Evolution of Oxidative Phosphorylation (OXPHOS) Genes Reflecting the Evolutionary and Life Histories of Fig Wasps (Hymenoptera, Chalcidoidea)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Determination
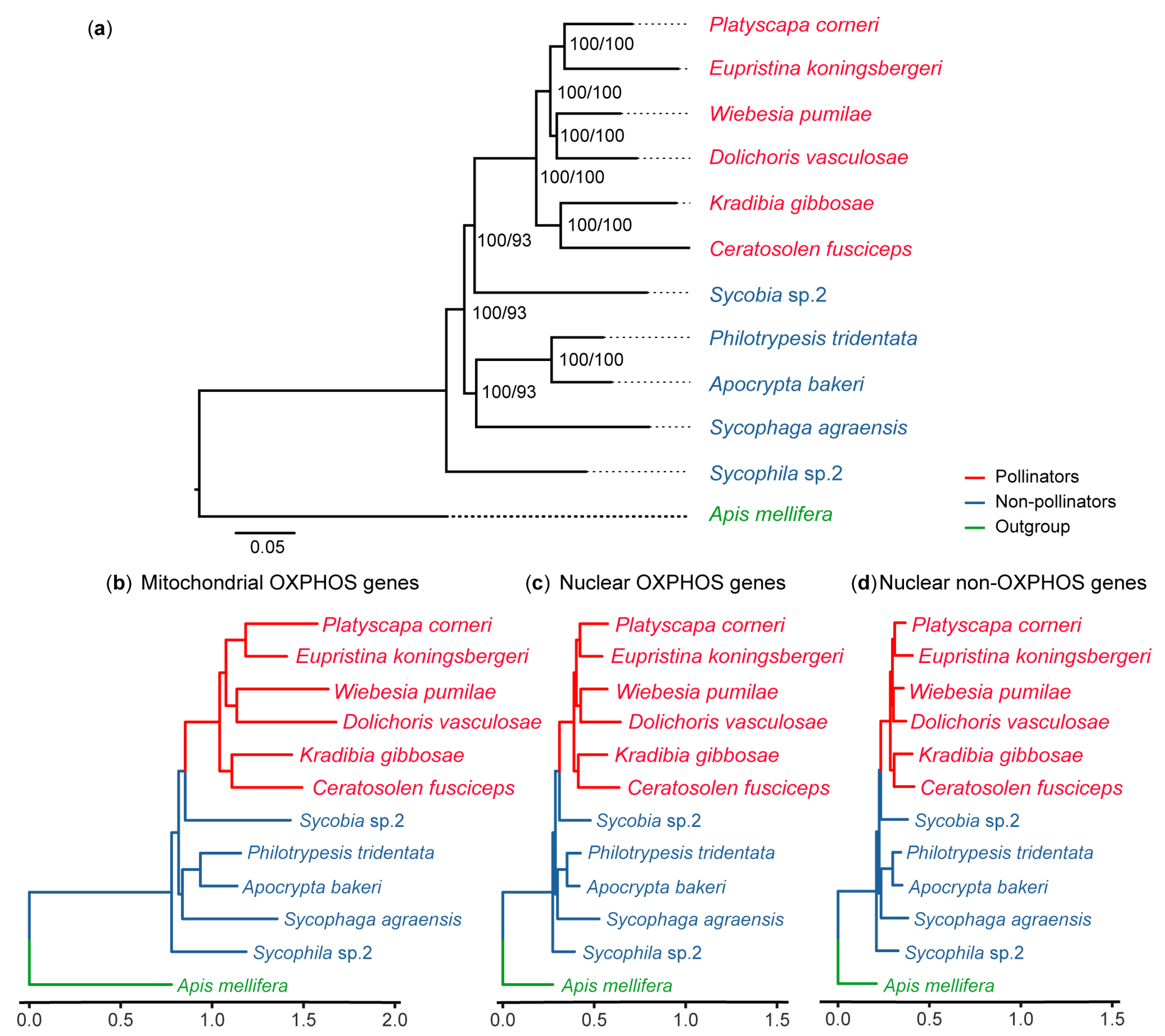
2.2. Phylogenetic Reconstruction
2.3. Estimation of the Amino Acid Substitution Rate
2.4. Natural Selection Analysis
3. Results
3.1. OXPHOS Gene Annotation
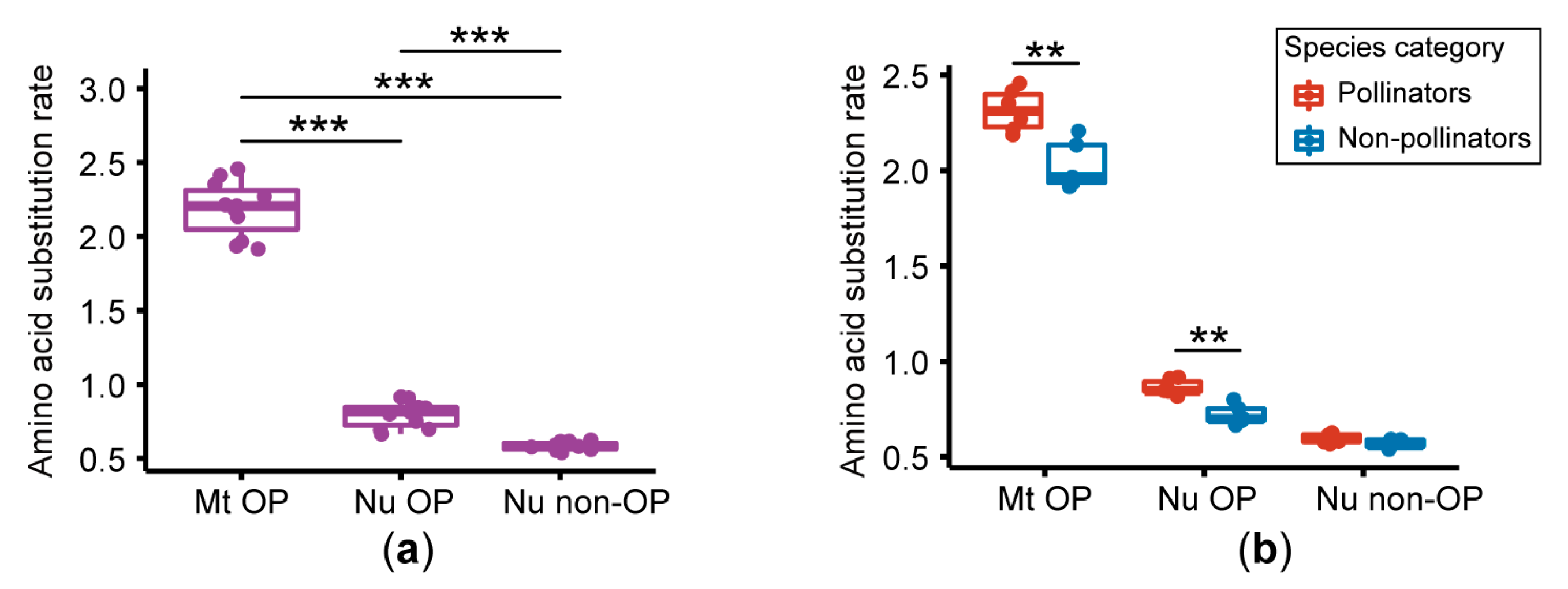
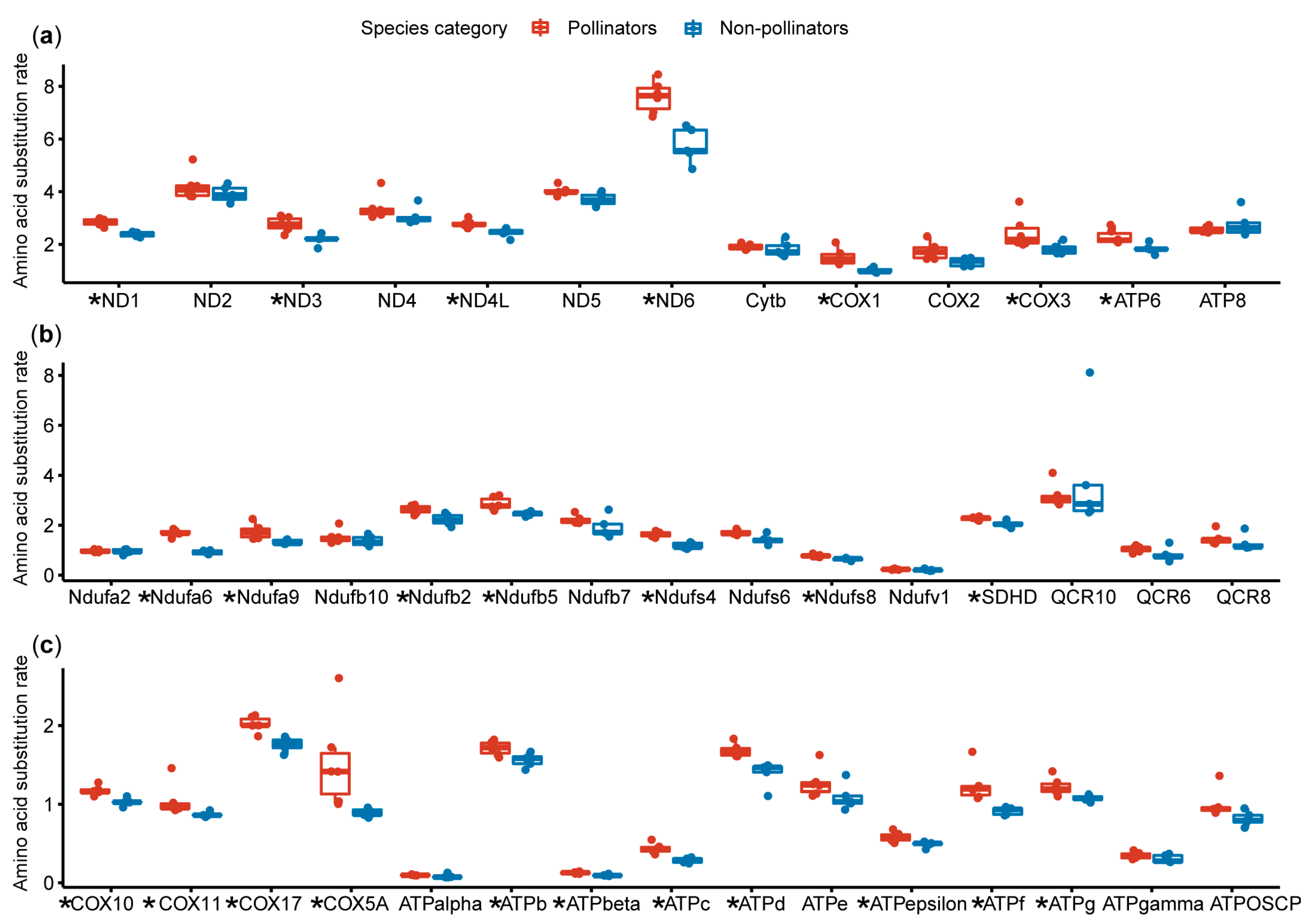
3.2. Amino Acid Substitution Rate Analysis
3.3. Natural Selection Analysis on the OXPHOS Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cruaud, A.; Ronsted, N.; Chantarasuwan, B.; Chou, L.S.; Clement, W.L.; Couloux, A.; Cousins, B.; Genson, G.; Harrison, R.D.; Hanson, P.E. An extreme case of plant-insect codiversification: Figs and fig-pollinating wasps. Syst. Biol. 2012, 61, 1029–1047. [Google Scholar] [CrossRef] [Green Version]
- Borges, R.M. How to be a fig wasp parasite on the fig-fig wasp mutualism. Curr. Opin. Insect Sci. 2015, 8, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Weiblen, G.D. How to be a fig wasp. Annu. Rev. Entomol. 2002, 47, 299–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, R.S.; Niehuis, O.; Gunkel, S.; Bläser, M.; Mayer, C.; Podsiadlowski, L.; Kozlov, A.; Donath, A.; van Noort, S.; Liu, S.; et al. Transcriptome sequence-based phylogeny of chalcidoid wasps (Hymenoptera: Chalcidoidea) reveals a history of rapid radiations, convergence, and evolutionary success. Mol. Phylogenet. Evol. 2018, 120, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Sunnucks, P.; Morales, H.E.; Lamb, A.M.; Pavlova, A.; Greening, C. Integrative approaches for studying mitochondrial and nuclear genome co-evolution in oxidative phosphorylation. Front. Genet. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Signes, A.; Fernandez-Vizarra, E. Assembly of mammalian oxidative phosphorylation complexes I–V and supercomplexes. Essays Biochem. 2018, 62, 255–270. [Google Scholar] [CrossRef]
- Hill, G.E. Mitonuclear ecology. Mol. Biol. Evol. 2015, 32, 1917–1927. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Broughton, R.E. Mitochondrial-nuclear interactions: Compensatory evolution or variable functional constraint among vertebrate oxidative phosphorylation genes? Genome Biol. Evol. 2013, 5, 1781–1791. [Google Scholar] [CrossRef] [Green Version]
- Tripoli, G.; D’Elia, D.; Barsanti, P.; Caggese, C. Comparison of the oxidative phosphorylation (OXPHOS) nuclear genes in the genomes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae. