Comparative Genomics and Functional Studies of Wheat BED-NLR Loci
Abstract
1. Introduction
2. Materials and Methods
2.1. Yr7 and Yr5 Alleles Identification in the Multiple Wheat Cultivar Genomes
2.2. Definition of Syntenic Regions across Wheat Cultivar Genomes
2.2.1. Definition of the Yr Region Gene Content in the Multiple Wheat Cultivar Genomes
2.2.2. Definition of the NLR Content of the Syntenic Regions in the Wheat Genomes
2.2.3. Annotation of nlr_11 in Cadenza
2.3. Identification of BED-NLRs and BED-proteins in Plant Genomes
2.4. Neighbor-net Analyses
2.5. Identification of Nuclear Localisation Signal (NLS) in BED-containing Proteins and Yr7 Cellular Localisation Experiments
2.5.1. Generation of the Constructs for Yr7 Cellular Localisation Experiments in N. Benthamiana
2.5.2. Generation of the Constructs for Yr7 Hyper-sensitive Response Assays in N. Benthamiana
2.5.3. Transient Assays in N. Benthamiana
3. Results
3.1. Identification of NLRs Across the Yr7/Yr5 Locus in the Multiple Wheat Cultivar Genomes
| Genome | Identity to Album-Yr5a on 2B (%) | Higher Identity to Album-Yr5a on Other Chromosomes (%) | Identity to Cadenza-Yr7 on 2B (%) | Higher Identity to Cadenza-Yr7 on Other Chromosomes (%) |
|---|---|---|---|---|
| Arina | - | 99.31 (2D) 1 | 86.5 | - |
| SY-Mattis | - | 99.31 (2D) 1 | - | - |
| Julius | 95.1 | 99.75 (2D) 2 | - | - |
| Jagger | - | 99.75 (2D) 2 | 86.5 | - |
| Lancer | 95.1 | - | - | - |
| Stanley | - | - | 99.98 | - |
| Mace | - | - | 99.98 | - |
| Landmark | - | - | 99.98 | - |
| Chinese Spring | - | - | 86.5 | - |
| Norin61 | - | - | 86.5 | - |
| Spelt PI190962 | 100 | - | - | - |
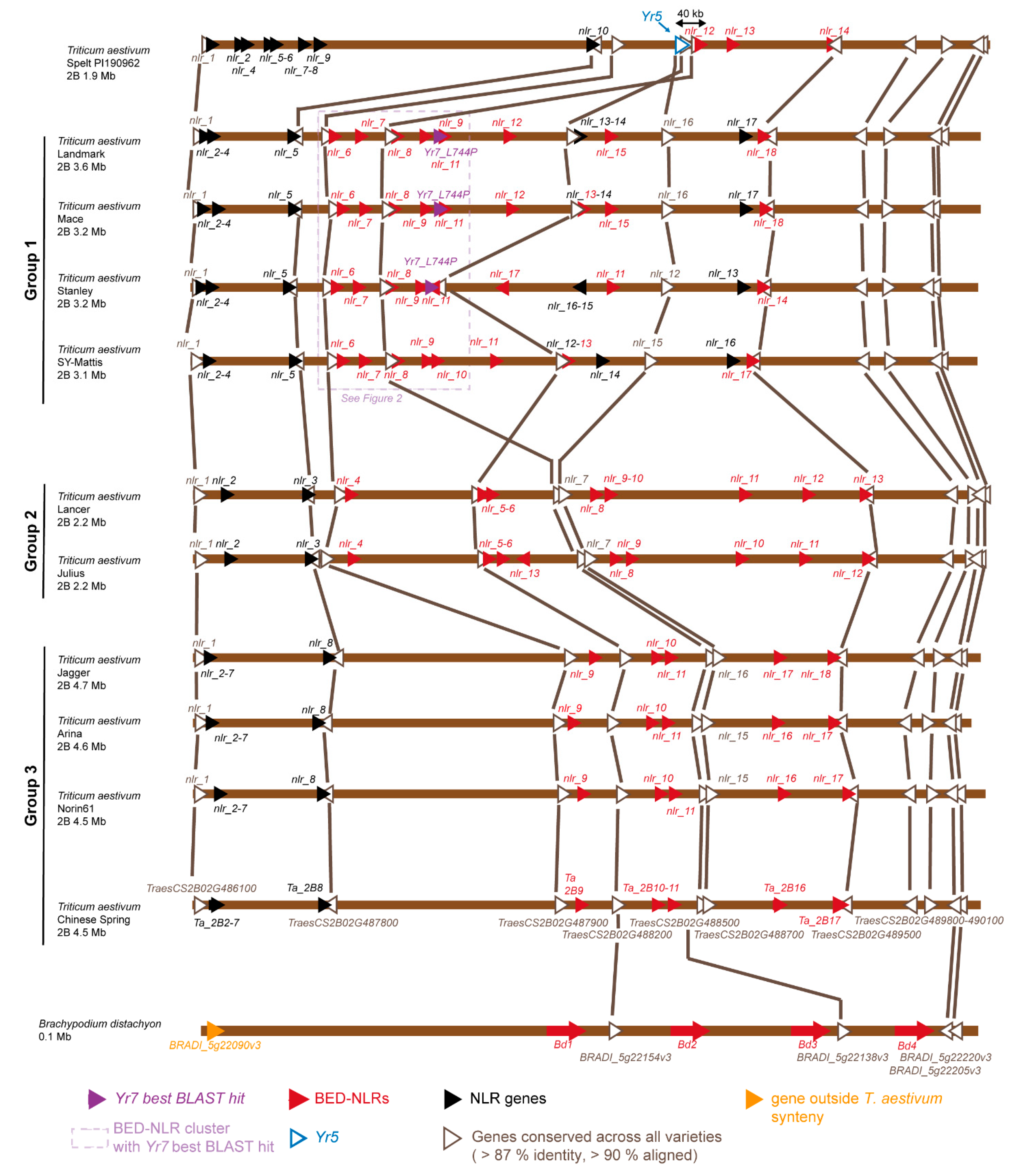
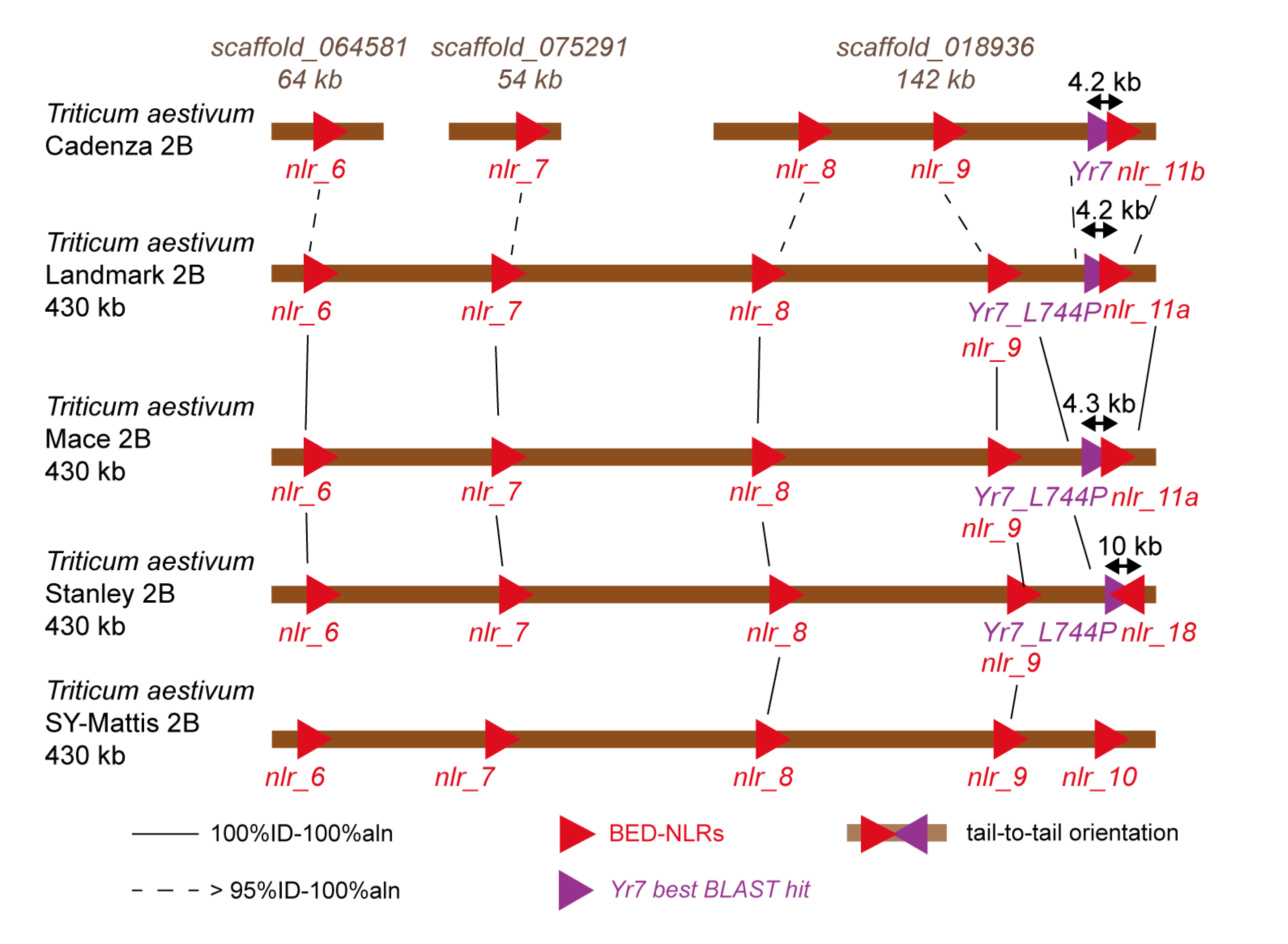
3.2. The Yr Region is Conserved across the Multiple Wheat Cultivar Genomes
| Genome | Region Size (Mb) | # Annotated Genes | # NLR Loci | # NLR Loci Overlapping with Gene Models | # BED-NLRs | # BED-NLRs Overlapping with Gene Models | |
|---|---|---|---|---|---|---|---|
| Spelt PI190962 | 1.9 | - | 13 | - | 4 | - | |
| Group 1 | Landmark | 3.6 | 40 | 18 | 7 | 9 | 2 |
| Mace | 3.2 | 39 | 18 | 5 | 10 | 2 | |
| Stanley | 3.2 | 39 | 18 | 5 | 9 | 1 | |
