Differential Gene Expression in Longissimus Dorsi Muscle of Hanwoo Steers—New Insight in Genes Involved in Marbling Development at Younger Ages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. RNA Extraction and Library Preparation
2.3. RNA-Seq Data Analysis
2.4. Protein–Protein Interaction Network
3. Results
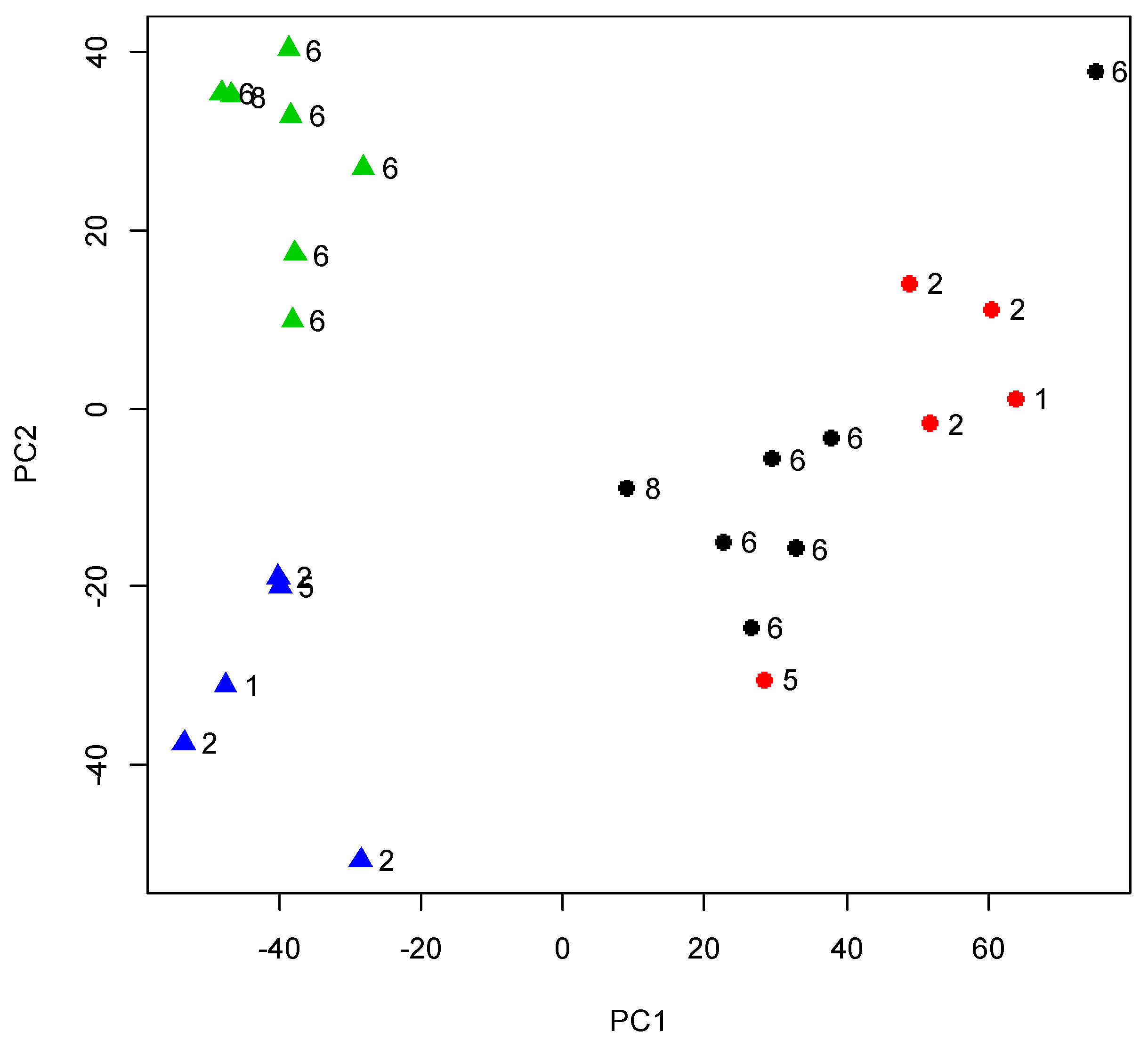
3.1. Gene Expression Analysis
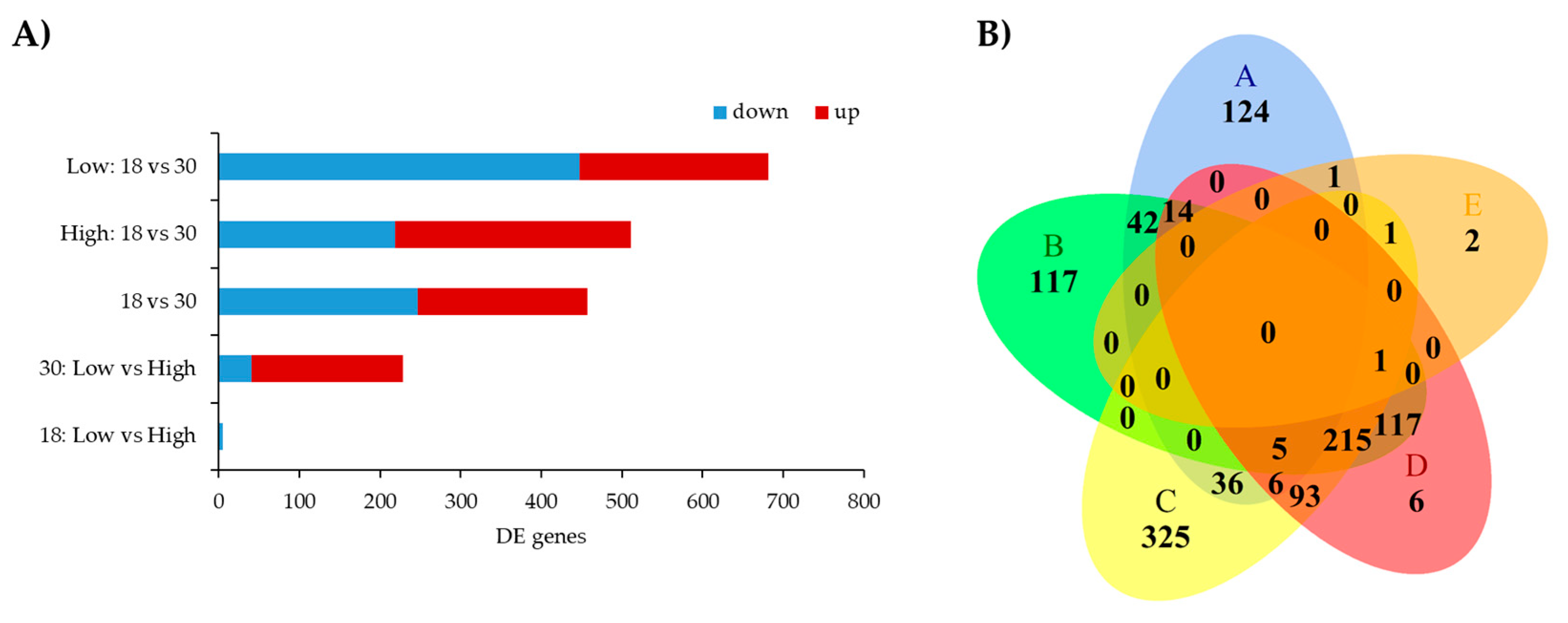
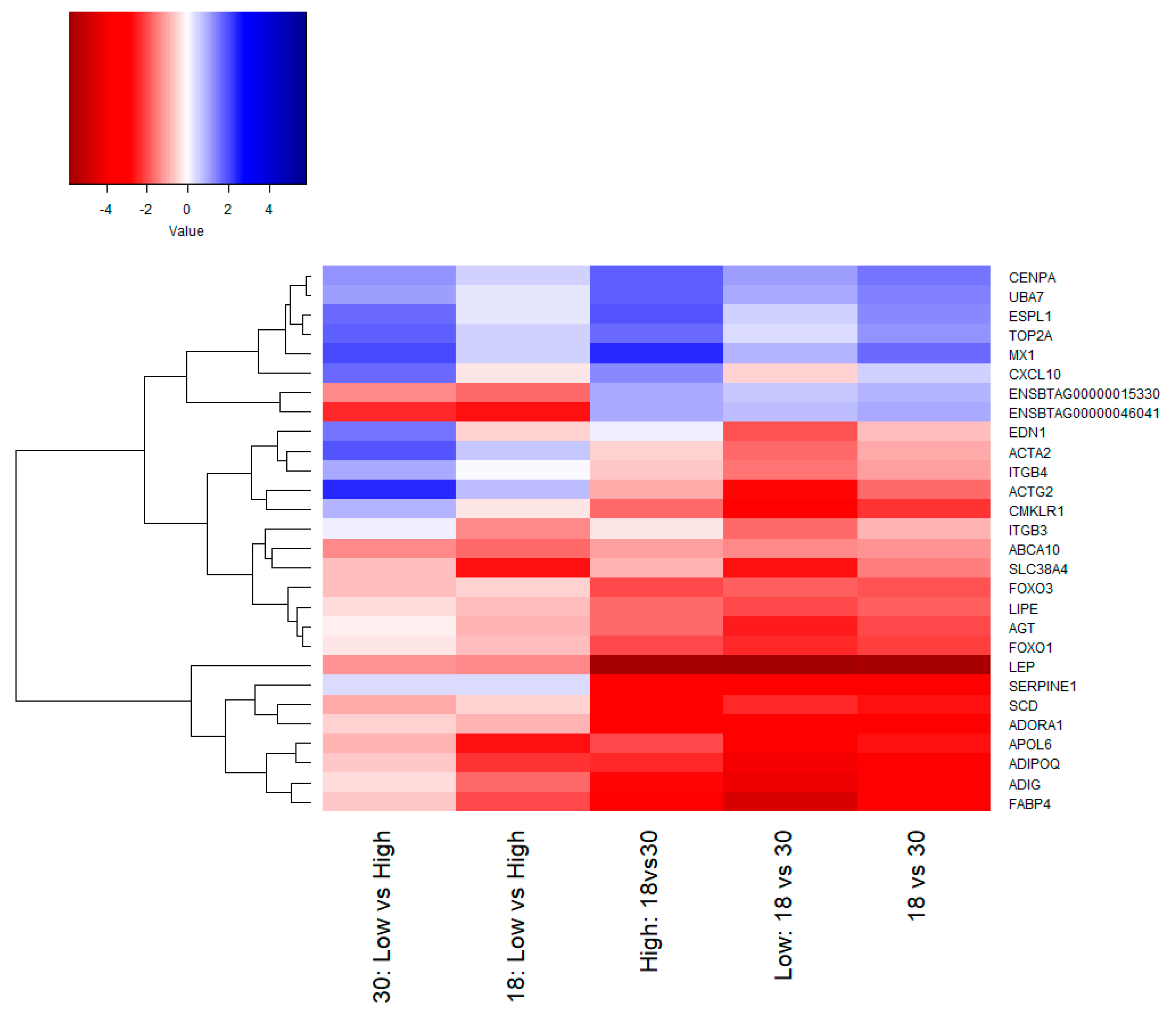
3.2. Functional Analysis of Differentially Expressed Genes
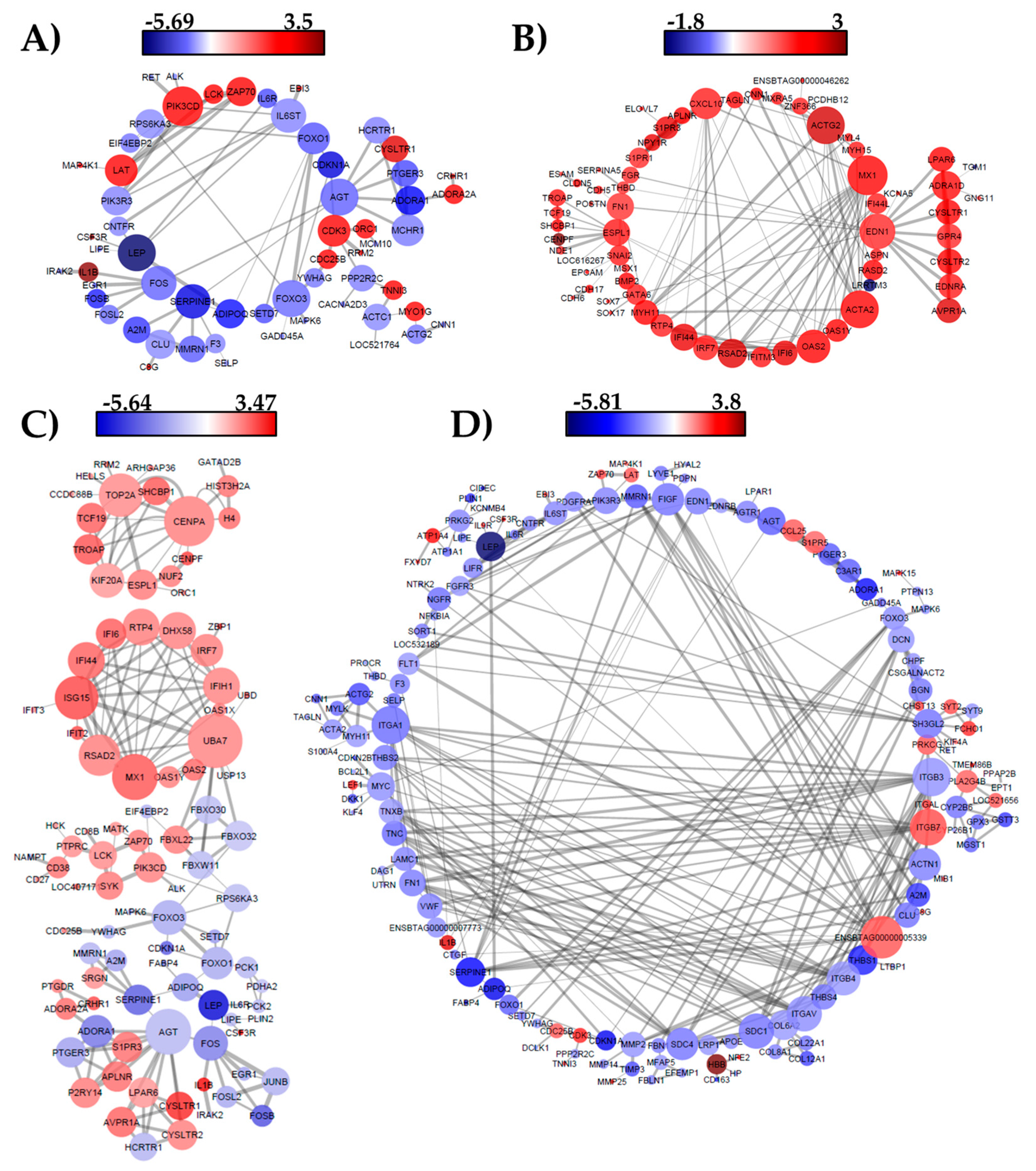
3.3. Network Analysis
4. Discussion
4.1. Transcriptomic Profile for Marbling Selection at an Early Age
4.2. Gene Ontology and Pathway Analysis
4.3. Interaction Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lim, D.; Gondro, C.; Park, H.S.; Cho, Y.M.; Chai, H.H.; Seong, H.H.; Yang, B.S.; Hong, S.K.; Chang, W.K.; Lee, S.H. Identification of recently selected mutations driven by artificial selection in Hanwoo (Korean cattle). Asian Australas. J. Anim. Sci. 2013, 26, 603. [Google Scholar] [CrossRef] [Green Version]
- Porto-Neto, L.R.; Lee, S.H.; Sonstegard, T.S.; Van Tassell, C.P.; Lee, H.K.; Gibson, J.P.; Gondro, C. Genome-wide detection of signatures of selection in Korean Hanwoo cattle. Anim. Genet. 2014, 45, 180–190. [Google Scholar] [CrossRef]
- Lee, S.H.; Park, B.H.; Sharma, A.; Dang, C.G.; Lee, S.S.; Choi, T.J.; Choy, Y.H.; Kim, H.C.; Jeon, K.J.; Kim, S.D. Hanwoo cattle: Origin, domestication, breeding strategies and genomic selection. J. Anim. Sci. Technol. 2014, 56, 2. [Google Scholar] [CrossRef] [Green Version]
- Jung, E.; Hwang, Y.; Joo, S. The relationship between chemical compositions, meat quality, and palatability of the 10 primal cuts from Hanwoo steer. Korean J. Food Sci. Anim. Resour. 2016, 36, 145. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.; Park, B.; Kim, J.; Hwang, I.; Kim, J.; Lee, J. Fatty acid profiles and sensory properties of longissimus dorsi, triceps brachii, and semimembranosus muscles from Korean Hanwoo and Australian Angus beef. Asian Australas. J. Anim. Sci. 2005, 18, 1786. [Google Scholar] [CrossRef]
- Smith, S.B.; Gill, C.A.; Lunt, D.K.; Brooks, M.A. Regulation of fat and fatty acid composition in beef cattle. Asian Australas. J. Anim. Sci. 2009, 22, 1225–1233. [Google Scholar] [CrossRef]
- Hwang, Y.H.; Joo, S.T. Fatty acid profiles, meat quality, and sensory palatability of grain-fed and grass-fed beef from Hanwoo, American, and Australian crossbred cattle. Korean J. Food Sci. Anim. Resour. 2017, 37, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.-J.; Park, H.-S.; Kim, W.; Yoon, D.; Seo, S. Comparison of metabolic network between muscle and intramuscular adipose tissues in Hanwoo beef cattle using a systems biology approach. Int. J. Genom. 2014, 2014, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hocquette, J.; Gondret, F.; Baéza, E.; Médale, F.; Jurie, C.; Pethick, D. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2010, 4, 303–319. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Lee, J.; Park, S. Effects of full-fat soybean diet on performance, carcass characteristics, and fatty acid composition of Hanwoo steers. Turk. J. Vet. Anim. Sci. 2016, 40, 451–458. [Google Scholar] [CrossRef]
- Dashdorj, D.; Oliveros, M.C.R.; Hwang, I.-H. Meat quality traits of Longissimus muscle of Hanwoo steers as a function of interaction between slaughter endpoint and chiller ageing. Korean J. Food Sci. Anim. Resour. 2012, 32, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Mehrban, H.; Lee, D.H.; Moradi, M.H.; IlCho, C.; Naserkheil, M.; Ibáñez-Escriche, N. Predictive performance of genomic selection methods for carcass traits in Hanwoo beef cattle: Impacts of the genetic architecture. Genet. Sel. Evol. 2017, 49, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; van der Werf, J.H.J.; Lee, S.H.; Park, E.W.; Gondro, C.; Yoon, D.; Oh, S.J.; Kim, O.H.; Gibson, J.; Thompson, J. Genome wide QTL mapping to identify candidate genes for carcass traits in Hanwoo (Korean Cattle). Genes Genom. 2012, 34, 43–49. [Google Scholar] [CrossRef]
- Shin, Y.; Jung, H.-j.; Jung, M.; Yoo, S.; Subramaniyam, S.; Markkandan, K.; Kang, J.-M.; Rai, R.; Park, J.; Kim, J.-J. Discovery of gene sources for economic traits in Hanwoo by whole-genome resequencing. Asian Australas. J. Anim. Sci. 2016, 29, 1353. [Google Scholar] [CrossRef] [Green Version]
- Strucken, E.M.; Al-Mamun, H.A.; de las Heras-Saldana, S.; Bedhane, M.N.; Lim, D.; Park, B.; Gondro, C. Finding the marble–The polygenic architecture of intramuscular fat. J. Anim. Breed. Genom. 2017, 1, 69–76. [Google Scholar]
