, P.; Pelkonen, S. Extraintestinal pathogenic Escherichia coli in poultry meat products on the Finnish retail market. Acta Vet. Scand. 2012, 54, 64. [Google Scholar] [CrossRef]
- Caruso, G.; Giammanco, A.; Cardamone, C.; Oliveri, G.; Mascarella, C.; Capra, G.; Fasciana, T. Extra-Intestinal Fluoroquinolone-Resistant Escherichia coli Strains Isolated from Meat. BioMed. Res. Int. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cross, A.S. What is a virulence factor? Crit. Care 2008, 12, 197. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, F.; Corsello, G.; Cricelli, C.; Ferrara, N.; Ghiselli, A.; Lucchin, L.; Poli, A. Role of poultry meat in a balanced diet aimed at maintaining health and wellbeing: An Italian consensus document. Food Nutr. Res. 2015, 59, 27606. [Google Scholar] [CrossRef] [PubMed]
- Bohrer, B.M. Review: Nutrient density and nutritional value of meat products and non-meat foods high in protein. Trends Food Sci. Technol. 2017, 65, 103–112. [Google Scholar] [CrossRef]
- Tay, M.Y.F.; Adzitey, F.; Sultan, S.A.; Tati, J.M.; Seow, K.L.G.; Schlundt, J. Whole-Genome Sequencing of Nontyphoidal Salmonella enterica Isolates Obtained from Various Meat Types in Ghana. Microbiol. Resour. Announc. 2019, 8, e00033-19. [Google Scholar] [CrossRef]
- Rwego, I.B.; Gillespie, T.R.; Isabirye-Basuta, G.; Goldberg, T.L. High Rates of Escherichia coli Transmission between Livestock and Humans in Rural Uganda. J. Clin. Microbiol. 2008, 46, 3187–3191. [Google Scholar] [CrossRef]
- Currie, A.; Honish, L.; Cutler, J.; Locas, A.; Lavoie, M.-C.; Gaulin, C.; Galanis, E.; Tschetter, L.; Chui, L.; Taylor, M.; et al. Outbreak of Escherichia coli O157:H7 Infections Linked to Mechanically Tenderized Beef and the Largest Beef Recall in Canada, 2012. J. Food Prot. 2019, 82, 1532–1538. [Google Scholar] [CrossRef]
- Ostroff, S.M.; Griffin, P.M.; Tauxe, R.V.; Shipman, L.D.; Greene, K.D.; Wells, J.G.; Lewis, J.H.; Blake, P.A.; Kobayashi, J.M. A statewide outbreak of Escherichia coli 0157: h7 infections in washington state. Am. J. Epidemiol. 1990, 132, 239–247. [Google Scholar] [CrossRef]
- Brzuszkiewicz, E.; Thürmer, A.; Schuldes, J.; Leimbach, A.; Liesegang, H.; Meyer, F.-D.; Boelter, J.; Petersen, H.; Gottschalk, G.; Daniel, R. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC). Arch. Microbiol. 2011, 193, 883–891. [Google Scholar] [CrossRef]
- Alonso, R.; Martín, A.; Peláez, T.; Marín, M.; Rodríguez-Creixéms, M.; Bouza, E. An improved protocol for pulsed-field gel electrophoresis typing of Clostridium difficile. J. Med. Microbiol. 2005, 54, 155–157. [Google Scholar] [CrossRef]
- Hollmén, T.E.; Debroy, C.; Flint, P.L.; Safine, D.E.; Schamber, J.L.; Riddle, A.E.; Trust, K.A. Molecular typing of Escherichia coli strains associated with threatened sea ducks and near-shore marine habitats of south-west Alaska. Environ. Microbiol. Rep. 2011, 3, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Adzitey, F.; Ali, G.R.R.; Huda, N.; Ahmad, R. Genotyping of Salmonella strains isolated from ducks, their rearing and processing environments in Penang, Malaysia, using RAPD. 3 Biotech 2013, 3, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Molechan, C.; Amoako, D.G.; Abia, A.L.K.; Somboro, A.M.; Bester, L.A.; Essack, S.Y. Molecular epidemiology of antibiotic-resistant Enterococcus spp. from the farm-to-fork continuum in intensive poultry production in KwaZulu-Natal, South Africa. Sci. Total Environ. 