| SY-Mattis | 3.1 | 41 | 15 | 5 | 8 | 1 | |
| Group 2 | Lancer | 2.2 | 32 | 13 | 4 | 9 | 2 |
| Julius | 2.2 | 34 | 13 | 5 | 9 | 2 | |
| Group 3 | Jagger | 4.7 | 54 | 14 | 9 | 6 | 3 |
| Arina | 4.6 | 54 | 14 | 8 | 6 | 3 | |
| Norin61 | 4.5 | 46 | 17 | 8 | 7 | 3 | |
| Chinese-Spring | 4.5 | 42 | 14 | 10 | 6 | 3 | |
| Related grasses | Ae. Tauschii * | 3.8 | 39 | 8 | 8 | 4 | 4 |
| Barley * | 3.7 | 29 | 2 | 2 | 0 | 0 | |
| B. distachyon * | 0.096 | 22 | 4 | 4 | 4 | 4 | |
| Rice * | 0.143 | 50 | 6 | 6 | 2 | 2 |
3.3. BED Domains from BED NLRs Share Similarities with BED Domains from Single-BED and BED-DUF-hAT Proteins
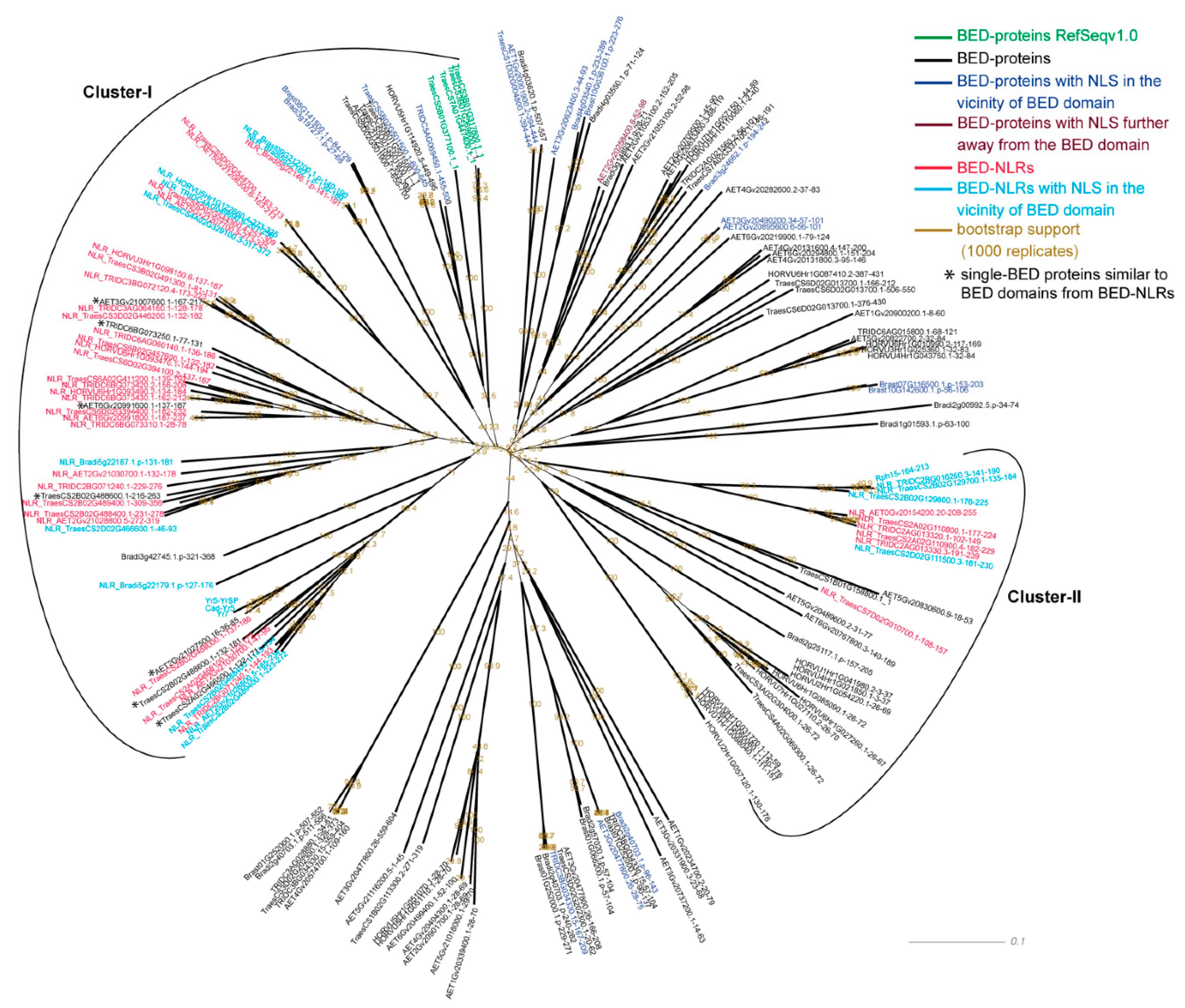
| Domain | Pfam | In Cluster with BED-NLRs | In all BED Containing Proteins | Fisher Exact Test (p-Value, Alternative Hypothesis = Greater) |
|---|---|---|---|---|
| Dimer Tnp hAT | PF05699 | 8 | 14 | 6.18 × 10−3 * |
| DUF4413 | PF14372 | 9 | 14 | 1.04 × 10−3 * |
| single-BED 1 | PF02892 | 22 | 49 | 1.56 × 10−4 * |
| All domains in BED-containing proteins (minus NLR-related domains) | - | 39 | 121 | - |
3.4. Functional BED-NLRs Carry a Nuclear Localisation Signal
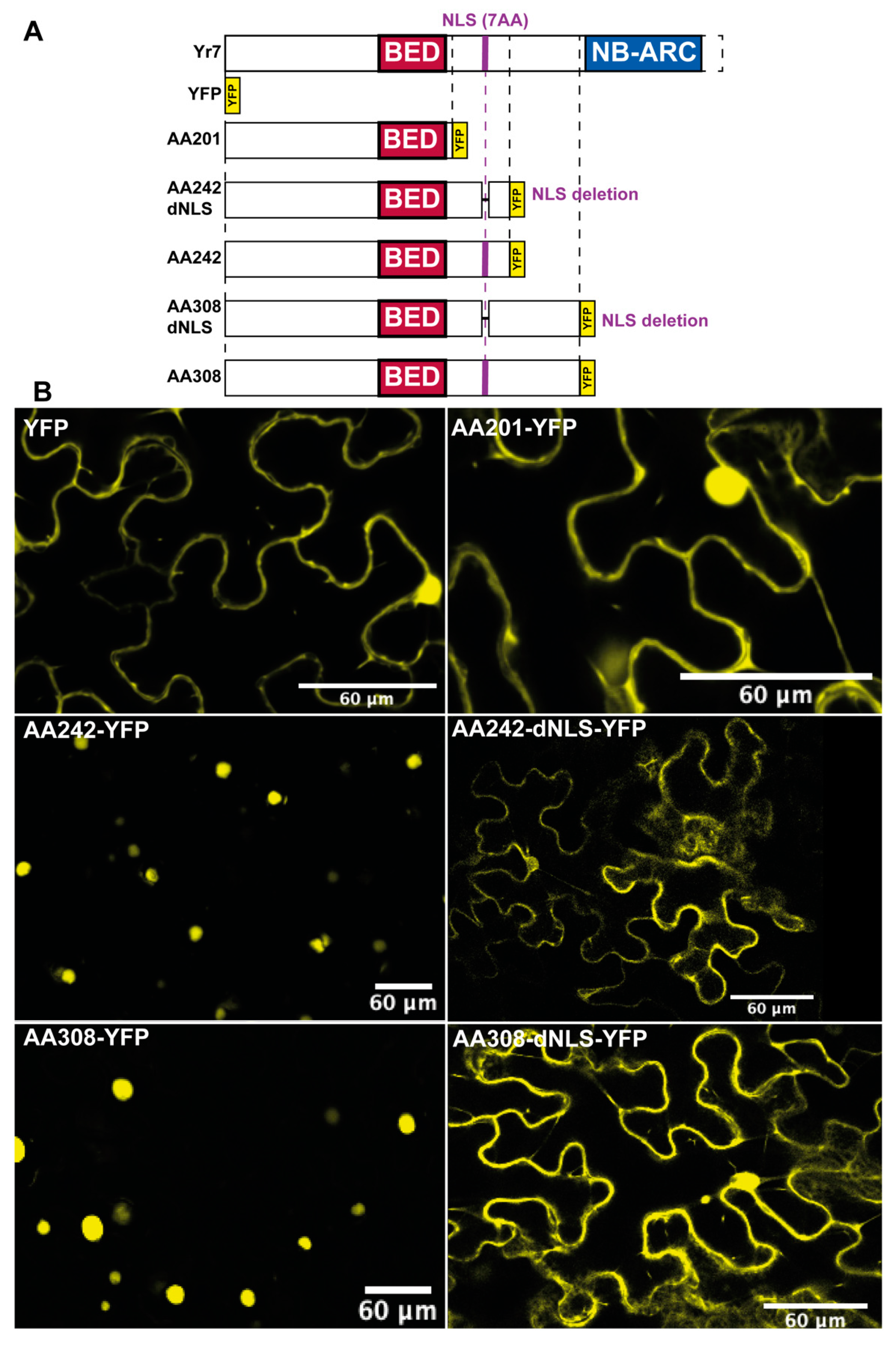
3.5. Yr7 and Its Variants do not Trigger Cell-death upon Overexpression in N. Benthamiana
4. Discussion
4.1. There is no Evidence for a NLR-partner in Yr7/Yr5 Locus: could Truncated nlr_11 be Functionaly Relevant?
4.2. Mode of Action of BED-NLRs may be Different that of Known NLR-IDs
4.3. N. benthamiana May not Be a Suitable Heterologous System to Study Signalling Mediated by BED-NLRs
4.4. BED Domains from BED-NLRs Share Similarities with BED Domains from Single-BED and BED-DUF4413/659-(hAT) Proteins
4.5. Nuclear Localization Signal is a Feature of BED-NLRs that Confer Resistance to Plant Pathogens
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BED | BED-type zinc finger domain named after the Drosophila proteins BEAF and DREF |
| CC | Coiled-coil |
| DUF | Domain of unknown function |
| Dimer_Tnp_hAT | hAT family C-terminal dimerization region |
| NLR | Nucleotide-binding Leucine-rich repeat |
| NLR-ID | NLR with integrated domain |
| NLS | Nuclear localization signal |
| RPW8 | Resistance to powdery mildew 8 |
| TIR | Toll and Interleukin-1 Receptor |
References
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Xue, J.Y.; Wu, P.; Zhang, Y.M.; Wu, Y.; Hang, Y.Y.; Wang, B.; Chen, J.Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016, 170, 2095–2109. [Google Scholar] [CrossRef]
- Kroj, T.; Chanclud, E.; Michel-Romiti, C.; Grand, X.; Morel, J.-B. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 2016, 210, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, C.; Huet, G.; Jauneau, A.; Camborde, L.; Tremousaygue, D.; Kraut, A.; Zhou, B.B.; Levaillant, M.; Adachi, H.; Yoshioka, H.; et al. A Receptor Pair with an Integrated Decoy Converts Pathogen Disabling of Transcription Factors to Immunity. Cell 2015, 161, 1074–1088. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Zhai, C.; Wang, W.; Zeng, X.; Xu, X.; Hu, H.; Lin, F.; Wang, L.; Pan, Q. The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor. Appl. Genet. 2011, 122, 1017–1028. [Google Scholar] [CrossRef]
- Cesari, S.; Bernoux, M.; Moncuquet, P.; Kroj, T.; Dodds, P.N. A novel conserved mechanism for plant NLR protein pairs: The integrated decoy hypothesis. Front. Plant Sci. 2014, 5, 606. [Google Scholar] [CrossRef]
- Fujisaki, K.; Abe, Y.; Kanzaki, E.; Ito, K.; Utsushi, H.; Saitoh, H.; Białas, A.; Banfield, M.; Kamoun, S.; Terauchi, R. An unconventional NOI/RIN4 domain of a rice NLR protein binds host EXO70 protein to confer fungal immunity. bioRxiv 2017. [Google Scholar] [CrossRef]
- Bailey, P.C.; Schudoma, C.; Jackson, W.; Baggs, E.; Dagdas, G.; Haerty, W.; Moscou, M.; Krasileva, K.V. Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 2018, 19, 23. [Google Scholar] [CrossRef]
- Sarris, P.F.; Cevik, V.; Dagdas, G.; Jones, J.D.G.; Krasileva, K.V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016, 14, 8. [Google Scholar] [CrossRef]
- Germain, H.; Séguin, A. Innate immunity: Has poplar made its BED? New Phytol. 2011, 189, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, S.; Yamanouchi, U.; Katayose, Y.; Toki, S.; Wang, Z.X.; Kono, I.; Kurata, N.; Yano, M.; Iwata, N.; Sasaki, T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. USA 1998, 95, 1663–1668. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Sengupta, S.; Prasad, M.; Ghose, T. Genetic diversity of the conserved motifs of six bacterial leaf blight resistance genes in a set of rice landraces. BMC Genet. 2014, 15, 82. [Google Scholar] [CrossRef] [PubMed]
- Read, A.C.; Hutin, M.; Moscou, M.J.; Rinaldi, F.C.; Bogdanove, A.I. Cloning of the Rice Xo1 Resistance Gene and Interaction of the Xo1 Protein with the Defense-Suppressing Xanthomonas Effector Tal2h. Mol. Plant-Microbe Interact. 2020, 33, 1189–1195. [Google Scholar]
- Chen, C.; Clark, B.; Martin, M.; Matny, O.; Steffenson, B.J.; Franckowiak, J.D.; Mascher, M.; Singh, D.; Perovic, D.; Richardson, T.; et al. Ancient BED-domain-containing immune receptor from wild barley confers widely effective resistance to leaf rust. bioRxiv 2020. [Google Scholar] [CrossRef]
- Marchal, C.; Zhang, J.; Zhang, P.; Fenwick, P.; Steuernagel, B.; Adamski, N.M.; Boyd, L.; McIntosh, R.; Wulff, B.B.H.; Berry, S.; et al. BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plants 2018, 4, 662–668. [Google Scholar] [CrossRef]
- Zuluaga, P.; Szurek, B.; Koebnik, R.; Kroj, T.; Morel, J.-B. Effector mimics and integrated decoys, the never-ending arms race between rice and Xanthomonas oryzae. Front. Plant Sci. 2017, 8, 431. [Google Scholar] [CrossRef]
- Aravind, L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem. Sci. 2000, 25, 421–423. [Google Scholar] [CrossRef]
- Rubin, E.; Lithwick, G.; Levy, A.A. Structure and evolution of the hAT transposon superfamily. Genetics 2001, 158, 949–957. [Google Scholar] [PubMed]
- Knip, M.; de Pater, S.; Hooykaas, P.J. The SLEEPER genes: A transposase-derived angiosperm-specific gene family. BMC Plant Biol. 2012, 12, 192. [Google Scholar] [CrossRef] [PubMed]
- Bundock, P.; Hooykaas, P. An Arabidopsis hAT-like transposase is essential for plant development. Nature 2005, 436, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.G. Integrated decoys and effector traps: How to catch a plant pathogen. BMC Biol. 2016, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.; Lawrence, G.; Ayliffe, M.; Anderson, P.; Collins, N.; Finnegan, J.; Frost, D.; Luck, J.; Pryor, T. Advances in the molecular genetic analysis of the flax-flax rust Interaction. Annu. Rev. Phytopathol. 1997, 35, 271–291. [Google Scholar] [CrossRef]
- Rose, L.E.; Bittner-Eddy, P.D.; Langley, C.H.; Holub, E.B.; Michelmore, R.W.; Beynon, J.L. The maintenance of extreme amino acid diversity at the disease resistance gene, Rpp13, in Arabidopsis thaliana. Genetics 2004, 166, 1517–1527. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Kaur, N.; Keller, B. Independent evolution of functional Pm3 resistance genes in wild tetraploid wheat and domesticated bread wheat. Plant J. 2009, 57, 846–856. [Google Scholar] [CrossRef]
- Srichumpa, P.; Brunner, S.; Keller, B.; Yahiaoui, N. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 2005, 139, 885–895. [Google Scholar] [CrossRef]
- Lu, X.; Kracher, B.; Saur, I.M.L.; Bauer, S.; Ellwood, S.R.; Wise, R.; Yaeno, T.; Maekawa, T.; Schulze-Lefert, P. Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl. Acad. Sci. USA 2016, 113, 6486–6495. [Google Scholar] [CrossRef]
