The Dark Side of UV-Induced DNA Lesion Repair
Abstract
1. Introduction
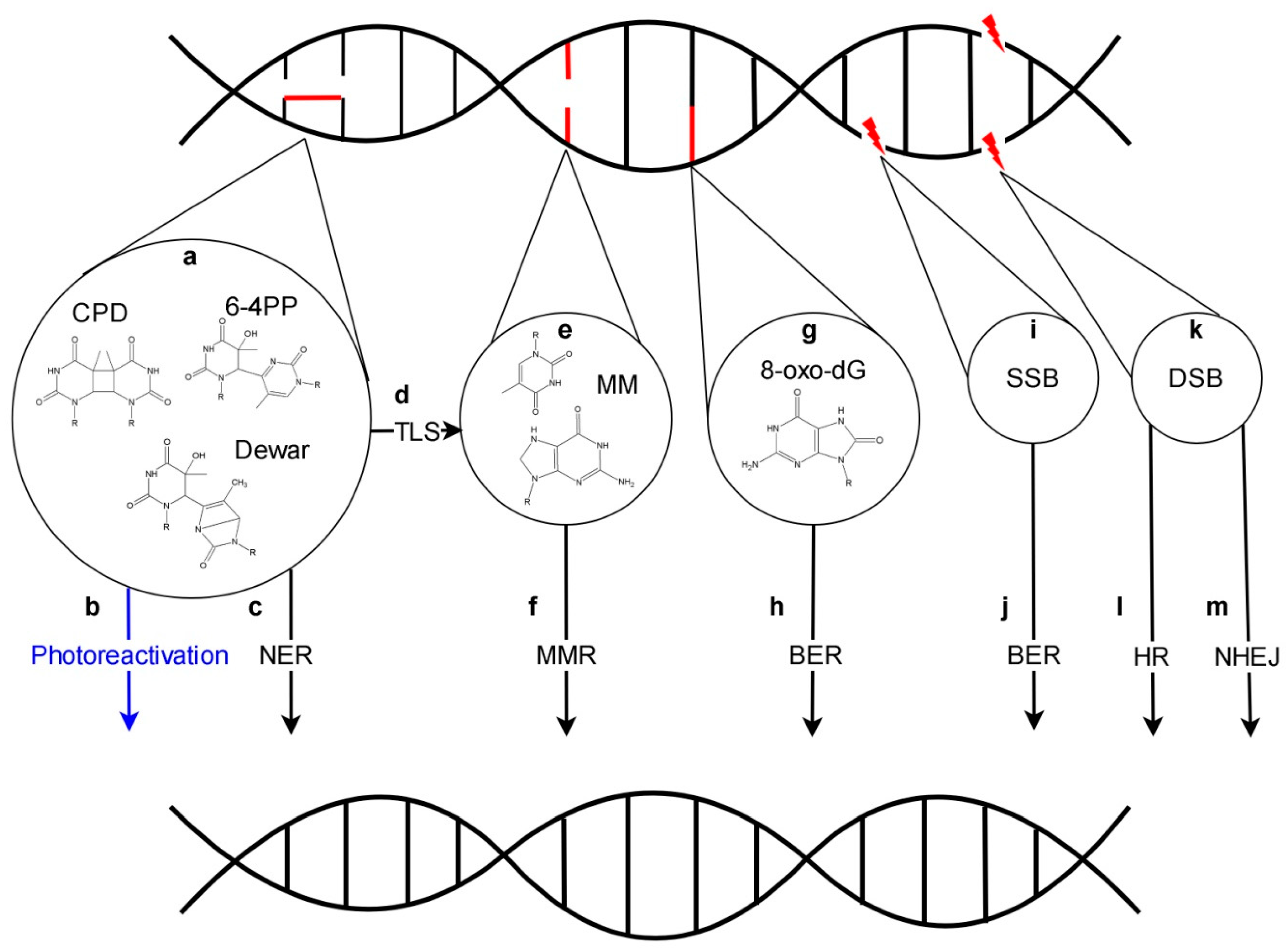
2. Dark DNA Repair
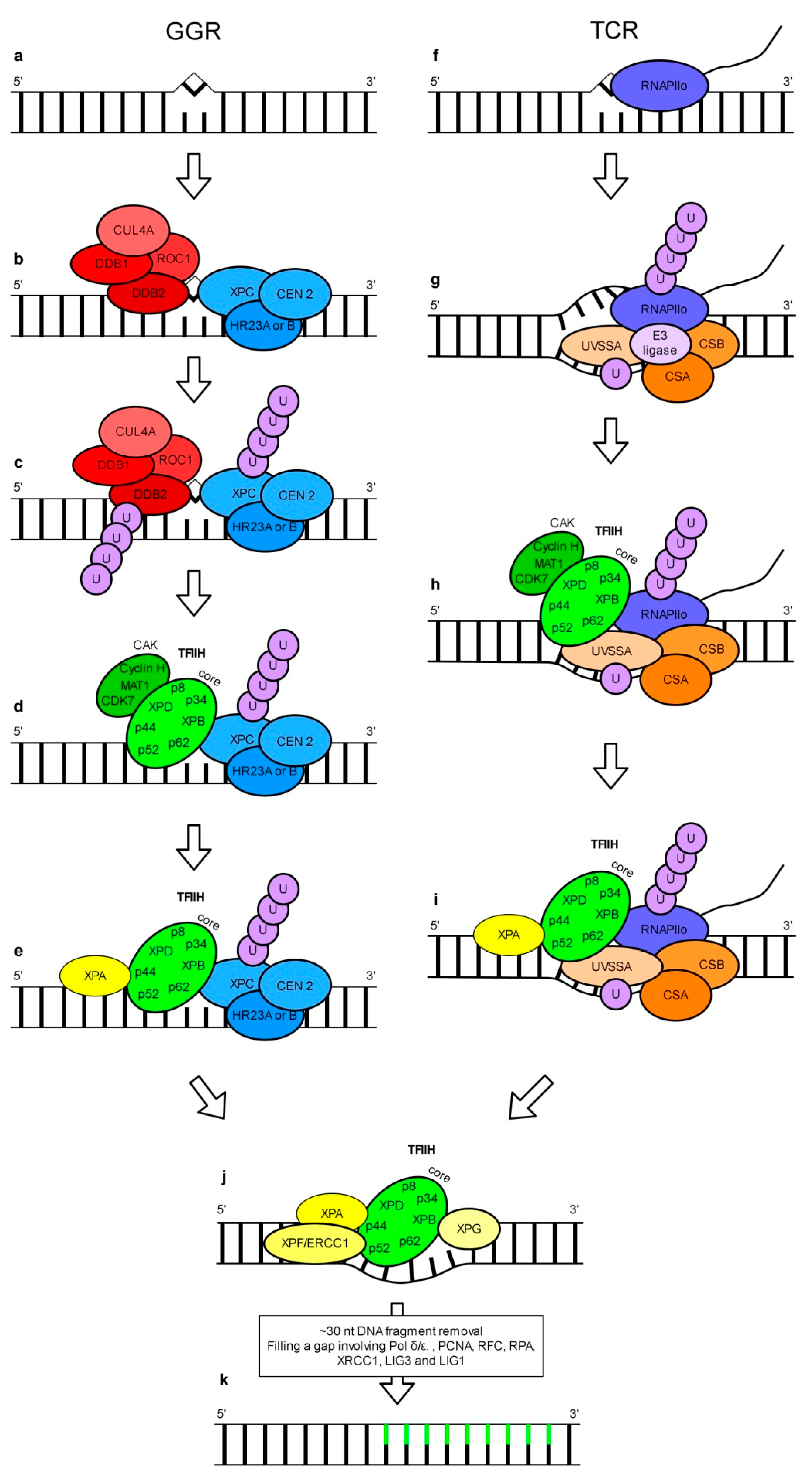
3. Nucleotide Excision Repair
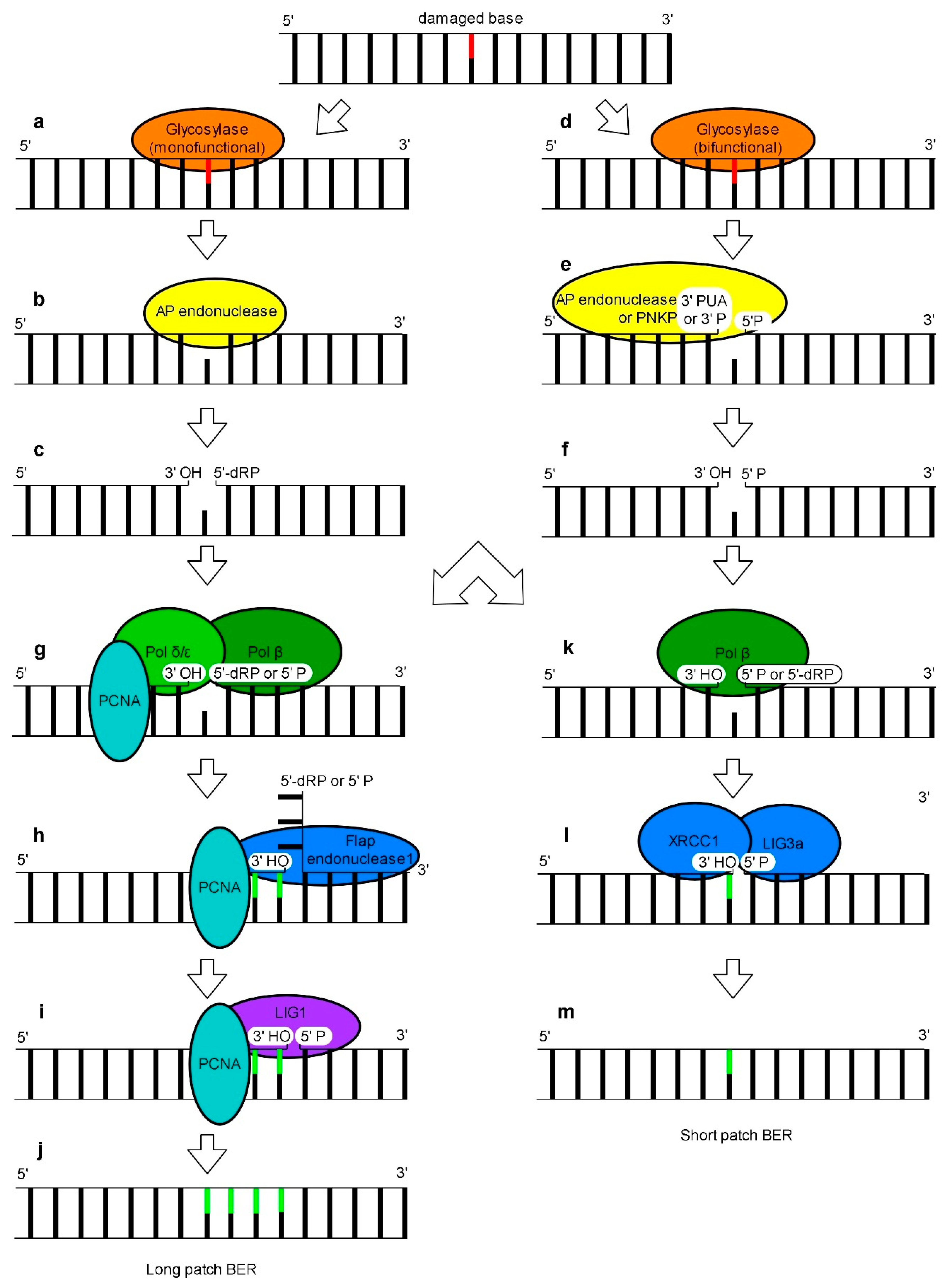
4. Base Excision Repair
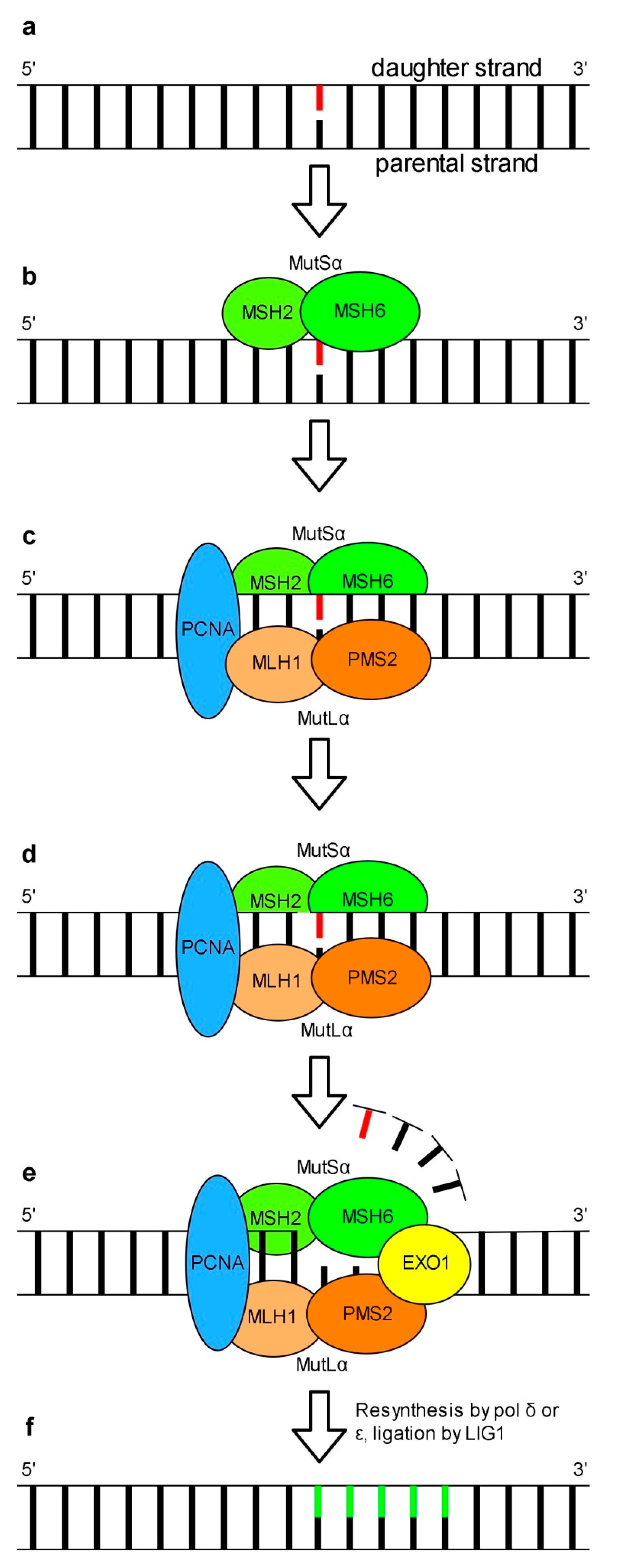
5. Mismatch Repair
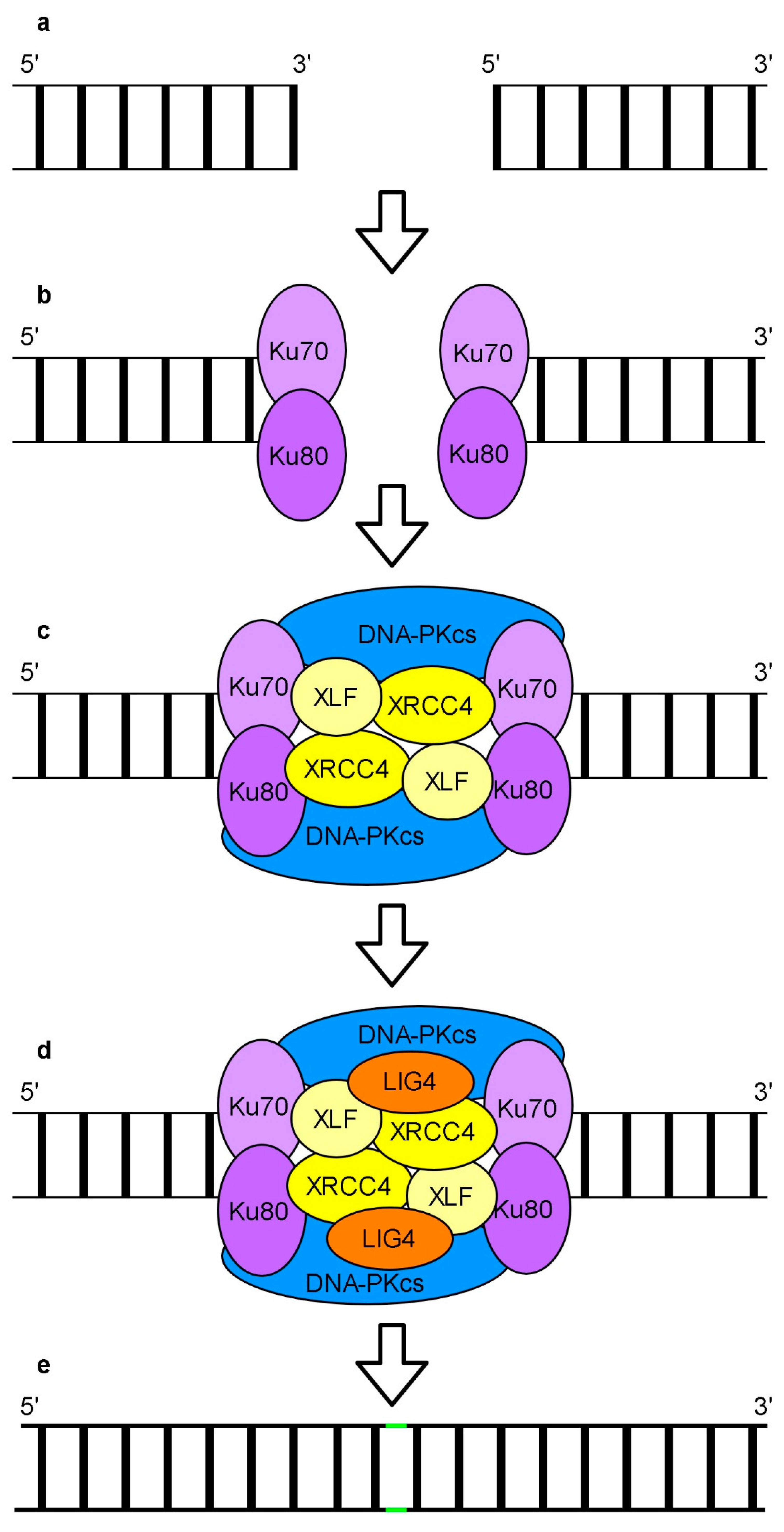
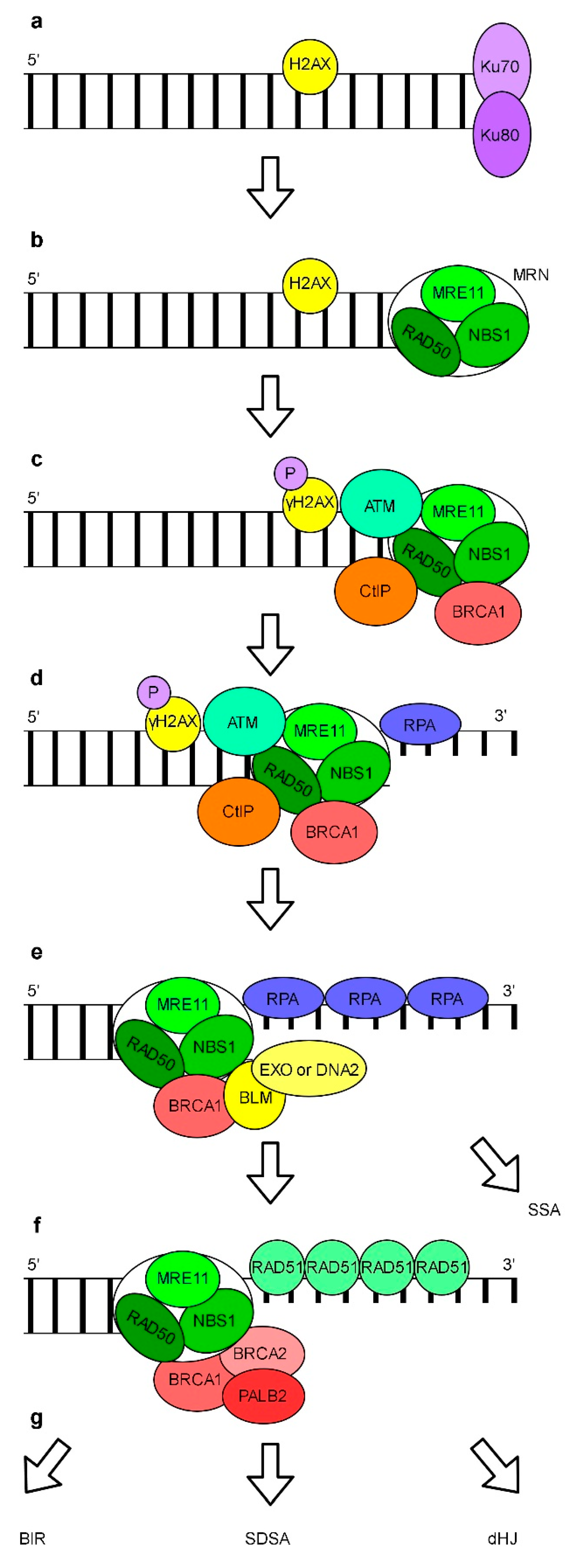
6. Repair of DNA Breaks
Repair of DSBs by Non-Homologous End Joining and Homologous Recombination
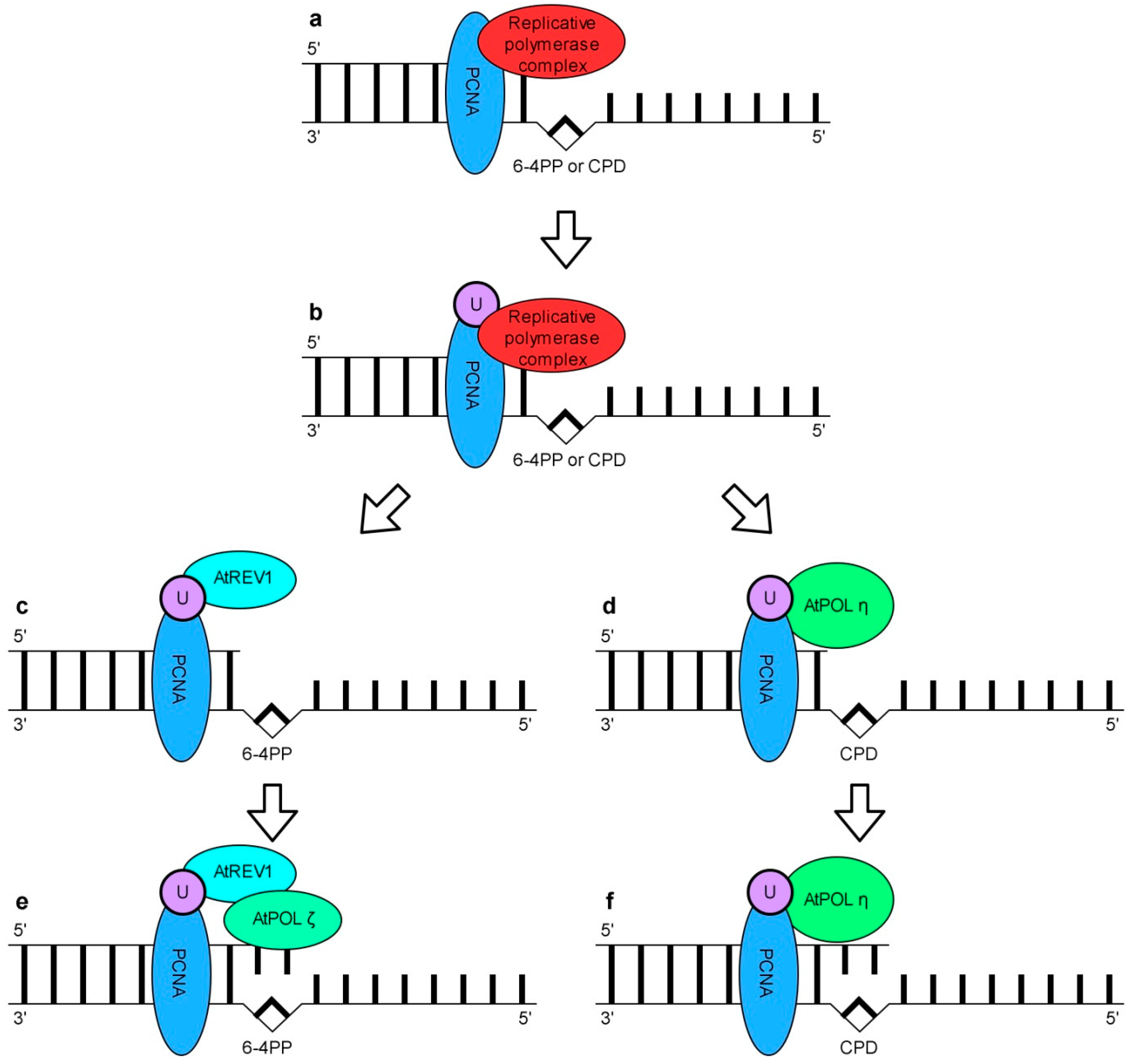
7. DNA Damage Tolerance
8. Perspectives and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- CIE. The Spectroradiometric Measurement of Light Sources. CIE 063-1984. 1984. Available online: http://cie.co.at/publications/spectroradiometric-measurement-light-sources (accessed on 1 January 1984).
- CIE. The Measurement of Absolute Luminous Intensity Distributions. CIE 070–1987. 1987. Available online: http://cie.co.at/publications/measurement-absolute-luminous-intensity-distributions (accessed on 1 January 1987).
- CIE. Standardization of the Terms UV-A1, UV-A2 and UV-B. 134/1 TC 6. 1999. Available online: http://cie.co.at/publications/cie-collection-photobiology-photochemistry-1999 (accessed on 1 January 1999).
- Moan, J. Visible Light and UV Radiation. In Radiation at Home, Outdoors and in the Workplace; Brune, D., Hellborg, R., Persson, B.R.R., Pääkkönen, R., Eds.; Scandinavian Science Publisher: Oslo, Norway, 2002; Volume 7, ISBN 8291833028. [Google Scholar]
- Kielbassa, C.; Roza, L.; Epe, B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis 1997, 18, 811–816. [Google Scholar] [CrossRef]
- Dany, A.L.; Tissier, A. A functional OGG1 homologue from Arabidopsis thaliana. Mol. Gen. Genet. 2001, 265, 293–301. [Google Scholar] [CrossRef]
- Douki, T.; Cadet, J. Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry 2001, 40, 2495–2501. [Google Scholar] [CrossRef]
- Douki, T.; Sage, E. Dewar valence isomers, the third type of environmentally relevant DNA photoproducts induced by solar radiation. Photochem. Photobiol. Sci. 2016, 15, 24–30. [Google Scholar] [CrossRef]
- Peak, J.G.; Peak, M.J. Ultraviolet light induces double-strand breaks in DNA of cultured human P3 cells as measured by neutral filter elution. Photochem. Photobiol. 1990, 52, 387–393. [Google Scholar] [CrossRef]
- Peng, W.; Shaw, B.R. Accelerated deamination of cytosine residues in UV-induced cyclobutane pyrimidine dimers leads to CC→TT transitions. Biochemistry 1996, 35, 10172–10181. [Google Scholar] [CrossRef]
- Doetsch, P.W.; Zastawny, T.H.; Martin, A.M.; Dizdaroglu, M. Monomeric base damage products from adenine, guanine, and thymine induced by exposure of DNA to ultraviolet radiation. Biochemistry 1995, 34, 737–742. [Google Scholar] [CrossRef]
- Cheng, K.C.; Cahill, D.S.; Kasai, H.; Nishimura, S.; Loeb, L.A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J. Biol. Chem. 1992, 267, 166–172. [Google Scholar]
- Datta, K.; Purkayastha, S.; Neumann, R.D.; Pastwa, E.; Winters, T.A. Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3′-termini. Radiat. Res. 2011, 175, 97–112. [Google Scholar] [CrossRef]
- Ai, Y.J.; Liao, R.Z.; Chen, S.L.; Hua, W.J.; Fang, W.H.; Luo, Y. Repair of DNA dewar photoproduct to (6-4) photoproduct in (6-4) photolyase. J. Phys. Chem. B 2011, 115, 10976–10982. [Google Scholar] [CrossRef]
- Sancar, A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 2003, 103, 2203–2237. [Google Scholar] [CrossRef]
- Banas, A.K.; Zglobicki, P.; Bazant, A.; Kowalska, E.; Dziga, D.; Strzalka, W. All you need is light. Photorepair of UV-induced pyrimidine dimers. Genes (Basel) 2020, 11, 1304. [Google Scholar] [CrossRef]
- Strzalka, W.; Ziemienowicz, A. Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation. Ann. Bot. 2011, 107, 1127–1140. [Google Scholar] [CrossRef]
- Kimura, S.; Tahira, Y.; Ishibashi, T.; Mori, Y.; Mori, T.; Hashimoto, J.; Sakaguchi, K. DNA repair in higher plants; photoreactivation is the major DNA repair pathway in non-proliferating cells while excision repair (nucleotide excision repair and base excision repair) is active in proliferating cells. Nucleic Acids Res. 2004, 32, 2760–2767. [Google Scholar] [CrossRef]
- Banas, A.K.; Hermanowicz, P.; Sztatelman, O.; Labuz, J.; Aggarwal, C.; Zglobicki, P.; Jagiello-Flasinska, D.; Strzalka, W. 6,4-PP Photolyase Encoded by AtUVR3 is Localized in Nuclei, Chloroplasts and Mitochondria and its Expression is Down-Regulated by Light in a Photosynthesis-Dependent Manner. Plant Cell Physiol. 2018, 59, 44–57. [Google Scholar] [CrossRef]
- Gillet, L.C.J.; Schärer, O.D. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006, 106, 253–276. [Google Scholar] [CrossRef]
- Araki, M.; Masutani, C.; Takemura, M.; Uchida, A.; Sugasawa, K.; Kondoh, J.; Ohkuma, Y.; Hanaoka, F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 2001, 276, 18665–18672. [Google Scholar] [CrossRef]
- Masutani, C.; Sugasawa, K.; Yanagisawa, J.; Sonoyama, T.; Ui, M.; Enomoto, T.; Takio, K.; Tanaka, K.; Van Der Spek, P.J.; Bootsma, D.; et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994, 13, 1831–1843. [Google Scholar] [CrossRef]
- Batty, D.; Rapic’-Otrin, V.; Levine, A.S.; Wood, R.D. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 2000, 300, 275–290. [Google Scholar] [CrossRef]
- Sugasawa, K.; Okamoto, T.; Shimizu, Y.; Masutani, C.; Iwai, S.; Hanaoka, F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001, 15, 507–521. [Google Scholar] [CrossRef]
- Sugasawa, K.; Shimizu, Y.; Iwai, S.; Hanaoka, F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair 2002, 1, 95–107. [Google Scholar] [CrossRef]
- Keeney, S.; Chang, G.J.; Linn, S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 1993, 268, 21293–21300. [Google Scholar]
- Takao, M.; Abramic, M.; Moos, M.; Rapic’ Otrin, V.; Wootton, J.C.; Mclenigan, M.; Levine, A.S.; Protic, M. A 127 kDa component of a UV-damaged DNA-binding complex, which is defective in some xeroderma pigmentosum group E patients, is homologous to a slime mold protein. Nucleic Acids Res. 1993, 21, 4111–4118. [Google Scholar] [CrossRef][Green Version]
- Fujiwara, Y.; Masutani, C.; Mizukoshi, T.; Kondo, J.; Hanaoka, F.; Iwai, S. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem. 1999, 274, 20027–20033. [Google Scholar] [CrossRef]
- Payne, A.; Chu, G. Xeroderma pigmentosum group E binding factor recognizes a broad spectrum of DNA damage. Mutat. Res. Regul. Pap. 1994, 310, 89–102. [Google Scholar] [CrossRef]
- Reardon, J.T.; Nichols, A.F.; Keeney, S.; Smith, C.A.; Taylor, J.S.; Linn, S.; Sancar, A. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6 -4]T, and T[Dewar]T. J. Biol. Chem. 1993, 268, 21301–21308. [Google Scholar]
- Sugasawa, K.; Okuda, Y.; Saijo, M.; Nishi, R.; Matsuda, N.; Chu, G.; Mori, T.; Iwai, S.; Tanaka, K.; Tanaka, K.; et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 2005, 121, 387–400. [Google Scholar] [CrossRef]
- Treiber, D.K.; Chen, Z.; Essigmann, J.M. An ultraviolet light-damaged DNA recognition protein absent in xeroderma pigmentosum group E cells binds selectively to pyrimidine (6-4) pyrimidone photoproducts. Nucleic Acids Res. 1992, 20, 5805–5810. [Google Scholar] [CrossRef]
- Matsumoto, S.; Cavadini, S.; Bunker, R.D.; Grand, R.S.; Potenza, A.; Rabl, J.; Yamamoto, J.; Schenk, A.D.; Schübeler, D.; Iwai, S.; et al. DNA damage detection in nucleosomes involves DNA register shifting. Nature 2019, 571, 79–84. [Google Scholar] [CrossRef]
- Hwang, B.J.; Toering, S.; Francke, U.; Chu, G. p48 Activates a UV-Damaged-DNA Binding Factor and Is Defective in Xeroderma Pigmentosum Group E Cells That Lack Binding Activity. Mol. Cell. Biol. 1998, 18, 4391–4399. [Google Scholar] [CrossRef]
- Tang, J.Y.; Hwang, B.J.; Ford, J.M.; Hanawalt, P.C.; Chu, G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 2000, 5, 737–744. [Google Scholar] [CrossRef]
- Hwang, B.J.; Ford, J.M.; Hanawalt, P.C.; Chu, G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA 1999, 96, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Fitch, M.E.; Nakajima, S.; Yasui, A.; Ford, J.M. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 2003, 278, 46906–46910. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.E.; Zhu, Q.; Wani, G.; Chen, J.; Wani, A.A. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis 2004, 25, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Reardon, J.T.; Sancar, A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 2003, 17, 2539–2551. [Google Scholar] [CrossRef]
- Wakasugi, M.; Shimizu, M.; Morioka, H.; Linn, S.; Nikaido, O.; Matsunaga, T. Damaged DNA-binding Protein DDB Stimulates the Excision of Cyclobutane Pyrimidine Dimers in Vitro in Concert with XPA and Replication Protein A. J. Biol. Chem. 2001, 276, 15434–15440. [Google Scholar] [CrossRef]
- Wakasugi, M.; Kawashima, A.; Morioka, H.; Linn, S.; Sancar, A.; Mori, T.; Nikaido, O.; Matsunaga, T. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 2002, 277, 1637–1640. [Google Scholar] [CrossRef]
- Groisman, R.; Polanowska, J.; Kuraoka, I.; Sawada, J.I.; Saijo, M.; Drapkin, R.; Kisselev, A.F.; Tanaka, K.; Nakatani, Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 2003, 113, 357–367. [Google Scholar] [CrossRef]
- Wang, H.; Zhai, L.; Xu, J.; Joo, H.Y.; Jackson, S.; Erdjument-Bromage, H.; Tempst, P.; Xiong, Y.; Zhang, Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 2006, 22, 383–394. [Google Scholar] [CrossRef]
- de Jesus, B.M.B.; Bjørås, M.; Coin, F.; Egly, J.M. Dissection of the molecular defects caused by pathogenic mutations in the DNA repair factor XPC. Mol. Cell. Biol. 2008, 28, 7225–7235. [Google Scholar] [CrossRef]
- Yokoi, M.; Masutani, C.; Maekawa, T.; Sugasawa, K.; Ohkuma, Y.; Hanaoka, F. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 2000, 275, 9870–9875. [Google Scholar] [CrossRef] [PubMed]
- Sandrock, B.; Egly, J.M. A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. J. Biol. Chem. 2001, 276, 35328–35333. [Google Scholar] [CrossRef] [PubMed]
- Arab, H.H.; Wani, G.; Ray, A.; Shah, Z.I.; Zhu, Q.; Wani, A.A. Dissociation of CAK from core TFIIH reveals a functional link between XP-G/CS and the TFIIH Disassembly State. PLoS ONE 2010, 5, e11007. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.J.; Wood, R.D. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry 1993, 32, 12096–12104. [Google Scholar] [CrossRef]
- Missura, M.; Buterin, T.; Hindges, R.; Hübscher, U.; Kaspárková, J.; Brabec, V.; Naegeli, H. Double-check probing of DNA bending and unwinding by XPA-RPA: An architectural function in DNA Repair. EMBO J 2001, 20, 3554–3564. [Google Scholar] [CrossRef]
- Coin, F.; Oksenych, V.; Mocquet, V.; Groh, S.; Blattner, C.; Egly, J.M. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell 2008, 31, 9–20. [Google Scholar] [CrossRef]
- Compe, E.; Egly, J.M. Nucleotide Excision Repair and Transcriptional Regulation: TFIIH and beyond. Annu. Rev. Biochem. 2016, 85, 265–290. [Google Scholar] [CrossRef]
- Coin, F.; Oksenych, V.; Egly, J.M. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol. Cell 2007, 26, 245–256. [Google Scholar] [CrossRef]
- Li, C.-L.; Golebiowski, F.M.; Onishi, Y.; Samara, N.L.; Sugasawa, K.; Yang, W. Tripartite DNA Lesion Recognition and Verification by XPC, TFIIH, and XPA in Nucleotide Excision Repair. Mol Cell 2015, 59, 1025–1034. [Google Scholar] [CrossRef]
- Kusakabe, M.; Onishi, Y.; Tada, H.; Kurihara, F.; Kusao, K.; Furukawa, M.; Iwai, S.; Yokoi, M.; Sakai, W.; Sugasawa, K. Mechanism and regulation of DNA damage recognition in nucleotide excision repair. Genes Environ. 2019, 41, 2. [Google Scholar] [CrossRef]
- Kokic, G.; Chernev, A.; Tegunov, D.; Dienemann, C.; Urlaub, H.; Cramer, P. Structural basis of TFIIH activation for nucleotide excision repair. Nat. Commun. 2019, 10, 2885. [Google Scholar] [CrossRef]
- Zhu, Q.; Wani, A.A. Nucleotide Excision Repair: Finely Tuned Molecular Orchestra of Early Pre-incision Events. Photochem. Photobiol. 2017, 93, 166–177. [Google Scholar] [CrossRef]
- Lafrance-Vanasse, J.; Arseneault, G.V.; Cappadocia, L.; Legault, P.; Omichinski, J.G. Structural and functional evidence that Rad4 competes with Rad2 for binding to the Tfb1 subunit of TFIIH in NER. Nucleic Acids Res. 2013, 41, 2736–2745. [Google Scholar] [CrossRef]
- Ito, S.; Kuraoka, I.; Chymkowitch, P.; Compe, E.; Takedachi, A.; Ishigami, C.; Coin, F.; Egly, J.M.; Tanaka, K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: Implications for Cockayne syndrome in XP-G/CS patients. Mol. Cell 2007, 26, 231–243. [Google Scholar] [CrossRef]
- O’Donovan, A.; Davies, A.A.; Moggs, J.G.; West, S.C.; Wood, R.D. XPG endonuclease makes the 3′ Incision in human DNA nucleotide excision repair. Nature 1994, 371, 432–435. [Google Scholar] [CrossRef]
- Sijbers, A.M.; De Laat, W.L.; Ariza, R.R.; Biggerstaff, M.; Wei, Y.F.; Moggs, J.G.; Carter, K.C.; Shell, B.K.; Evans, E.; De Jong, M.C.; et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 1996, 86, 811–822. [Google Scholar] [CrossRef]
- Huang, J.C.; Svoboda, D.L.; Reardon, J.T.; Sancar, A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl. Acad. Sci. USA 1992, 89, 3664–3668. [Google Scholar] [CrossRef]
- Moggs, J.G.; Yarema, K.J.; Essigmann, J.M.; Wood, R.D. Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J. Biol. Chem. 1996, 271, 7177–7186. [Google Scholar] [CrossRef]
- Shivji, M.K.K.; Wood, R.D.; Podust, V.N.; Hübscher, U. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry 1995, 34, 5011–5017. [Google Scholar] [CrossRef]
- Moser, J.; Kool, H.; Giakzidis, I.; Caldecott, K.; Mullenders, L.H.F.; Fousteri, M.I. Sealing of Chromosomal DNA Nicks during Nucleotide Excision Repair Requires XRCC1 and DNA Ligase IIIα in a Cell-Cycle-Specific Manner. Mol. Cell 2007, 27, 311–323. [Google Scholar] [CrossRef]
- Xu, J.; Lahiri, I.; Wang, W.; Wier, A.; Cianfrocco, M.A.; Chong, J.; Hare, A.A.; Dervan, P.B.; DiMaio, F.; Leschziner, A.E.; et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA Repair. Nature 2017, 551, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Sasaki, K.; Mitsutake, N.; Matsuse, M.; Shimada, M.; Nardo, T.; Takahashi, Y.; Ohyama, K.; Ito, K.; Mishima, H.; et al. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat. Genet. 2012, 44, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Nakazawa, Y.; Guo, C.; Ogi, T.; Nishimura, Y. Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways. Nucleic Acids Res. 2017, 45, 13043–13055. [Google Scholar] [CrossRef]
- van der Weegen, Y.; Golan-Berman, H.; Mevissen, T.E.T.; Apelt, K.; González-Prieto, R.; Goedhart, J.; Heilbrun, E.E.; Vertegaal, A.C.O.; van den Heuvel, D.; Walter, J.C.; et al. The cooperative action of CSB, CSA, and UVSSA target TFIIH to DNA damage-stalled RNA polymerase II. Nat. Commun. 2020, 11, 2104. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Hara, Y.; Oka, Y.; Komine, O.; van den Heuvel, D.; Guo, C.; Daigaku, Y.; Isono, M.; He, Y.; Shimada, M.; et al. Ubiquitination of DNA Damage-Stalled RNAPII Promotes Transcription-Coupled Repair. Cell 2020, 180, 1228–1244.e24. [Google Scholar] [CrossRef] [PubMed]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef]
- Kunz, B.A.; Anderson, H.J.; Osmond, M.J.; Vonarx, E.J. Components of nucleotide excision repair and DNA damage tolerance in Arabidopsis thaliana. Environ. Mol. Mutagen. 2005, 45, 115–127. [Google Scholar] [CrossRef]
- Kimura, S.; Sakaguchi, K. DNA repair in plants. Chem. Rev. 2006, 106, 753–766. [Google Scholar] [CrossRef]
- Koga, A.; Ishibashi, T.; Kimura, S.; Uchiyama, Y.; Sakaguchi, K. Characterization of T-DNA insertion mutants and RNAi silenced plants of Arabidopsis thaliana UV-damaged DNA binding protein 2 (AtUV-DDB2). Plant Mol. Biol. 2006, 61, 227–240. [Google Scholar] [CrossRef]
- Molinier, J.; Lechner, E.; Dumbliauskas, E.; Genschik, P. Regulation and Role of Arabidopsis CUL4-DDB1A-DDB2 in Maintaining Genome Integrity upon UV Stress. PLoS Genet. 2008, 4, e1000093. [Google Scholar] [CrossRef]
- Al Khateeb, W.M.; Schroeder, D.F. Overexpression of Arabidopsis damaged DNA binding protein 1A (DDB1A) enhances UV tolerance. Plant Mol. Biol. 2009, 70, 371–383. [Google Scholar] [CrossRef]
- Castells, E.; Molinier, J.; Benvenuto, G.; Bourbousse, C.; Zabulon, G.; Zalc, A.; Cazzaniga, S.; Genschik, P.; Barneche, F.; Bowler, C. integrity upon UV stress. EMBO J. 2011, 30, 1162–1172. [Google Scholar] [CrossRef] [PubMed]
- Molinier, J. CENTRIN2 Modulates Homologous Recombination and Nucleotide Excision Repair in Arabidopsis. Plant Cell Online 2004, 16, 1633–1643. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.M.A.; Morgante, P.G.; Berra, C.M.; Nakabashi, M.; Bruneau, D.; Bouchez, D.; Sweder, K.S.; Van Sluys, M.A.; Menck, C.F.M. The participation of AtXPB1, the XPB/RAD25 homologue gene from Arabidopsis thaliana, in DNA repair and plant development. Plant J. 2001, 28, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Morgante, P.G.; Berra, C.M.; Nakabashi, M.; Costa, R.M.; Menck, C.F.M.; Sluys, M.A. Van Functional XPB / RAD25 redundancy in Arabidopsis genome: Characterization of AtXPB2 and expression analysis. Gene 2005, 344, 93–103. [Google Scholar] [CrossRef]
- Liu, Z. Arabidopsis UVH6, a Homolog of Human XPD and Yeast RAD3 DNA Repair Genes, Functions in DNA Repair and Is Essential for Plant Growth. Plant Physiol. 2003, 132, 1405–1414. [Google Scholar] [CrossRef]
- Liu, Z.; Hall, J.D.; Mount, D.W. Arabidopsis UVH3 gene is a homolog of the Saccharomyces cerevisiae RAD2 and human XPG DNA repair genes. Plant J. 2001, 26, 329–338. [Google Scholar] [CrossRef]
- Fidantsef, A.L.; Mitchell, D.L.; Britt, A.B. The Arabidopsis UVH1 gene is a homolog of the yeast repair endonuclease RAD1. Plant Physiol. 2000, 124, 579–586. [Google Scholar] [CrossRef]
- Gallego, F.; Fleck, O.; Li, A.; Wyrzykowska, J.; Tinland, B. AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonucleases, is involved in dark repair of UV damages and recombination. Plant J. 2000, 21, 507–518. [Google Scholar] [CrossRef]
- Hefner, E.; Preuss, S.B.; Britt, A.B. Arabidopsis mutants sensitive to γ radiation include the homologue of the human repair gene ERCC1. J. Exp. Bot. 2003, 54, 669–680. [Google Scholar] [CrossRef]
- Xu, H.; Swoboda, I.; Bhalla, P.L.; Sijbers, A.M.; Zhao, C.; Ong, E.; Hoeijmakers, J.H.J.; Singh, M.B. Plant homologue of human excision repair gene ERCC1 points to conservation of DNA repair mechanisms. Plant J. 1998, 13, 823–829. [Google Scholar] [CrossRef]
- Fidantsef, A.L.; Britt, A.B. Preferential repair of the transcribed DNA strand in plants. Front. Plant Sci. 2012, 2, 105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Guo, H.; Zhang, J.; Guo, G.; Schumaker, K.S.; Guo, Y. Arabidopsis cockayne syndrome A-like proteins 1A and 1B form a complex with CULLIN4 and damage DNA binding protein 1A and regulate the response to UV irradiation. Plant Cell 2010, 22, 2353–2369. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, S.; Hellmann, H.; Genetik, A.; Universita, F. The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B tolerance and genomic integrity in Arabidopsis thaliana. Plant J. 2010, 62, 404–415. [Google Scholar] [CrossRef]
- Canturk, F.; Karaman, M.; Selby, C.P.; Kemp, M.G.; Kulaksiz-Erkmen, G.; Hu, J.; Li, W.; Lindsey-Boltz, L.A.; Sancar, A. Nucleotide excision repair by dual incisions in plants. Proc. Natl. Acad. Sci. USA 2016, 113, 4706–4710. [Google Scholar] [CrossRef]
- Oztas, O.; Selby, C.P.; Sancar, A.; Adebali, O. Genome-wide excision repair in Arabidopsis is coupled to transcription and reflects circadian gene expression patterns. Nat. Commun. 2018, 9, 1503. [Google Scholar] [CrossRef]
- Geijer, M.E.; Marteijn, J.A. What happens at the lesion does not stay at the lesion: Transcription-coupled nucleotide excision repair and the effects of DNA damage on transcription in cis and trans. DNA Repair 2018, 71, 56–68. [Google Scholar] [CrossRef]
- Al Khateeb, W.M.; Sher, A.A.; Marcus, J.M.; Schroeder, D.F. UVSSA, UBP12, and RDO2/TFIIS contribute to Arabidopsis UV tolerance. Front. Plant Sci. 2019, 10, 516. [Google Scholar] [CrossRef]
- Borsos, B.N.; Majoros, H.; Pankotai, T. Emerging Roles of Post-Translational Modifications in Nucleotide Excision Repair. Cells 2020, 9, 1466. [Google Scholar] [CrossRef]
- Dutta, A.; Babbarwal, V.; Fu, J.; Brunke-reese, D.; Libert, D.M.; Willis, J.; Reese, J.C. Ccr4-Not and TFIIS Function Cooperatively To Rescue Arrested RNA. Mol. Cell. Biol. 2015, 35, 1915–1925. [Google Scholar] [CrossRef]
- Roy, S.; Choudhury, S.R.; Singh, S.K.; Das, K.P. AtPolλ, a homolog of mammalian DNA polymerase λ in Arabidopsis thaliana, is involved in the repair of UV-B induced DNA damage through the dark repair pathway. Plant Cell Physiol 2011, 52, 448–467. [Google Scholar] [CrossRef]
- Chevigny, N.; Schatz-Daas, D.; Lotfi, F.; Gualberto, J.M. DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 2020, 21, 328. [Google Scholar] [CrossRef]
- Cooke, M.S.; Mistry, N.; Ladapo, A.; Herbert, K.E.; Lunec, J. Immunochemical quantitation of UV-induced oxidative and dimeric DNA damage to human keratinocytes. Free Radic. Res. 2000, 33, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Dalhus, B.; Laerdahl, J.K.; Backe, P.H.; Bjørås, M. DNA base repair—Recognition and initiation of catalysis. FEMS Microbiol. Rev. 2009, 33, 1044–1078. [Google Scholar] [CrossRef]
- Stivers, J.T.; Jiang, Y.L. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem. Rev. 2003, 103, 2729–2759. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.J.; Ellenberger, T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J. Biol. Chem. 2004, 279, 9750–9757. [Google Scholar] [CrossRef]
- Fortini, P.; Dogliotti, E. Base damage and single-strand break repair: Mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair 2007, 6, 398–409. [Google Scholar] [CrossRef]
- Krokan, H.E.; Bjoras, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef]
- Hegde, M.L.; Hazra, T.K.; Mitra, S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008, 18, 27–47. [Google Scholar] [CrossRef]
- Roldán-Arjona, T.; Ariza, R.R.; Córdoba-Cañero, D. DNA Base Excision Repair in Plants: An Unfolding Story with Familiar and Novel Characters. Front. Plant Sci. 2019, 10, 1055. [Google Scholar] [CrossRef]
- Frosina, G.; Fortini, P.; Rossi, O.; Carrozzino, F.; Raspaglio, G.; Cox, L.S.; Lane, D.P.; Abbondandolo, A.; Dogliotti, E. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 1996, 271, 9573–9578. [Google Scholar] [CrossRef]
- Fortini, P.; Parlanti, E.; Sidorkina, O.M.; Laval, J.; Dogliotti, E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J. Biol. Chem. 1999, 274, 15230–15236. [Google Scholar] [CrossRef]
- Srivastava, D.K.; Vande Berg, B.J.; Prasad, R.; Molina, J.T.; Beard, W.A.; Tomkinson, A.E.; Wilson, S.H. Mammalian abasic site base excision repair: Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 1998, 273, 21203–21209. [Google Scholar] [CrossRef]
- Allinson, S.L.; Dianova, I.I.; Dianov, G.L. DNA polymerase β is the major dRP lyase involved in repair of oxidative base lesions in DNA by mammalian cell extracts. EMBO J. 2001, 20, 6919–6926. [Google Scholar] [CrossRef]
- Dianov, G.; Price, A.; Lindahl, T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell. Biol. 1992, 12, 1605–1612. [Google Scholar] [CrossRef]
- Nash, R.A.; Caldecott, K.W.; Barnes, D.E.; Lindahl, T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry 1997, 36, 5207–5211. [Google Scholar] [CrossRef]
- Podlutsky, A.J.; Dianova, I.I.; Podust, V.N.; Bohr, V.A.; Dianov, G.L. Human DNA polymerase β initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO J. 2001, 20, 1477–1482. [Google Scholar] [CrossRef] [PubMed]
- Woodrick, J.; Gupta, S.; Camacho, S.; Parvathaneni, S.; Choudhury, S.; Cheema, A.; Bai, Y.; Khatkar, P.; Erkizan, H.V.; Sami, F.; et al. A new sub-pathway of long-patch base excision repair involving 5′ gap formation. EMBO J. 2017, 36, 1605–1622. [Google Scholar] [CrossRef] [PubMed]
- Gutman, B.L.; Niyogi, K.K. Evidence for base excision repair of oxidative DNA damage in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 2009, 284, 17006–17012. [Google Scholar] [CrossRef]
- Córdoba-Cañero, D.; Dubois, E.; Ariza, R.R.; Doutriaux, M.P.; Roldán-Arjona, T. Arabidopsis uracil DNA glycosylase (UNG) is required for base excision repair of uracil and increases plant sensitivity to 5-fluorouracil. J. Biol. Chem. 2010, 285, 7475–7483. [Google Scholar] [CrossRef]
- Boesch, P.; Ibrahim, N.; Paulus, F.; Cosset, A.; Tarasenko, V.; Dietrich, A. Plant mitochondria possess a short-patch base excision DNA repair pathway. Nucleic Acids Res. 2009, 37, 5690–5700. [Google Scholar] [CrossRef]
- Bensen, R.J.; Warner, H.R. The Partial Purification and Characterization of Nuclear and Mitochondrial Uracil-DNA Glycosylase Activities from Zea mays Seedlings. Plant Physiol. 1987, 83, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, T.; Matsuda, O.; Iba, K.; Terashima, I.; Sekiguchi, M.; Nakabeppu, Y. Molecular cloning of AtMMH, an Arabidopsis thaliana ortholog of the Escherichia coli mutM gene, and analysis of functional domains of its product. Mol. Gen. Genet. 1998, 259, 577–590. [Google Scholar] [CrossRef]
- Gao, M.-J.; Murphy, T.M. Alternative Forms of Formamidopyrimidine-DNA Glycosylase from Arabidopsis thaliana. Photochem. Photobiol. 2001, 73, 128–134. [Google Scholar] [CrossRef]
- García-Ortiz, M.V.; Ariza, R.R.; Roldán-Arjona, T. An OGG1 orthologue encoding a functional 8-oxoguanine DNA glycosylase/lyase in Arabidopsis thaliana. Plant Mol. Biol. 2001, 47, 795–804. [Google Scholar] [CrossRef]
- Cõrdoba-Cañero, D.; Roldán-Arjona, T.; Ariza, R.R. Arabidopsis ZDP DNA 3′-phosphatase and ARP endonuclease function in 8-oxoG repair initiated by FPG and OGG1 DNA glycosylases. Plant J. 2014, 79, 824–834. [Google Scholar] [CrossRef]
- Cõrdoba-Cañero, D.; Roldán-Arjona, T.; Ariza, R.R. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011, 68, 693–702. [Google Scholar] [CrossRef]
- Ferrando, B.; Furlanetto, A.L.D.M.; Gredilla, R.; Havelund, J.F.; Hebelstrup, K.H.; Møller, I.M.; Stevnsner, T. DNA repair in plant mitochondri—A complete base excision repair pathway in potato tuber mitochondria. Physiol. Plant. 2019, 166, 494–512. [Google Scholar] [CrossRef]
- Roldan-Arjona, T.; Garcia-Ortiz, M.V.; Ruiz-Rubio, M.; Ariza, R.R. cDNA cloning, expression and functional characterization of an Arabidopsis thaliana homologue of the Escherichia coli DNA repair enzyme endonuclease III. Plant Mol. Biol. 2000, 44, 43–52. [Google Scholar] [CrossRef]
- Furlanetto, A.L.D.M.; Cadena, S.M.S.C.; Martinez, G.R.; Ferrando, B.; Stevnsner, T.; Møller, I.M. Short-term high temperature treatment reduces viability and inhibits respiration and DNA repair enzymes in Araucaria angustifolia cells. Physiol. Plant. 2019, 166, 513–524. [Google Scholar] [CrossRef]
- Zhang, Y.; Wen, C.; Liu, S.; Zheng, L.; Shen, B.; Tao, Y. Shade avoidance 6 encodes an arabidopsis flap endonuclease required for maintenance of genome integrity and development. Nucleic Acids Res. 2015, 44, 1271–1284. [Google Scholar] [CrossRef]
- Uchiyama, Y.; Kimura, S.; Yamamoto, T.; Ishibashi, T.; Sakaguchi, K. Plant DNA polymerase λ, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur. J. Biochem. 2004, 271, 2799–2807. [Google Scholar] [CrossRef]
- Mori, Y.; Kimura, S.; Saotome, A.; Kasai, N.; Sakaguchi, N.; Uchiyama, Y.; Ishibashi, T.; Yamamoto, T.; Chiku, H.; Sakaguchi, K. Plastid DNA polymerases from higher plants, Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2005, 334, 43–50. [Google Scholar] [CrossRef]
- Elo, A.; Lyznik, A.; Gonzalez, D.O.; Kachman, S.D.; Mackenzie, S.A. Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 2003, 15, 1619–1631. [Google Scholar] [CrossRef]
- Ono, Y.; Sakai, A.; Takechi, K.; Takio, S.; Takusagawa, M.; Takano, H. NtPolI-like1 and NtPolI-like2, bacterial DNA polymerase I homologs isolated from BY-2 cultured tobacco cells, encode DNA polymerases engaged in DNA replication in both plastids and mitochondria. Plant Cell Physiol. 2007, 48, 1679–1692. [Google Scholar] [CrossRef]
- Trasviña-Arenas, C.H.; Baruch-Torres, N.; Cordoba-Andrade, F.J.; Ayala-García, V.M.; García-Medel, P.L.; Díaz-Quezada, C.; Peralta-Castro, A.; Ordaz-Ortiz, J.J.; Brieba, L.G. Identification of a unique insertion in plant organellar DNA polymerases responsible for 5′-dRP lyase and strand-displacement activities: Implications for Base Excision Repair. DNA Repair (Amst.) 2018, 65, 1–10. [Google Scholar] [CrossRef]
- Sunderland, P.A.; West, C.E.; Waterworth, W.M.; Bray, C.M. An evolutionarily conserved translation initiation mechanism regulates nuclear or mitochondrial targeting of DNA ligase 1 in Arabidopsis thaliana. Plant J. 2006, 47, 356–367. [Google Scholar] [CrossRef]
- Young, L.C.; Hays, J.B.; Tron, V.A.; Andrew, S.E. DNA mismatch repair proteins: Potential guardians against genomic instability and tumorigenesis induced by ultraviolet photoproducts. J. Investig. Dermatol. 2003, 121, 435–440. [Google Scholar] [CrossRef][Green Version]
- Tsaalbi-Shtylik, A.; Ferrás, C.; Pauw, B.; Hendriks, G.; Temviriyanukul, P.; Carlée, L.; Calléja, F.; Van Hees, S.; Akagi, J.I.; Iwai, S.; et al. Excision of translesion synthesis errors orchestrates responses to helix-distorting DNA lesions. J. Cell Biol. 2015, 209, 33–46. [Google Scholar] [CrossRef]
- Pećina-Šlaus, N.; Kafka, A.; Salamon, I.; Bukovac, A. Mismatch Repair Pathway, Genome Stability and Cancer. Front. Mol. Biosci. 2020, 7, 122. [Google Scholar] [CrossRef]
- Mazur, D.J.; Mendillo, M.L.; Kolodner, R.D. Inhibition of Msh6 ATPase Activity by Mispaired DNA Induces a Msh2(ATP)-Msh6(ATP) State Capable of Hydrolysis-Independent Movement along DNA. Mol. Cell 2006, 22, 39–49. [Google Scholar] [CrossRef]
- Gupta, S.; Gellert, M.; Yang, W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 2012, 19, 72–78. [Google Scholar] [CrossRef]
- Bradford, K.C.; Wilkins, H.; Hao, P.; Li, Z.M.; Wang, B.; Burke, D.; Wu, D.; Smith, A.E.; Spaller, L.; Du, C.; et al. Dynamic human MutSα–MutLα complexes compact mismatched DNA. Proc. Natl. Acad. Sci. USA 2020, 117, 16302–16312. [Google Scholar] [CrossRef]
- Pluciennik, A.; Dzantiev, L.; Iyer, R.R.; Constantin, N.; Kadyrov, F.A.; Modrich, P. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc. Natl. Acad. Sci. USA 2010, 107, 16066–16071. [Google Scholar] [CrossRef]
- Genschel, J.; Kadyrova, L.Y.; Iyer, R.R.; Dahal, B.K.; Kadyrov, F.A.; Modrich, P. Interaction of proliferating cell nuclear antigen with PMS2 is required for MutLaα activation and function in mismatch repair. Proc. Natl. Acad. Sci. USA 2017, 114, 4930–4935. [Google Scholar] [CrossRef]
- Genschel, J.; Bazemore, L.R.; Modrich, P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 2002, 277, 13302–13311. [Google Scholar] [CrossRef]
- Genschel, J.; Modrich, P. Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell 2003, 12, 1077–1086. [Google Scholar] [CrossRef]
- Zhang, Y.; Yuan, F.; Presnell, S.R.; Tian, K.; Gao, Y.; Tomkinson, A.E.; Gu, L.; Li, G.-M. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 2005, 122, 693–705. [Google Scholar] [CrossRef]
- Kadyrov, F.A.; Genschel, J.; Fang, Y.; Penland, E.; Edelmann, W.; Modrich, P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl. Acad. Sci. USA 2009, 106, 8495–8500. [Google Scholar] [CrossRef]
- Goellner, E.M.; Putnama, C.D.; Kolodnera, R.D. Exonuclease 1-dependent and independent mismatch repair. DNA Repair (Amst.) 2015, 32, 24–32. [Google Scholar] [CrossRef]
- Culligan, K.M.; Hays, J.B. Arabidopsis MutS homologs—AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7—Form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell 2000, 12, 991–1002. [Google Scholar] [CrossRef]
- Wu, S.Y.; Culligan, K.; Lamers, M.; Hays, J. Dissimilar mispair-recognition spectra of Arabidopsis DNA-mismatch-repair proteins MSH2·MSH6 (MutSα) and MSH2·MSH7 (MutSγ). Nucleic Acids Res. 2003, 31, 6027–6034. [Google Scholar] [CrossRef]
- Lario, L.D.; Ramirez-Parra, E.; Gutierrez, C.; Casati, P.; Spampinato, C.P. Regulation of plant MSH2 and MSH6 genes in the UV-B-induced DNA damage response. J. Exp. Bot. 2011, 62, 2925–2937. [Google Scholar] [CrossRef]
- Lario, L.D.; Botta, P.; Casati, P.; Spampinato, C.P. Role of AtMSH7 in UV-B-induced DNA damage recognition and recombination. J. Exp. Bot. 2015, 66, 3019–3026. [Google Scholar] [CrossRef]
- Xu, Y.Z.; Arrieta-Montiel, M.P.; Virdi, K.S.; de Paula, W.B.M.; Widhalm, J.R.; Basset, G.J.; Davila, J.I.; Elthon, T.E.; Elowsky, C.G.; Sato, S.J.; et al. Muts homolog1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 2011, 23, 3428–3441. [Google Scholar] [CrossRef]
- Wu, Z.; Waneka, G.; Broz, A.K.; King, C.R.; Sloan, D.B. MSH1 is required for maintenance of the low mutation rates in plant mitochondrial and plastid genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 16448–16455. [Google Scholar] [CrossRef]
- Khakhlova, O.; Bock, R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006, 46, 85–94. [Google Scholar] [CrossRef]
- Caldecott, K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008, 9, 619–631. [Google Scholar] [CrossRef]
- Kuzminov, A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. USA 2001, 98, 8241–8246. [Google Scholar] [CrossRef]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef]
- Puchta, H. The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 2005, 56, 1–14. [Google Scholar] [CrossRef]
- Puchta, H.; Fauser, F. Synthetic nucleases for genome engineering in plants: Prospects for a bright future. Plant J. 2014, 78, 727–741. [Google Scholar] [CrossRef]
- Wang, H.; Xu, X. Microhomology-mediated end joining: New players join the team. Cell Biosci. 2017, 7, 6. [Google Scholar] [CrossRef]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Marechal, A.; Zou, L. DNA Damage Sensing by the ATM and ATR Kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef]
- Mah, L.J.; El-Osta, A.; Karagiannis, T.C. γh2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef]
- Fillingham, J.; Keogh, M.C.; Krogan, N.J. γH2AX and its role in DNA double-strand break repair. Biochem. Cell Biol. 2006, 84, 568–577. [Google Scholar] [CrossRef]
- Sancar, A.; Lindsey-Boltz, L.A.; Ünsal-Kaçmaz, K.; Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef]
- Ruff, S.E.; Logan, S.K.; Garabedian, M.J.; Huang, T.T. Roles for MDC1 in cancer development and treatment. DNA Repair (Amst.) 2020, 95, 102948. [Google Scholar] [CrossRef]
- Konecny, G.E.; Kristeleit, R.S. PARP inhibitors for BRCA1/2-mutated and sporadic ovarian cancer: Current practice and future directions. Br. J. Cancer 2016, 115, 1157–1173. [Google Scholar] [CrossRef]
- Garcia, V.; Bruchet, H.; Camescasse, D.; Granier, F.; Bouchez, D.; Tissier, A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 2003, 15, 119–132. [Google Scholar] [CrossRef]
- Culligan, K.; Tissier, A.; Britt, A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 2004, 16, 1091–1104. [Google Scholar] [CrossRef]
- Jia, Q.; Dulk-Ras, A.D.; Shen, H.; Hooykaas, P.J.J.; de Pater, S. Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana. Plant Mol. Biol. 2013, 82, 339–351. [Google Scholar] [CrossRef]
- Binz, S.K.; Sheehan, A.M.; Wold, M.S. Replication Protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst.) 2004, 3, 1015–1024. [Google Scholar] [CrossRef]
- Mladenov, E.; Iliakis, G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2011, 711, 61–72. [Google Scholar] [CrossRef]
- Davis, A.J.; Chen, D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013, 2, 130–143. [Google Scholar] [CrossRef]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef]
- Sfeir, A.; Symington, L.S. Microhomology-mediated end joining: A back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 2015, 40, 701–714. [Google Scholar] [CrossRef]
- Kelley, M.R.; Fishel, M.L. DNA Repair in Cancer Therapy: Molecular Targets and Clinical Applications, 2nd ed.; Academic Press Elsevier: Amsterdam, The Netherlands, 2016; ISBN 9780128035825. [Google Scholar]
- Kusumoto, R.; Dawut, L.; Marchetti, C.; Wan Lee, J.; Vindigni, A.; Ramsden, D.; Bohr, V.A. Werner Protein Cooperates with the XRCC4-DNA Ligase IV Complex in End-Processing. Biochemistry 2008, 47, 7548–7556. [Google Scholar] [CrossRef]
- Shamanna, R.A.; Lu, H.; De Freitas, J.K.; Tian, J.; Croteau, D.L.; Bohr, V.A. WRN regulates pathway choice between classical and alternative non-homologous end joining. Nat. Commun. 2016, 7, 13785. [Google Scholar] [CrossRef]
- Wang, Q.; Goldstein, M.; Alexander, P.; Wakeman, T.P.; Sun, T.; Feng, J.; Lou, Z.; Kastan, M.B.; Wang, X.F. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 2014, 33, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Nimonkar, A.V.; Genschel, J.; Kinoshita, E.; Polaczek, P.; Campbell, J.L.; Wyman, C.; Modrich, P.; Kowalczykowski, S.C. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011, 25, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Trenner, A.; Sartori, A.A. Harnessing DNA Double-Strand Break Repair for Cancer Treatment. Front. Oncol. 2019, 9, 1388. [Google Scholar] [CrossRef] [PubMed]
- Heyer, W.-D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Steinert, J.; Schiml, S.; Puchta, H. Homology-based double-strand break-induced genome engineering in plants. Plant Cell Rep. 2016, 35, 1429–1438. [Google Scholar] [CrossRef]
- Aleksandrov, R.; Hristova, R.; Stoynov, S.; Gospodinov, A. The Chromatin Response to Double-Strand DNA Breaks and Their Repair. Cells 2020, 9, 1853. [Google Scholar] [CrossRef]
- Sun, Y.; McCorvie, T.J.; Yates, L.A.; Zhang, X. Structural basis of homologous recombination. Cell. Mol. Life Sci. 2020, 77, 3–18. [Google Scholar] [CrossRef]
- Osakabe, K.; Osakabe, Y.; Toki, S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 12034–12039. [Google Scholar] [CrossRef]
- García-Medel, P.L.; Baruch-Torres, N.; Peralta-Castro, A.; Trasviña-Arenas, C.H.; Torres-Larios, A.; Brieba, L.G. Plant organellar DNA polymerases repair double-stranded breaks by microhomology-mediated end-joining. Nucleic Acids Res. 2019, 47, 3028–3044. [Google Scholar] [CrossRef]
- Ishibashi, T.; Koga, A.; Yamamoto, T.; Uchiyama, Y.; Mori, Y.; Hashimoto, J.; Kimura, S.; Sakaguchi, K. Two types of replication protein A in seed plants: Characterization of their functions in vitro and in vivo. FEBS J. 2005, 272, 3270–3281. [Google Scholar] [CrossRef]
- Takashi, Y.; Kobayashi, Y.; Tanaka, K.; Tamura, K. Arabidopsis replication protein A 70a is required for DNA damage response and telomere length homeostasis. Plant Cell Physiol. 2009, 50, 1965–1976. [Google Scholar] [CrossRef]
- Tamura, K.; Adachi, Y.; Chiba, K.; Oguchi, K.; Takahashi, H. Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: Evidence for a role in the repair of DNA double-strand breaks. Plant J. 2002, 29, 771–781. [Google Scholar] [CrossRef] [PubMed]
- West, C.E.; Waterworth, W.M.; Jiang, Q.; Bray, C.M. Arabidopsis DNA ligase IV is induced by γ-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 2000, 24, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef] [PubMed]
- West, C.E.; Waterworth, W.M.; Story, G.W.; Sunderland, P.A.; Jiang, Q.; Bray, C.M. Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 2002, 31, 517–528. [Google Scholar] [CrossRef] [PubMed]
- van Attikum, H.; Bundock, P.; Overmeer, R.M.; Lee, L.Y.; Gelvin, S.B.; Hooykaas, P.J.J. The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 2003, 31, 4247–4255. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Angelis, K.J.; Britt, A.B. Arabidopsis DNA polymerase lambda mutant is mildly sensitive to DNA double strand breaks but defective in integration of a transgene. Front. Plant Sci. 2015, 6, 357. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Kozak, J.; Provost, C.M.; Bray, C.M.; Angelis, K.J.; West, C.E. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009, 9, 79. [Google Scholar] [CrossRef]
- Li, B.; Conway, N.; Navarro, S.; Comai, L.; Comai, L. A conserved and species-specific functional interaction between the Werner syndrome-like exonuclease at WEX and the Ku heterodimer in Arabidopsis. Nucleic Acids Res. 2005, 33, 6861–6867. [Google Scholar] [CrossRef]
- Charbonnel, C.; Gallego, M.E.; White, C.I. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 2010, 64, 280–290. [Google Scholar] [CrossRef]
- Siebert, R.; Puchta, H. Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 2002, 14, 1121–1131. [Google Scholar] [CrossRef]
- Puchta, H. Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J. 1998, 13, 331–339. [Google Scholar] [CrossRef]
- Shultz, R.W.; Tatineni, V.M.; Hanley-Bowdoin, L.; Thompson, W.F. Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 2007, 144, 1697–1714. [Google Scholar] [CrossRef]
- Daoudal-Cotterell, S.; Gallego, M.E.; White, C.I.Ã. The plant Rad50-Mre11 protein complex. FEBS Lett. 2002, 516, 164–166. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Altun, C.; Armstrong, S.J.; Roberts, N.; Dean, P.J.; Young, K.; Weil, C.F.; Bray, C.M.; West, C.E. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 2007, 52, 41–52. [Google Scholar] [CrossRef]
- Amiard, S.; Charbonnel, C.; Allain, E.; Depeiges, A.; White, C.I.; Gallego, M.E. Distinct Roles of the ATR Kinase and the Mre11-Rad50-Nbs1 Complex in the Maintenance of Chromosomal Stability in Arabidopsis. Plant Cell 2010, 22, 3020–3033. [Google Scholar] [CrossRef]
- Samach, A.; Melamed-Bessudo, C.; Avivi-Ragolski, N.; Pietrokovski, S.; Levy, A.A. Identification of Plant RAD52 Homologs and Characterization of the Arabidopsis thaliana RAD52 -Like Genes. Plant Cell 2011, 23, 4266–4279. [Google Scholar] [CrossRef]
- Seeliger, K.; Dukowic-Schulze, S.; Wurz-Wildersinn, R.; Pacher, M.; Puchta, H. BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol. 2012, 193, 364–375. [Google Scholar] [CrossRef]
- Osakabe, K.; Abe, K.; Yoshioka, T.; Osakabe, Y.; Todoriki, S.; Ichikawa, H.; Hohn, B.; Toki, S. Isolation and characterization of the RAD54 gene from Arabidopsis thaliana. Plant J. 2006, 48, 827–842. [Google Scholar] [CrossRef]
- Dubest, S.; Gallego, M.E.; White, C.I. Roles of the AtErcc1 protein in recombination. Plant J. 2004, 39, 334–342. [Google Scholar] [CrossRef]
- Jia, N.; Liu, X.; Gao, H. A DNA2 Homolog Is Required for DNA Damage Repair, Cell Cycle Regulation, and Meristem Maintenance in Plants. Plant Physiol. 2016, 171, 318–333. [Google Scholar] [CrossRef]
- Hartung, F.; Suer, S.; Puchta, H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 18836–18841. [Google Scholar] [CrossRef]
- Serra, H.; da Ines, O.; Degroote, F.; Gallego, M.; White, C. RAD51-Independent SSA Recombination. PLoS Genet. 2013, 9, e1003971. [Google Scholar] [CrossRef]
- Abe, K.; Osakabe, K.; Nakayama, S.; Endo, M.; Tagiri, A.; Todoriki, S.; Ichikawa, H.; Toki, S. Arabidopsis RAD51C Gene Is Important for Homologous Recombination in Meiosis and Mitosis. Plant Physiol. 2005, 139, 896–908. [Google Scholar] [CrossRef]
- Johnson, R.A.; Hellens, R.P.; Love, D.R. A transient assay for recombination demonstrates that Arabidopsis SNM1 and XRCC3 enhance non-homologous recombination. Genet. Mol. Res. 2011, 10, 2104–2132. [Google Scholar] [CrossRef]
- Reidt, W.; Wurz, R.; Wanieck, K.; Chu, H.H.; Puchta, H. A homologue of the breast cancer-associated gene BARD1 is involved in DNA repair in plants. EMBO J. 2006, 25, 4326–4337. [Google Scholar] [CrossRef]
- Heitzeberg, F.; Chen, I.-P.; Hartung, F.; Orel, N.; Angelis, K.J.; Puchta, H. The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J. 2004, 38, 954–968. [Google Scholar] [CrossRef]
- Tsai, F.-L.; Kai, M. The checkpoint clamp protein Rad9 facilitates DNA-end resection and prevents alternative non-homologous end joining. Cell Cycle 2014, 13, 3460–3464. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Block-Schmidt, A.S.; Dukowic-Schulze, S.; Wanieck, K.; Reidt, W.; Puchta, H. BRCC36A is epistatic to BRCA1 in DNA crosslink repair and homologous recombination in Arabidopsis thaliana. Nucleic Acids Res. 2011, 39, 146–154. [Google Scholar] [CrossRef]
- Weimer, A.K.; Biedermann, S.; Harashima, H.; Roodbarkelari, F.; Foreman, J.; Guan, Y.; Pochon, G.; Heese, M.; Damme, D. Van The plant-specific CDKB 1 -CYCB 1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016, 35, 2068–2086. [Google Scholar] [CrossRef]
- Miller-Messmer, M.; Kuhn, K.; Bichara, M.; le Ret, M.; Imbault, P.; Gualberto, J.M. RecA-dependent DNA repair results in increased heteroplasmy of the arabidopsis mitochondrial genome. Plant Physiol. 2012, 159, 211–226. [Google Scholar] [CrossRef]
- Maréchal, A.; Brisson, N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010, 186, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Huq, E.; Herrin, D.L. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 13954–13959. [Google Scholar] [CrossRef] [PubMed]
- Le Ret, M.; Belcher, S.; Graindorge, S.; Wallet, C.; Koechler, S.; Erhardt, M.; Williams-Carrier, R.; Barkan, A.; Gualberto, J.M. Efficient replication of the plastid genome requires an organellar thymidine kinase. Plant Physiol. 2018, 178, 1643–1656. [Google Scholar] [CrossRef]
- Pedroza-Garcia, J.-A.; De Veylder, L.; Raynaud, C. Plant DNA Polymerases. Int. J. Mol. Sci. 2019, 20, 4814. [Google Scholar] [CrossRef]
- Tourrière, H.; Pasero, P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst.) 2007, 6, 900–913. [Google Scholar] [CrossRef]
- Mirkin, E.V.; Mirkin, S.M. Replication Fork Stalling at Natural Impediments. Microbiol. Mol. Biol. Rev. 2007, 71, 13–35. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Stelter, P.; Ulrich, H.D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 2003, 425, 188–191. [Google Scholar] [CrossRef]
- Giannattasio, M.; Zwicky, K.; Follonier, C.; Foiani, M.; Lopes, M.; Branzei, D. Visualization of recombination-mediated damage bypass by template switching. Nat. Struct. Mol. Biol. 2014, 21, 884–892. [Google Scholar] [CrossRef]
- Vaisman, A.; Woodgate, R. Translesion DNA polymerases in eukaryotes: What makes them tick? Crit. Rev. Biochem. Mol. Biol. 2017, 52, 274–303. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, A.N. Translesion Synthesis in Plants: Ultraviolet Resistance and Beyond. Front. Plant Sci. 2019, 10, 1208. [Google Scholar] [CrossRef]
- Kannouche, P.L.; Wing, J.; Lehmann, A.R. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 2004, 14, 491–500. [Google Scholar] [CrossRef]
- Brown, S.; Niimi, A.; Lehmann, A.R. Ubiquitination and deubiquitination of PCNA in response to stalling of the replication fork. Cell Cycle 2009, 8, 689–692. [Google Scholar] [CrossRef]
- Hendel, A.; Krijger, P.H.L.; Diamant, N.; Goren, Z.; Langerak, P.; Kim, J.; Reißner, T.; Lee, K.; Geacintov, N.E.; Carell, T.; et al. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011, 7, e1002262. [Google Scholar] [CrossRef]
- Pozo, F.M.; Oda, T.; Sekimoto, T.; Murakumo, Y.; Masutani, C.; Hanaoka, F.; Yamashita, T. Molecular Chaperone Hsp90 Regulates REV1-Mediated Mutagenesis. Mol. Cell. Biol. 2011, 31, 3396–3409. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, A.; Thuong Lan, V.T.; Hase, Y.; Shikazono, N.; Matsunaga, T.; Tanaka, A. Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and γ-rays in Arabidopsis: Implication of the presence of a translesion synthesis mechanism in plants. Plant Cell 2003, 15, 2042–2057. [Google Scholar] [CrossRef]
- Ioki, M.; Takahashi, S.; Nakajima, N.; Saji, H.; Fujikura, K.; Tamaoki, M.; Aono, M.; Kanna, M.; Ogawa, D.; Watanabe, M.; et al. Wavelength dependency of the light-driven transcriptional activation of the cucumber CPD photolyase gene. Phyt. Ann. Rei Bot. 2005, 45, 177–184. [Google Scholar]
- Anderson, H.J.; Vonarx, E.J.; Pastushok, L.; Nakagawa, M.; Katafuchi, A.; Gruz, P.; Di Rubbo, A.; Grice, D.M.; Osmond, M.J.; Sakamoto, A.N.; et al. Arabidopsis thaliana Y-family DNA polymerase η catalyses translesion synthesis and interacts functionally with PCNA2. Plant J. 2008, 55, 895–908. [Google Scholar] [CrossRef]
- Amoroso, A.; Concia, L.; Maggio, C.; Raynaud, C.; Bergounioux, C.; Crespan, E.; Cella, R.; Maga, G. Oxidative DNA Damage Bypass in Arabidopsis thaliana Requires DNA Polymerase λ and Proliferating Cell Nuclear Antigen 2. Plant Cell 2011, 23, 806–822. [Google Scholar] [CrossRef]
- García-Ortiz, M.V.; Ariza, R.R.; Hoffman, P.D.; Hays, J.B.; Roldán-Arjona, T. Arabidopsis thaliana AtPOLK encodes a DinB-like DNA polymerase that extends mispaired primer termini and is highly expressed in a variety of tissues. Plant J. 2004, 39, 84–97. [Google Scholar] [CrossRef]
- Hoffman, P.D.; Curtis, M.J.; Iwai, S.; Hays, J.B. Biochemical evolution of DNA polymerase η: Properties of plant, human, and yeast proteins. Biochemistry 2008, 47, 4583–4596. [Google Scholar] [CrossRef]
- Takahashi, S.; Sakamoto, A.N.; Tanaka, A.; Shimizu, K. AtREV1, a Y-family DNA polymerase in Arabidopsis, has deoxynucleotidyl transferase activity in vitro. Plant Physiol. 2007, 145, 1052–1060. [Google Scholar] [CrossRef][Green Version]
- Nakagawa, M.; Takahashi, S.; Tanaka, A.; Narumi, I.; Sakamoto, A.N. Role of AtPolζ, AtRev1, and AtPolη in UV light-induced mutagenesis in Arabidopsis. Plant Physiol. 2011, 155, 414–420. [Google Scholar] [CrossRef]
- Sakamoto, A.N.; Kaya, H.; Endo, M. Deletion of TLS polymerases promotes homologous recombination in Arabidopsis. Plant Signal. Behav. 2018, 13, e1483673. [Google Scholar] [CrossRef]
- Wang, S.; Wen, R.; Shi, X.; Lambrecht, A.; Wang, H.; Xiao, W. RAD5a and REV3 function in two alternative pathways of DNA-damage tolerance in Arabidopsis. DNA Repair (Amst.) 2011, 10, 620–628. [Google Scholar] [CrossRef]
- Gulbis, J.M.; Kelman, Z.; Hurwitz, J.; O’Donnell, M.; Kuriyan, J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 1996, 87, 297–306. [Google Scholar] [CrossRef]
- Strzalka, W.; Oyama, T.; Tori, K.; Morikawa, K. Crystal structures of the Arabidopsis thaliana proliferating cell nuclear antigen 1 and 2 proteins complexed with the human p21 C-terminal segment. Protein Sci. 2009, 18, 1072–1080. [Google Scholar] [CrossRef]
- Strzalka, W.; Ziemienowicz, A. Molecular cloning of Phaseolus vulgaris cDNA encoding proliferating cell nuclear antigen. J. Plant Physiol. 2007, 164, 209–213. [Google Scholar] [CrossRef]
- Strzalka, W.; Kaczmarek, A.; Naganowska, B.; Ziemienowicz, A. Identification and functional analysis of PCNA1 and PCNA-like1 genes of Phaseolus coccineus. J. Exp. Bot. 2010, 61, 873–888. [Google Scholar] [CrossRef]
- Strzalka, W.; Bartnicki, F.; Pels, K.; Jakubowska, A.; Tsurimoto, T.; Tanaka, K. RAD5a ubiquitin ligase is involved in ubiquitination of Arabidopsis thaliana proliferating cell nuclear antigen. J. Exp. Bot. 2013, 64, 859–869. [Google Scholar] [CrossRef]
- Janku, M.; Luhová, L.; Petrivalský, M. On the origin and fate of reactive oxygen species in plant cell compartments. Antioxidants 2019, 8, 105. [Google Scholar] [CrossRef]
- Sarangi, P.; Zhao, X. SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem. Sci. 2015, 40, 233–242. [Google Scholar] [CrossRef]
- Garvin, A.J.; Morris, J.R. SUMO, a small, but powerful, regulator of double-strand break repair. Phil. Trans. R. Soc. B 2017, 372, 20160281. [Google Scholar] [CrossRef]
- Leung, W.; Baxley, R.M.; Moldovan, G.L.; Bielinsky, A.K. Mechanisms of DNA damage tolerance: Post-translational regulation of PCNA. Genes (Basel) 2019, 10, 10. [Google Scholar] [CrossRef]
- Mengiste, T.; Revenkova, E.; Bechtold, N.; Paszkowski, J. An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 1999, 18, 4505–4512. [Google Scholar] [CrossRef]
- Diaz, M.; Pecinka, A. Scaffolding for repair: Understanding molecular functions of the SMC5/6 complex. Genes 2018, 9, 36. [Google Scholar] [CrossRef]
- Yuan, D.; Lai, J.; Xu, P.; Zhang, S.; Zhang, J.; Li, C.; Wang, Y.; Du, J.; Liu, Y.; Yang, C. AtMMS21 regulates DNA damage response and homologous recombination repair in Arabidopsis. DNA Repair (Amst.) 2014, 21, 140–147. [Google Scholar] [CrossRef]
- Rytz, T.C.; Miller, M.J.; McLoughlin, F.; Augustine, R.C.; Marshall, R.S.; Juan, Y.T.; Charng, Y.Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. Plant Cell 2018, 30, 1077–1099. [Google Scholar] [CrossRef]
- Elrouby, N.; Coupland, G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA 2010, 107, 17415–17420. [Google Scholar] [CrossRef]
- Miller, M.J.; Barrett-Wilt, G.A.; Hua, Z.; Vierstra, R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 16512–16517. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.J.; Vierstra, R.D. Mass spectrometric identification of SUMO substrates provides insights into heat stress-induced SUMOylation in plants. Plant Signal. Behav. 2011, 6, 130–133. [Google Scholar] [CrossRef]
- Strzalka, W.; Labecki, P.; Bartnicki, F.; Aggarwal, C.; Rapala-Kozik, M.; Tani, C.; Tanaka, K.; Gabrys, H. Arabidopsis thaliana proliferating cell nuclear antigen has several potential sumoylation sites. J. Exp. Bot. 2012, 63, 2971–2983. [Google Scholar] [CrossRef] [PubMed]
- Elrouby, N. Analysis of small ubiquitin-like modifier (SUMO) targets reflects the essential nature of protein SUMOylation and provides insight to elucidate the role of SUMO in plant development. Plant Physiol. 2015, 169, 1006–1017. [Google Scholar] [CrossRef]
- Arroyo-Mateos, M.; Sabarit, B.; Maio, F.; Sánchez-Durán, M.A.; Rosas-Díaz, T.; Prins, M.; Ruiz-Albert, J.; Luna, A.P.; van den Burg, H.A.; Bejarano, E.R. Geminivirus Replication Protein Impairs SUMO Conjugation of Proliferating Cellular Nuclear Antigen at Two Acceptor Sites. J. Virol. 2018, 92, e00611-18. [Google Scholar] [CrossRef]
- Pfander, B.; Moldovan, G.L.; Sacher, M.; Hoege, C.; Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005, 436, 428–433. [Google Scholar] [CrossRef]
- Halas, A.; Podlaska, A.; Derkacz, J.; Mcintyre, J.; Skoneczna, A.; Sledziewska-Gojska, E. The roles of PCNA SUMOylation, Mms2-Ubc13 and Rad5 in translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 2011, 80, 786–797. [Google Scholar] [CrossRef]
- Mohiuddin, M.; Evans, T.J.; Rahman, M.M.; Keka, I.S.; Tsuda, M.; Sasanuma, H.; Takeda, S. SUMOylation of PCNA by PIAS1 and PIAS4 promotes template switch in the chicken and human B cell lines. Proc. Natl. Acad. Sci. USA 2018, 115, 12793–12798. [Google Scholar] [CrossRef]
- Liang, L.; Flury, S.; Kalck, V.; Hohn, B.; Molinier, J. CENTRIN2 interacts with the arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions. Plant Mol. Biol. 2006, 61, 345–356. [Google Scholar] [CrossRef]
- Wang, Q.E.; Zhu, Q.; Wani, G.; El-Mahdy, M.A.; Li, J.; Wani, A.A. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005, 33, 4023–4034. [Google Scholar] [CrossRef]
- Emmert, S.; Kobayashi, N.; Khan, S.G.; Kraemer, K.H. The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6-4 photoproducts. Proc. Natl. Acad. Sci. USA 2000, 97, 2151–2156. [Google Scholar] [CrossRef]
- Lahari, T.; Lazaro, J.; Schroeder, D.F. RAD4 and RAD23/HMR contribute to arabidopsis UV tolerance. Genes 2018, 9, 8. [Google Scholar] [CrossRef]
- Han, C.; Zhao, R.; Kroger, J.; He, J.; Wani, G.; Wang, Q.E.; Wani, A.A. UV radiation-induced SUMOylation of DDB2 regulates nucleotide excision repair. Carcinogenesis 2017, 38, 976–985. [Google Scholar] [CrossRef] [PubMed]
| Protein | Type of DNA Repair Pathway in Which the Arabidopsis Proteins or Their Human Homologs Is Involved |
|---|---|
| AtPCNA1 [255,258]/AtPCNA2 [258] | NER, BER, MMR, HR |
| AtKU80 [257] | NHEJ |
| AtRAD4 [254] | NER |
| AtXRCC1 [254] | NER, BER, NHEJ |
| AtLIG1 [254] | NER, BER, MMR, NHEJ |
| AtPOL D3 [254] | NER, BER, MMR, HR |
| AtRCF1 [259] | NER, BER, MMR, HR |
| AtCUL4 [255] | NER |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strzałka, W.; Zgłobicki, P.; Kowalska, E.; Bażant, A.; Dziga, D.; Banaś, A.K. The Dark Side of UV-Induced DNA Lesion Repair. Genes 2020, 11, 1450. https://doi.org/10.3390/genes11121450
Strzałka W, Zgłobicki P, Kowalska E, Bażant A, Dziga D, Banaś AK. The Dark Side of UV-Induced DNA Lesion Repair. Genes. 2020; 11(12):1450. https://doi.org/10.3390/genes11121450
Chicago/Turabian StyleStrzałka, Wojciech, Piotr Zgłobicki, Ewa Kowalska, Aneta Bażant, Dariusz Dziga, and Agnieszka Katarzyna Banaś. 2020. "The Dark Side of UV-Induced DNA Lesion Repair" Genes 11, no. 12: 1450. https://doi.org/10.3390/genes11121450
APA StyleStrzałka, W., Zgłobicki, P., Kowalska, E., Bażant, A., Dziga, D., & Banaś, A. K. (2020). The Dark Side of UV-Induced DNA Lesion Repair. Genes, 11(12), 1450. https://doi.org/10.3390/genes11121450