2019, 692, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Pillay, S.; Amoako, D.G.; Abia, A.L.K.; Somboro, A.M.; Shobo, C.O.; Perrett, K.; Bester, L.A.; Essack, S.Y. Characterisation of Campylobacter spp. Isolated from Poultry in KwaZulu-Natal, South Africa. Antibiotics 2020, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Nutman, A.; Marchaim, D. How to: Molecular investigation of a hospital outbreak. Clin. Microbiol. Infect. 2019, 25, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.; Petkau, A.; Knox, N.; Graham, M.; Van Domselaar, G. A Primer on Infectious Disease Bacterial Genomics. Clin. Microbiol. Rev. 2016, 29, 881–913. [Google Scholar] [CrossRef]
- Osei Sekyere, J.; Amoako, D.G. Genomic and phenotypic characterisation of fluoroquinolone resistance mechanisms in Enterobacteriaceae in Durban, South Africa. PLoS ONE 2017, 12, e0178888. [Google Scholar] [CrossRef]
- Neher, R.A.; Bedford, T. Real-Time Analysis and Visualization of Pathogen Sequence Data. J. Clin. Microbiol. 2018, 56, 1–15. [Google Scholar] [CrossRef]
- Petkau, A.; Stuart-Edwards, M.; Stothard, P.; Van Domselaar, G. Interactive microbial genome visualization with GView. Bioinformatics 2010, 26, 3125–3126. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Feil, E.J.; Li, B.C.; Aanensen, D.M.; Hanage, W.P.; Spratt, B.G. eBURST: Inferring Patterns of Evolutionary Descent among Clusters of Related Bacterial Genotypes from Multilocus Sequence Typing Data. J. Bacteriol. 2004, 186, 1518–1530. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing Friend from Foe Using Bacterial Whole Genome Sequence Data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Kleinheinz, K.A.; Joensen, K.G.; Larsen, M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 2014, 4, e27943. [Google Scholar] [CrossRef] [PubMed]
- Ahrenfeldt, J.; Skaarup, C.; Hasman, H.; Pedersen, A.G.; Aarestrup, F.M.; Lund, O. Bacterial whole genome-based phylogeny: Construction of a new benchmarking dataset and assessment of some existing methods. BMC Genom. 2017, 18, 19. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; AbuDahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- Adzitey, F.; Assoah-Peprah, P.; Teye, G.A. Whole-genome sequencing of Escherichia coli isolated from contaminated meat samples collected from the Northern Region of Ghana reveals the presence of multiple antimicrobial resistance genes. J. Glob. Antimicrob. Resist. 2019, 18, 179–182. [Google Scholar] [CrossRef]
- O’Donoghue, S.I.; Baldi, B.F.; Clark, S.J.; Darling, A.E.; Hogan, J.M.; Kaur, S.; Maier-Hein, L.; McCarthy, D.J.; Moore, W.J.; Stenau, E.; et al. Visualization of Biomedical Data. Annu. Rev. Biomed. Data Sci. 2018, 1, 275–304. [Google Scholar] [CrossRef]
- Castellanos, L.R.; Donado-Godoy, P.; León, M.; Clavijo, V.; Arevalo, A.; Bernal, J.F.; Timmerman, A.J.; Mevius, D.J.; Wagenaar, J.A.; Hordijk, J. High Heterogeneity of Escherichia coli Sequence Types Harbouring ESBL/AmpC Genes on IncI1 Plasmids in the Colombian Poultry Chain. PLoS ONE 2017, 12, e0170777. [Google Scholar] [CrossRef]
- Yamaji, R.; Friedman, C.R.; Rubin, J.; Suh, J.; Thys, E.; McDermott, P.; Hung-Fan, M.; Riley, L.W. A Population-Based Surveillance Study of Shared Genotypes of Escherichia coli Isolates from Retail Meat and Suspected Cases of Urinary Tract Infections. mSphere 2018, 3, e00179-18. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, D.A.; Li, C.; Mukherjee, S.; Hsu, C.-H.; Bodeis Jones, S.; Gaines, S.A.; Kabera, C.; Loneragan, G.H.; Torrence, M.; Harhay, D.M.; et al. Whole-Genome Sequence Analysis of CTX-M Containing Escherichia coli Isolates from Retail Meats and Cattle in the United States. Microb. Drug Resist. 2018, 24, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.M.; Opintan, J.A.; Frimodt-Møller, N.; Styrishave, B. Beta-Lactamase Producing Escherichia coli Isolates in Imported and Locally Produced Chicken Meat from Ghana. PLoS ONE 2015, 10, e0139706. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Day, M.; Ciesielczuk, H.; Hope, R.; Underwood, A.; Reynolds, R.; Wain, J.; Livermore, D.M.; Woodford, N. Rapid Identification of Major Escherichia coli Sequence Types Causing Urinary Tract and Bloodstream Infections. J. Clin. Microbiol. 2015, 53, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Ogura, Y.; Gotoh, Y.; Itoh, T.; Sato, M.P.; Seto, K.; Yoshino, S.; Isobe, J.; Etoh, Y.; Kurogi, M.; Kimata, K.; et al. Population structure of Escherichia coli O26: H11 with recent and repeated stx2 acquisition in multiple lineages. Microb. Genom. 2017, 3. [Google Scholar] [CrossRef] [PubMed]
- Bakr Shabbir, M.A.; Hao, H.; Shabbir, M.Z.; Hussain, H.I.; Iqbal, Z.; Ahmed, S.; Sattar, A.; Iqbal, M.; Li, J.; Yuan, Z. Survival and evolution of CRISPR-Cas system in prokaryotes and its applications. Front. Immunol. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas Systems: Prokaryotes Upgrade to Adaptive Immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef]
- Klompe, S.E.; Sternberg, S.H. Harnessing “A Billion Years of Experimentation”: The Ongoing Exploration and Exploitation of CRISPR–Cas Immune Systems. Cris. J. 2018, 1, 141–158. [Google Scholar] [CrossRef]
- Bozic, B.; Repac, J.; Djordjevic, M. Endogenous Gene Regulation as a Predicted Main Function of Type I-E CRISPR/Cas System in E. coli. Molecules 2019, 24, 784. [Google Scholar] [CrossRef]
- Chang, Y.; Su, T.; Qi, Q.; Liang, Q. Easy regulation of metabolic flux in Escherichia coli using an endogenous type I-E CRISPR-Cas system. Microb. Cell Factories 2016, 15, 195. [Google Scholar] [CrossRef]
- Vasu, K.; Nagaraja, V. Diverse Functions of Restriction-Modification Systems in Addition to Cellular Defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Pleška, M.; Qian, L.; Okura, R.; Bergmiller, T.; Wakamoto, Y.; Kussell, E.; Guet, C.C. Bacterial Autoimmunity Due to a Restriction-Modification System. Curr. Biol. 2016, 26, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Grayson, P.; Schulten, K. Glycerol Conductance and Physical Asymmetry of the Escherichia coli Glycerol Facilitator GlpF. Biophys. J. 2003, 85, 2977–2987. [Google Scholar] [CrossRef][Green Version]
- Borgnia, M.J.; Kozono, D.; Calamita, G.; Maloney, P.C.; Agre, P. Functional reconstitution and characterization of AqpZ, the E. coli water channel protein. J. Mol. Biol. 1999, 291, 1169–1179. [Google Scholar] [CrossRef]
- Figueroa-Soto, C.G.; Valenzuela-Soto, E.M. Glycine betaine rather than acting only as an osmolyte also plays a role as regulator in cellular metabolism. Biochimie 2018, 147, 89–97. [Google Scholar] [CrossRef]
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.-F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef]
- Bearson, S.; Bearson, B.; Foster, J.W. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 2006, 147, 173–180. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef]
- Kroos, L.; Akiyama, Y. Biochemical and structural insights into intramembrane metalloprotease mechanisms. Biochim. Biophys. Acta Biomembr. 2013, 1828, 2873–2885. [Google Scholar] [CrossRef]
- Turner, R.J.; Weiner, J.H.; Taylor, D.E. Selenium metabolism in Escherichia coli. BioMetals 1998, 11, 223–227. [Google Scholar] [CrossRef]
- Choudhury, H.G.; Beis, K. Tellurite-Resistance Protein TehA from Escherichia coli. In Encyclopedia of Metalloproteins; Springer: New York, NY, USA, 2013; pp. 2157–2160. [Google Scholar]
- Amoako, D.G.; Somboro, A.M.; Abia, A.L.K.; Allam, M.; Ismail, A.; Bester, L.A.; Essack, S.Y. Genome Mining and Comparative Pathogenomic Analysis of An Endemic Methicillin-Resistant Staphylococcus aureus (MRSA) Clone, ST612-CC8-t1257-SCCmec_IVd(2B), Isolated in South Africa. Pathogens 2019, 8, 166. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Martinez, J.L. Friends or foes: Can we make a distinction between beneficial and harmful strains of the Stenotrophomonas maltophilia complex? Front. Microbiol. 2015, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Deneke, C.; Rentzsch, R.; Renard, B.Y. PaPrBaG: A machine learning approach for the detection of novel pathogens from NGS data. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ratledge, C. Iron Metabolism and Infection. Food Nutr. Bull. 2007, 28, S515–S523. [Google Scholar] [CrossRef]
- Cassat, J.E.; Skaar, E.P. Iron in Infection and Immunity. Cell Host Microbe 2013, 13, 509–519. [Google Scholar] [CrossRef]
- Li, Y.; Dai, J.; Zhuge, X.; Wang, H.; Hu, L.; Ren, J.; Chen, L.; Li, D.; Tang, F. Iron-regulated gene ireA in avian pathogenic Escherichia coli participates in adhesion and stress-resistance. BMC Vet. Res. 2016, 12, 167. [Google Scholar] [CrossRef][Green Version]
- Okeke, I.N.; Scaletsky, I.C.A.; Soars, E.H.; Macfarlane, L.R.; Torres, A.G. Molecular Epidemiology of the Iron Utilization Genes of Enteroaggregative Escherichia coli. J. Clin. Microbiol. 2004, 42, 36–44. [Google Scholar] [CrossRef]
- Duquesne, S.; Destoumieux-Garzón, D.; Peduzzi, J.; Rebuffat, S. Microcins, gene-encoded antibacterial peptides from Enterobacteria. Nat. Prod. Rep. 2007, 24, 708. [Google Scholar] [CrossRef]
- Rebuffat, S. Microcins in action: Amazing defence strategies of Enterobacteria. Biochem. Soc. Trans. 2012, 40, 1456–1462. [Google Scholar] [CrossRef]
- Cid, D.; Sanz, R.; Marín, I.; De Greve, H.; Ruiz-Santa-Quiteria, J.A.; Amils, R.; De La Fuente, R. Characterization of Nonenterotoxigenic Escherichia coli Strains Producing F17 Fimbriae Isolated from Diarrheic Lambs and Goat Kids. J. Clin. Microbiol. 1999, 37, 1370–1375. [Google Scholar] [CrossRef]
- Bihannic, M.; Ghanbarpour, R.; Auvray, F.; Cavalié, L.; Châtre, P.; Boury, M.; Brugère, H.; Madec, J.-Y.; Oswald, E. Identification and detection of three new F17 fimbrial variants in Escherichia coli strains isolated from cattle. Vet. Res. 2014, 45, 76. [Google Scholar] [CrossRef] [PubMed]
- Do Vale, A.; Cabanes, D.; Sousa, S. Bacterial Toxins as Pathogen Weapons Against Phagocytes. Front. Microbiol. 2016, 7, 42. [Google Scholar] [CrossRef]
- Krüger, A.; Lucchesi, P.M.A.; Sanso, A.M.; Etcheverría, A.I.; Bustamante, A.V.; Burgán, J.; Fernández, L.; Fernandez, D.; Leotta, G.; Friedrich, A.W.; et al. Genetic characterization of Shiga toxin-producing Escherichia coli O26:H11 strains isolated from animal, food, and clinical samples. Front. Cell. Infect. Microbiol. 2015, 5, 74. [Google Scholar] [CrossRef] [PubMed]
- Faïs, T.; Delmas, J.; Serres, A.; Bonnet, R.; Dalmasso, G. Impact of CDT Toxin on Human Diseases. Toxins 2016, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Amoako, D.G.; Somboro, A.M.; Abia, A.L.K.; Allam, M.; Ismail, A.; Bester, L.; Essack, S.Y. Genomic analysis of methicillin-resistant Staphylococcus aureus isolated from poultry and occupational farm workers in Umgungundlovu District, South Africa. Sci. Total Environ. 2019, 670, 704–716. [Google Scholar] [CrossRef]
- Ramsamy, Y.; Mlisana, K.P.; Amoako, D.G.; Allam, M.; Ismail, A.; Singh, R.; Abia, A.L.K.; Essack, S.Y. Pathogenomic Analysis of a Novel Extensively Drug-Resistant Citrobacter freundii Isolate Carrying a blaNDM-1 Carbapenemase in South Africa. Pathogens 2020, 9, 89. [Google Scholar] [CrossRef]
- Robins-Browne, R.M.; Holt, K.E.; Ingle, D.J.; Hocking, D.M.; Yang, J.; Tauschek, M. Are Escherichia coli Pathotypes Still Relevant in the Era of Whole-Genome Sequencing? Front. Cell. Infect. Microbiol. 2016, 6, 141. [Google Scholar] [CrossRef] [PubMed]
- Tagini, F.; Greub, G. Bacterial genome sequencing in clinical microbiology: A pathogen-oriented review. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2007–2020. [Google Scholar] [CrossRef]
| 1 D | Demographic Information | In Silico Typing | Molecular Defence Systems | Pathogenic Determinants | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source | Area | MLST | Serotype 1 | CHTyper | CRISPRs (Cas) (Cluster) | Restriction-Modification (R-M) System | Pathogenicity Score (Pathogenic Families) | Virulence Factors | ||
| FumC | FimH | |||||||||
| AB1 | Beef | Aboabo | 540 | O11 | fumC7 | fimH54 | 7 (1) CAS-TypeIE | Type II (M.EcoGVI, M.Eco4255 Dcm) Type IV (EcoAPECGmrSD) | 0.923 (525) | gad, iss |
| CB1 | Beef | Central Market | 6646 | H39 | fumC203 | fimH562 | 3 (0) | Type I (S.Sso30807 I) Type II (M.EcoKII, M.EcoGI, M.Eco3609 Dcm) | 0.938 (698) | air, cdtB, eilA, gad, iss |
| NB12 | Beef | Nyorni | 7483 | O8 | fumC23 | fimH38 | 3 (1) CAS-TypeIE | Type I (S.EcoNIH1 II, EcoO127 I, M.EcoNIH1 II) Type II (M.EcoKII, M.Eco3740 Dcm) | 0.942 (710) | astA, gad, lpfA |
| AC1 | Chevon | Aboabo | 44 | O162 | fumC11 | fimH54 | 8 (1) CAS-TypeIE | Type I (M.Eco6409 I, S.Eco3325 I, M.Eco2747 II) Type II (M.EcoGVII, M.EcoKII, M.EcoJA03 PDcm) | 0.938 (698) | gad |
| CC6 | Chevon | Central Market | 469 | O8 | fumC65 | fimH68 | 6 (1) CAS-TypeIE | Type I (M.Sso30807 I), Type II (M.EcoKII, M.Eco3740 Dcm) | 0.940 (678) | celb, f17 A, f17 G, gad, iss, lpfA mchB, mchC, mchF, mcmA |
| NC3 | Chevon | Nyorni | 1727 | O88 | fumC19 | fimH31 | 3 (1) CAS-TypeIE | Type II (M.EcoKII, M.EcoGVII, M.EcoJA03 PDcm) | 0.940 (686) | f17 A, f17 G, gad, iss, lpfA |
| SG6 | Guinea fowl | Victory Cinema | 69 | O15 | fumC35 | fimH27 | 6 (2) CAS-TypeIE | Type I (EcoKI, M.EcoJA69 PI, S.EcoJA69 PI), Type II (M.EcoGI, M.EcoKII, M.Eco3317 Dcm) Type IV (EcoKMrr) | 0.930 (1001) | air, eilA, gad, iha, ireA, iroN, iss, IpfA, sat, senB |
| TG1 | Guinea fowl | Tishegu | 540 | O9 | fumC7 | fimH54 | 8 (1) CAS-TypeIE | Type II (M.EcoGVII, M.EcoJA03 PDcm) | 0.934 (564) | gad, iss |
| TG5 | Guinea fowl | Tishegu | 7473 | O61 | fumC7 | fimH54 | 10 (0) | Type II (M.EcoKII, M.EcoJA03 PDcm) | 0.929 (604) | astA, gad |
| SLC2 | Local Chicken | Victory Cinema | 155 | H9 | fumC4 | fimH32 | 4 (1) CAS-TypeIE | Type II (M.EcoKII, M.EcoGVII, M.EcoJA03 PDcm) | 0.943 (651) | gad, lpfA |
| TLC1 | Local Chicken | Tishegu | 297 | H9 | fumC65 | fimH38 | 8 (1) CAS-TypeIE | Type I (M.Eco3609 I) Type II (M.EcoKII, M.EcoGI, M.Eco3609 Dcm) | 0.943 (691) | gad, lpfA |
| TLC13 | Local Chicken | Tishegu | 155 | O132 | fumC4 | fimH32 | 4 (1) CAS-TypeIE | Type II (M.EcoKII, M.EcoGVII, M.EcoJA03 PDcm) | 0.943 (648) | gad, lpfA |
| CM4 | Mutton | Central Market | 155 | H40 | fumC4 | fimH366 | 6 (1) CAS-TypeIE | Type I (M.Eco2747 II, EcoR124 II), Type II (M.EcoE1140 Dcm, M.EcoKII) | 0.940 (655) | gad, iss, lpfA |
| NM11 | Mutton | Nyorni | 1141 | O113 | fumC11 | fimH25 | 9 (1) CAS-TypeIE | Type I (M.EcoJA65 PI), Type II (M.EcoJA03 PDcm, M.EcoKII) | 0.936 (623) | gad, iss |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adzitey, F.; Asante, J.; Kumalo, H.M.; Khan, R.B.; Somboro, A.M.; Amoako, D.G. Genomic Investigation into the Virulome, Pathogenicity, Stress Response Factors, Clonal Lineages, and Phylogenetic Relationship of Escherichia coli Strains Isolated from Meat Sources in Ghana. Genes 2020, 11, 1504. https://doi.org/10.3390/genes11121504
Adzitey F, Asante J, Kumalo HM, Khan RB, Somboro AM, Amoako DG. Genomic Investigation into the Virulome, Pathogenicity, Stress Response Factors, Clonal Lineages, and Phylogenetic Relationship of Escherichia coli Strains Isolated from Meat Sources in Ghana. Genes. 2020; 11(12):1504. https://doi.org/10.3390/genes11121504
Chicago/Turabian StyleAdzitey, Frederick, Jonathan Asante, Hezekiel M. Kumalo, Rene B. Khan, Anou M. Somboro, and Daniel G. Amoako. 2020. "Genomic Investigation into the Virulome, Pathogenicity, Stress Response Factors, Clonal Lineages, and Phylogenetic Relationship of Escherichia coli Strains Isolated from Meat Sources in Ghana" Genes 11, no. 12: 1504. https://doi.org/10.3390/genes11121504
APA StyleAdzitey, F., Asante, J., Kumalo, H. M., Khan, R. B., Somboro, A. M., & Amoako, D. G. (2020). Genomic Investigation into the Virulome, Pathogenicity, Stress Response Factors, Clonal Lineages, and Phylogenetic Relationship of Escherichia coli Strains Isolated from Meat Sources in Ghana. Genes, 11(12), 1504. https://doi.org/10.3390/genes11121504