- Lee, S.-H.; Cho, Y.-M.; Lee, S.-H.; Kim, B.-S.; Kim, N.-K.; Choy, Y.-H.; Kim, K.-H.; Yoon, D.-H.; Im, S.-K.; Oh, S.-J. Identification of marbling-related candidate genes in M. longissimus dorsi of high-and low marbled Hanwoo (Korean Native Cattle) steers. BMB Rep. 2008, 41, 846–851. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhao, Y.; Ning, Y.; Wang, H.; Zan, L. Ectopical expression of FABP4 gene can induce bovine muscle-derived stem cells adipogenesis. Biochem. Biophys. Res. Commun. 2017, 482, 352–358. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Gondro, C.; van der Werf, J.H.J.; Kim, N.K.; Lim, D.J.; Park, E.W.; Oh, S.J.; Gibson, J.P.; Thompson, J.M. Use of a bovine genome array to identify new biological pathways for beef marbling in Hanwoo (Korean Cattle). BMC Genom. 2010, 11, 623. [Google Scholar] [CrossRef] [Green Version]
- Lim, D.; Chai, H.H.; Lee, S.H.; Cho, Y.M.; Choi, J.W.; Kim, N.K. Gene expression patterns associated with peroxisome proliferator-activated receptor (PPAR) signaling in the longissimus dorsi of Hanwoo (Korean Cattle). Asian Australas. J. Anim. Sci. 2015, 28, 1075–1083. [Google Scholar] [CrossRef] [Green Version]
- Seong, J.; Yoon, H.; Kong, H.S. Identification of microRNA and target gene associated with marbling score in Korean cattle (Hanwoo). Genes Genom. 2016, 38, 529–538. [Google Scholar] [CrossRef]
- Lim, D.; Kim, N.K.; Park, H.S.; Lee, S.H.; Cho, Y.M.; Oh, S.J.; Kim, T.H.; Kim, H. Identification of candidate genes related to bovine marbling using protein-protein interaction networks. Int. J. Biol. Sci. 2011, 7, 992. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Y.; Zhang, X.; Wang, D.; Jin, G.; Li, B.; Xu, F.; Cheng, J.; Zhang, F.; Wu, S. The comprehensive liver transcriptome of two cattle breeds with different intramuscular fat content. Biochem. Biophys. Res. Commun. 2017, 490, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, K.; Lee, S.H.; Chung, K.Y.; Park, J.E.; Jang, G.W.; Park, M.R.; Kim, N.Y.; Kim, T.H.; Chai, H.H.; Park, W.C. A Gene-Set Enrichment and Protein–Protein Interaction Network-Based GWAS with Regulatory SNPs Identifies Candidate Genes and Pathways Associated with Carcass Traits in Hanwoo Cattle. Genes 2020, 11, 316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Bower, N.; Reverter, A.; Tan, S.; De Jager, N.; Wang, R.; McWilliam, S.; Cafe, L.; Greenwood, P.; Lehnert, S. Gene expression patterns during intramuscular fat development in cattle. J. Anim. Sci. 2009, 87, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Pampusch, M.; White, M.; Hathaway, M.; Baxa, T.; Chung, K.; Parr, S.; Johnson, B.; Weber, W.; Dayton, W. Effects of implants of trenbolone acetate, estradiol, or both, on muscle insulin-like growth factor-I, insulin-like growth factor-I receptor, estrogen receptor-α, and androgen receptor messenger ribonucleic acid levels in feedlot steers. J. Anim. Sci. 2008, 86, 3418–3423. [Google Scholar] [CrossRef]
- Chung, K.; Baxa, T.; Parr, S.; Luqué, L.; Johnson, B. Administration of estradiol, trenbolone acetate, and trenbolone acetate/estradiol implants alters adipogenic and myogenic gene expression in bovine skeletal muscle. J. Anim. Sci. 2012, 90, 1421–1427. [Google Scholar] [CrossRef]
- Chung, K.Y.; Lee, S.H.; Cho, S.H.; Kwon, E.G.; Lee, J.H. Current situation and future prospects for beef production in South Korea—A review. Asian Australas. J. Anim. Sci. 2018, 31, 951. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, M.; Huber, W.; Pages, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wenzel, J.J.; Kaminski, W.E.; Piehler, A.; Heimerl, S.; Langmann, T.; Schmitz, G. ABCA10, a novel cholesterol-regulated ABCA6-like ABC transporter. Biochem. Biophys. Res. Commun. 2003, 306, 1089–1098. [Google Scholar] [CrossRef]
- Dean, M.; Hamon, Y.; Chimini, G. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res. 2001, 42, 1007–1017. [Google Scholar] [CrossRef]
- Gai, J.; Ji, M.; Shi, C.; Li, W.; Chen, S.; Wang, Y.; Li, H. FoxO regulates expression of ABCA6, an intracellular ATP-binding-cassette transporter responsive to cholesterol. Int. J. Biochem. Cell Biol. 2013, 45, 2651–2659. [Google Scholar] [CrossRef] [PubMed]
- Iwayama, T.; Steele, C.; Yao, L.; Dozmorov, M.G.; Karamichos, D.; Wren, J.D.; Olson, L.E. PDGFRα signaling drives adipose tissue fibrosis by targeting progenitor cell plasticity. Genes Dev. 2015, 29, 1106–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lui, J.C.; Finkielstain, G.P.; Barnes, K.M.; Baron, J. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R189–R196. [Google Scholar] [CrossRef] [PubMed]
- Ambele, M.A.; Dessels, C.; Durandt, C.; Pepper, M.S. Genome-wide analysis of gene expression during adipogenesis in human adipose-derived stromal cells reveals novel patterns of gene expression during adipocyte differentiation. Stem Cell Res. 2016, 16, 725–734. [Google Scholar] [CrossRef] [Green Version]
- Menssen, A.; Häupl, T.; Sittinger, M.; Delorme, B.; Charbord, P.; Ringe, J. Differential gene expression profiling of human bone marrow-derived mesenchymal stem cells during adipogenic development. BMC Genom. 2011, 12, 461. [Google Scholar] [CrossRef] [Green Version]
- Dalrymple, B.; Guo, B.; Zhou, G.; Zhang, W. Using muscle gene expression to estimate triacylglyceride deposition, and relative contributions of fatty acid synthesis and fatty acid import in intramuscular fat in cattle. Anim. Prod. Sci. 2014, 54, 1436–1442. [Google Scholar] [CrossRef]
- Reddy, K.E.; Jeong, J.Y.; Lee, S.D.; Baek, Y.C.; Oh, Y.K.; Kim, M.; So, K.M.; Kim, D.; Kim, J.H.; Park, S. Effect of different early weaning regimens for calves on adipogenic gene expression in Hanwoo loin at the fattening stage. Livest. Sci. 2017, 195, 87–98. [Google Scholar] [CrossRef]
- Suzuki, Y.; Hong, Y.; Song, S.; Ardiyanti, A.; Kato, D.; So, K.; Katoh, K.; Roh, S.-G. The regulation of chemerin and CMKLR1 genes expression by TNF-α, adiponectin, and chemerin analog in bovine differentiated adipocytes. Asian Australas. J. Anim. Sci. 2012, 25, 1316. [Google Scholar] [CrossRef] [Green Version]
- Graugnard, D.E.; Piantoni, P.; Bionaz, M.; Berger, L.L.; Faulkner, D.B.; Loor, J.J. Adipogenic and energy metabolism gene networks in longissimus lumborum during rapid post-weaning growth in Angus and Angus× Simmental cattle fed high-starch or low-starch diets. BMC Genom. 2009, 10, 142. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Guo, Y.; Du, W.; Zhang, X.; Li, A.; Miao, X. Global transcriptome analysis identifies differentially expressed genes related to lipid metabolism in Wagyu and Holstein cattle. Sci. Rep. 2017, 7, 5278. [Google Scholar] [CrossRef] [Green Version]
- Mihaylova, M.M.; Shaw, R.J. The AMP-activated protein kinase (AMPK) signaling pathway coordinates cell growth, autophagy, & metabolism. Nat. Cell Biol. 2011, 13, 1016. [Google Scholar] [PubMed]
- Wu, G.S. The functional interactions between the MAPK and p53 signaling pathways. Cancer Biol. Ther. 2004, 3, 156–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.H.; Reverter, A.; Wang, Y.; Byrne, K.A.; McWilliam, S.M.; Lehnert, S.A. Gene expression profiling of bovine in vitro adipogenesis using a cDNA microarray. Funct. Integr. Genom. 2006, 6, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; White, M.F. Targeting Forkhead box O1 from the concept to metabolic diseases: Lessons from mouse models. Antioxid. Redox Signal. 2011, 14, 649–661. [Google Scholar] [CrossRef] [Green Version]
- Bastie, C.C.; Nahlé, Z.; McLoughlin, T.; Esser, K.; Zhang, W.; Unterman, T.; Abumrad, N.A. FoxO1 stimulates fatty acid uptake and oxidation in muscle cells through CD36-dependent and-independent mechanisms. J. Biol. Chem. 2005, 280, 14222–14229. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, K.; Ogawa, Y. FoxOs: The biology and pathophysiological roles in diabetes. J. Diabetes Investig. 2017, 8, 726–734. [Google Scholar] [CrossRef] [Green Version]
- Nakae, J.; Cao, Y.; Oki, M.; Orba, Y.; Sawa, H.; Kiyonari, H.; Iskandar, K.; Suga, K.; Lombes, M.; Hayashi, Y. Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes 2008, 57, 563–576. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, Y.; Itoh, T.; Yamada, T.; Sasaki, Y. Genomic structure and promoter analysis of the bovine leptin gene. IUBMB Life 2002, 53, 131–135. [Google Scholar] [CrossRef]
- Yonekura, S.; Oka, A.; Noda, M.; Uozumi, N.; Yonezawa, T.; Katoh, K.; Obara, Y. Relationship between serum leptin concentrations and the marbling scores in Japanese Black Cattle. Anim. Sci. J. 2002, 73, 51–57. [Google Scholar] [CrossRef]
- Geary, T.W.; McFadin, E.L.; MacNeil, M.D.; Grings, E.E.; Short, R.E.; Funston, R.N.; Keisler, D.H. Leptin as a predictor of carcass composition in beef cattle 1. J. Anim. Sci. 2003, 81, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Jin, Q.; Zhang, C.; Fang, X.; Gu, C.; Lei, C.; Wang, J.; Chen, H. Polymorphisms in the bovine ghrelin precursor (GHRL) and Syndecan-1 (SDC1) genes that are associated with growth traits in cattle. Mol. Biol. Rep. 2011, 38, 3153–3160. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Olson, E.N.; Moore, S.S.; Basarab, J.A.; Basu, U.; Guan, L.L. Transcriptome analysis of subcutaneous adipose tissues in beef cattle using 3′ digital gene expression-tag profiling. J. Anim. Sci. 2012, 90, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.S.; Kang, L.; Zheng, J.; Grueter, C.; Bracy, D.P.; James, F.D.; Pozzi, A.; Wasserman, D.H. Integrin α1-null mice exhibit improved fatty liver when fed a high fat diet despite severe hepatic insulin resistance. J. Biol. Chem. 2015, 290, 6546–6557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.J.; Jang, M.; Kim, H.; Kwak, W.; Park, W.C.; Hwang, J.Y.; Lee, C.K.; Jang, G.W.; Park, M.N.; Kim, H.C. Comparative transcriptome analysis of adipose tissues reveals that ECM-receptor interaction is involved in the depot-specific adipogenesis in cattle. PLoS ONE 2013, 8, e66267. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.M.; Zhou, X.; Shao, H.; Kong, X.F.; Yin, Y.L.; Huang, R. Genotyping of five Chinese local pig breeds focused on meat quality by using PCR-RFLP based on halothane and Mx1. J. Food Agric. Environ. 2012, 10, 840–845. [Google Scholar]
- Iqbal, A.; Kim, Y.-S.; Kang, J.-M.; Lee, Y.-M.; Rai, R.; Jung, J.-H.; Oh, D.-Y.; Nam, K.-C.; Lee, H.-K.; Kim, J.-J. Genome-wide Association Study to Identify Quantitative Trait Loci for Meat and Carcass Quality Traits in Berkshire. Asian Australas. J. Anim. Sci. 2015, 28, 1537. [Google Scholar] [CrossRef] [Green Version]
- Wicik, Z.; Sadkowski, T.; Jank, M.; Motyl, T. The transcriptomic signature of myostatin inhibitory influence on the differentiation of mouse C2C12 myoblasts. Pol. J. Vet. Sci. 2011, 14, 643–652. [Google Scholar] [CrossRef] [Green Version]

| 18 Months | 30 Months | |||
|---|---|---|---|---|
| High MS | Low MS | High MS | Low MS | |
| Total reads | 66,334,797 | 68,966,322 | 67,403,192 | 70,088,156 |
| Pair reads mapped | 32,628,467 | 33,654,496 | 33,488,986 | 33,644,083 |
| Unique matched | 15,865,630 | 16,234,604 | 15,904,205 | 16,064,734 |
| Multimatched reads | 3,722,653 | 4,588,413 | 4,211,752 | 4,346,990 |
| Overall map | 80.8% | 80.6% | 79.9% | 80.3% |
| Gene | logFC | Average Expression | p-Value | FDR |
|---|---|---|---|---|
| ABCA10 | −1.64 | 2.94 | 1.97 × 10−5 | 0.046 |
| ENSBTAG00000015330 | −1.68 | 1.43 | 1.75 × 10−5 | 0.046 |
| ENSBTAG00000046041 | −2.57 | −0.17 | 1.79 × 10−5 | 0.046 |
| APOL6 | −2.59 | −1.08 | 2.51 × 10−5 | 0.049 |
| SLC38A4 | −2.62 | 0.14 | 1.06 × 10−5 | 0.046 |
| Pathway | 18 vs. 30 | High 18 vs. 30 | Low 18 vs. 30 | Genes |
|---|---|---|---|---|
| AMPK signaling pathway (bta04152) | 12 ** | 12 * | 13 ** | LIPE, FOXO3, LEP, LOC101906058, SCD, FOXO1, PPP2R2C, PFKFB3, ADIPOQ, PCK2, PCK1*, PIK3CD*, PIK3R3+, PIK3CD+, MLYCD#, CPT1A#, PIK3R3# |
| Regulation of lipolysis in adipocytes (bta04923) | 7 * | 7 * | LIPE, ADORA1, PIK3R3, PLIN1, FABP4, PTGER3, PIK3CD*, PRKG2# | |
| p53 signaling pathway (bta04115) | 7 * | 9 * | CDKN1A, THBS1, SERPINE1, SESN3, GADD45A, TP73, RRM2*, IGFBP3#, ZMAT3#, PERP# | |
| PPAR signaling pathway (bta03320) | 8 * | 9 * | LOC101906058, SCD, PLIN1, FABP4, ADIPOQ, PCK2, PCK1+, PLIN2+, CPT1A#, SLC27A5#, ANGPTL4# | |
| PI3K-Akt signaling pathway (bta04151) | 3 * | 32 *** | FOXO3, FOXO1+, PIK3CD+, THBS4#, CDKN1A#, BCL2L1#, COL6A2#, VWF#, LAMC1#, PDGFRA#, THBS1#, ITGB4#, ITGAV#, VEGFA#, FLT1#, IL6R#, ITGA1#, TNXB#, LPAR1#, FN1#, YWHAG#, VEGFD#, TNC#, ITGB3#, PIK3R3#, THBS2#, MYC#, RELN#, NGFR#, PPP2R2C#, PCK2#, ITGB7#, FGFR3#, CSF3R# | |
| MAPK signaling pathway (bta04010) | 7 * | PDGFRA, MAP3K2, IL1B, CD14, PLA2G4B, NTRK2, TAOK1, ELK4, HSPA1A, MYC, PRKCG, GADD45A, RASGRP2, FGFR3, MAP4K1, CDC25B, HSPA6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de las Heras-Saldana, S.; Chung, K.Y.; Kim, H.; Lim, D.; Gondro, C.; van der Werf, J.H.J. Differential Gene Expression in Longissimus Dorsi Muscle of Hanwoo Steers—New Insight in Genes Involved in Marbling Development at Younger Ages. Genes 2020, 11, 1381. https://doi.org/10.3390/genes11111381
de las Heras-Saldana S, Chung KY, Kim H, Lim D, Gondro C, van der Werf JHJ. Differential Gene Expression in Longissimus Dorsi Muscle of Hanwoo Steers—New Insight in Genes Involved in Marbling Development at Younger Ages. Genes. 2020; 11(11):1381. https://doi.org/10.3390/genes11111381
Chicago/Turabian Stylede las Heras-Saldana, Sara, Ki Yong Chung, Hyounju Kim, Dajeong Lim, Cedric Gondro, and Julius H. J. van der Werf. 2020. "Differential Gene Expression in Longissimus Dorsi Muscle of Hanwoo Steers—New Insight in Genes Involved in Marbling Development at Younger Ages" Genes 11, no. 11: 1381. https://doi.org/10.3390/genes11111381
APA Stylede las Heras-Saldana, S., Chung, K. Y., Kim, H., Lim, D., Gondro, C., & van der Werf, J. H. J. (2020). Differential Gene Expression in Longissimus Dorsi Muscle of Hanwoo Steers—New Insight in Genes Involved in Marbling Development at Younger Ages. Genes, 11(11), 1381. https://doi.org/10.3390/genes11111381