- Narusaka, M.; Shirasu, K.; Noutoshi, Y.; Kubo, Y.; Shiraishi, T.; Iwabuchi, M.; Narusaka, Y. RRS1 and RPS4 provide a dual Resistance- gene system against fungal and bacterial pathogens. Plant J. 2009, 60, 218–226. [Google Scholar] [CrossRef]
- Wu, C.-H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, V.; Tang, S.; Stallmann, A.; Roberts, M.; Cherkis, K.; Dangl, J.L. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA 2011, 108, 16463–16468. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, M.; Dong, O.X.; Xia, S.; Liang, W.; Bao, Y.; Wasteneys, G.; Li, X. Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol. 2019, 222, 938–953. [Google Scholar] [CrossRef] [PubMed]
- Castel, B.; Ngou, P.M.; Cevik, V.; Redkar, A.; Kim, D.S.; Yang, Y.; Ding, P.; Jones, J.D.G. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 2019, 222, 966–980. [Google Scholar] [CrossRef] [PubMed]
- Peart, J.R.; Mestre, P.; Lu, R.; Malcuit, I.; Baulcombe, D.C. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 2005, 15, 968–973. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.L.; Monata, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 1–7. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Choulet, F.; Dostelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2009; ISBN 9780387981413. [Google Scholar]
- Steuernagel, B.; Witek, K.; Krattinger, S.G.; Ramirez-Gonzalez, R.H.; Schoonbeek, H.; Yu, G.; Baggs, E.; Witek, A.; Yadav, I.; Krasileva, K.V.; et al. The NLR-Annotator tool enables annotation of the intracellular immune receptor repertoire. Plant Physiol. 2020. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, 636–641. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.; Moulton, V. Neighbor-Net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2003, 21, 255–265. [Google Scholar] [CrossRef]
- Kloepper, T.H.; Huson, D.H. Drawing explicit phylogenetic networks and their integration into SplitsTree. BMC Evol. Biol. 2008, 8, 22. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef]
- Luo, M.-C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef]
- Vogel, J.P.; Garvin, D.F.; Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Steuernagel, B.; Witek, K.; Krattinger, S.G.; Ramirez-Gonzalez, R.H.; Schoonbeek, H.; Yu, G.; Baggs, E.; Witek, A.; Yadav, I.; Krasileva, K.V.; et al. Physical and transcriptional organisation of the bread wheat intracellular immune receptor repertoire. bioRxiv 2018. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Patron, N.J.; Orzaez, D.; Marillonnet, S.; Warzecha, H.; Matthewman, C.; Youles, M.; Raitskin, O.; Leveau, A.; Farré, G.; Rogers, C.; et al. Standards for plant synthetic biology: A common syntax for exchange of DNA parts. New Phytol. 2015, 208, 13–19. [Google Scholar] [CrossRef]
- Engler, C.; Youles, M.; Gruetzner, R.; Ehnert, T.M.; Werner, S.; Jones, J.D.G.; Patron, N.J.; Marillonnet, S. A Golden Gate modular cloning toolbox for plants. ACS Synth. Biol. 2014, 3, 839–843. [Google Scholar] [CrossRef]
- Bendahmane, A.; Farnham, G.; Moffett, P.; Baulcombe, D.C. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 2002, 32, 195–204. [Google Scholar] [CrossRef]
- Williams, S.J.; Sornaraj, P.; DeCourcy-Ireland, E.; Menz, R.I.; Kobe, B.; Ellis, J.G.; Dodds, P.N.; Anderson, P.A. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol. Plant-Microbe Interact. 2011, 24, 897–906. [Google Scholar] [CrossRef]
- Sueldo, D.J.; Shimels, M.; Spiridon, L.N.; Caldararu, O.; Petrescu, A.; Joosten, M.H.A.J.; Tameling, W.I.L. Random mutagenesis of the nucleotide-binding domain of NRC1 (NB-LRR Required for Hypersensitive Response-Associated Cell Death-1), a downstream signalling nucleotide-binding, leucine-rich repeat (NB-LRR) protein, identifies gain-of-function mutations in the nucleotide-binding pocket. New Phytol. 2015, 208, 210–223. [Google Scholar]
- Bai, S.; Liu, J.; Chang, C.; Zhang, L.; Maekawa, T.; Wang, Q.; Xiao, W.; Liu, Y.; Chai, J.; Takken, F.L.W.; et al. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef]
- Maqbool, A.; Saitoh, H.; Franceschetti, M.; Stevenson, C.; Uemura, A.; Kanzaki, H.; Kamoun, S.; Terauchi, R.; Banfield, M. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Lindbo, J.A.; Falk, B.W. The Impact of “Coat Protein-Mediated Virus Resistance” in Applied Plant Pathology and Basic Research. Phytopathology 2017, 107, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Derevnina, L.; Kamoun, S. NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 2019, 50, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Hu, M.; Wu, S.; Qi, J.; Wang, G.; Han, Z.; Qi, Y.; Gao, N.; Wang, H.-W.; et al. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 2019, 364, eaav5868. [Google Scholar] [CrossRef]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.-W.; Zhou, J.-M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, eaav5870. [Google Scholar] [CrossRef]
- Casey, L.W.; Lavrencic, P.; Bentham, A.R.; Cesari, S.; Ericsson, D.J.; Croll, T.; Turk, D.; Anderson, P.A.; Mark, A.E.; Dodds, P.N.; et al. The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins. Proc. Natl. Acad. Sci. USA 2016, 113, 12856–12861. [Google Scholar] [CrossRef]
- Nishimura, M.T.; Anderson, R.G.; Cherkis, K.A.; Law, T.F.; Liu, Q.L.; Machius, M.; Nimchuk, Z.L.; Yang, L.; Chung, E.-H.; El Kasmi, F.; et al. TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E2053–E2062. [Google Scholar] [CrossRef]
- Zhao, T.; Rui, L.; Li, J.; Nishimura, M.T.; Vogel, J.P.; Liu, N.; Liu, S.; Zhao, Y.; Dangl, J.L.; Tang, D. A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 2015, 11, e1004945. [Google Scholar] [CrossRef]
- Roth, C.; Lüdke, D.; Klenke, M.; Quathamer, A.; Valerius, O.; Braus, G.H.; Wiermer, M. The truncated NLR protein TIR-NBS13 is a MOS6/IMPORTIN-α3 interaction partner and required for plant immunity. Plant J. 2017. [Google Scholar] [CrossRef]
- Xiao, S.; Calis, O.; Patrick, E.; Zhang, G.; Charoenwattana, P.; Muskett, P.; Parker, J.E.; Turner, J.G. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 2005, 42, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Castel, B.; Wu, Y.; Xiao, S.; Jones, J.D.G. An rpw8 quadruple mutant of Arabidopsis Col-0 is partially compromised in bacterial and fungal resistance. bioRxiv 2019. [Google Scholar] [CrossRef]
- Read, A.C.; Moscou, M.J.; Zimin, A.V.; Pertea, G.; Meyer, R.S.; Purugganan, M.D.; Leach, J.E.; Triplett, L.R.; Salzberg, S.L.; Bogdanove, A.J. Genome assembly and characterization of a complex zfBED-NLR gene-containing disease resistance locus in Carolina Gold Select rice with Nanopore sequencing. PLoS Genet. 2020, 16, e1008571. [Google Scholar]
- Shi, X.; Dong, S.; Liu, W. Structures of plant resistosome reveal how NLR immune receptors are activated. aBIOTECH 2019, 1, 147–150. [Google Scholar] [CrossRef]
- Adachi, H.; Contreras, M.P.; Harant, A.; Wu, C.H.; Derevnina, L.; Sakai, T.; Duggan, C.; Moratto, E.; Bozkurt, T.O.; Maqbool, A.; et al. An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. eLife 2019, 8, 693291. [Google Scholar] [CrossRef]
- Chen, J.; Upadhyaya, N.M.; Ortiz, D.; Sperschneider, J.; Li, F.; Bouton, C.; Breen, S.; Dong, C.; Xu, B.; Zhang, X.; et al. Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 2017, 358, 1607–1610. [Google Scholar] [CrossRef]
- Bombarely, A.; Rosli, H.G.; Vrebalov, J.; Moffett, P.; Mueller, L.A.; Martin, G.B. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant-Microbe Interact. 2012, 25, 1523–1530. [Google Scholar] [CrossRef]
- Saur, I.M.L.; Bauer, S.; Lu, X.; Schulze-Lefert, P. A cell death assay in barley and wheat protoplasts for identification and validation of matching pathogen AVR effector and plant NLR immune receptors. Plant Methods 2019, 15, 118. [Google Scholar] [CrossRef]
- De la Concepcion, J.C.; Franceschetti, M.; Maqbool, A.; Saitoh, H.; Terauchi, R.; Kamoun, S.; Banfield, M.J. Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nat. Plants 2018, 4, 576–585. [Google Scholar] [CrossRef]
- Sato, M.; Sasagawa, A.; Tochio, N.; Koshiba, S.; Inoue, M.; Kigawa, T.; Yokoyama, S. Solution structures of the C2H2 type zinc finger domain of human zinc finger BED domain containing protein 2. RCSB 2006. [Google Scholar] [CrossRef]
- Shen, Q.-H.; Saijo, Y.; Mauch, S.; Biskup, C.; Bieri, S.; Keller, B.; Seki, H.; Ulker, B.; Somssich, I.E.; Schulze-Lefert, P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007, 315, 1098–1103. [Google Scholar] [CrossRef]
- Wirthmueller, L.; Zhang, Y.; Jones, J.D.G.; Parker, J.E. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 2007, 17, 2023–2029. [Google Scholar] [CrossRef] [PubMed]
| Cultivar | Name Used in This Study | Region of Origin |
|---|---|---|
| Julius | Julius | Germany |
| Jagger | Jagger | USA |
| Norin61 | Norin | Japan |
| CDC Landmark | Landmark | Canada |
| CDC Stanley | Stanley | Canada |
| ArinaLrFor | Arina | Switzerland |
| Mace | Mace | Australia |
| LongReach Lancer | Lancer | Australia |
| SY-Mattis | SY-Mattis | France |
| Spelt PI190962 | Spelt PI190962 | Europe |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchal, C.; 10+ Wheat Genome Project; Haberer, G.; Spannagl, M.; Uauy, C. Comparative Genomics and Functional Studies of Wheat BED-NLR Loci. Genes 2020, 11, 1406. https://doi.org/10.3390/genes11121406
Marchal C, 10+ Wheat Genome Project, Haberer G, Spannagl M, Uauy C. Comparative Genomics and Functional Studies of Wheat BED-NLR Loci. Genes. 2020; 11(12):1406. https://doi.org/10.3390/genes11121406
Chicago/Turabian StyleMarchal, Clemence, 10+ Wheat Genome Project, Georg Haberer, Manuel Spannagl, and Cristobal Uauy. 2020. "Comparative Genomics and Functional Studies of Wheat BED-NLR Loci" Genes 11, no. 12: 1406. https://doi.org/10.3390/genes11121406
APA StyleMarchal, C., 10+ Wheat Genome Project, Haberer, G., Spannagl, M., & Uauy, C. (2020). Comparative Genomics and Functional Studies of Wheat BED-NLR Loci. Genes, 11(12), 1406. https://doi.org/10.3390/genes11121406




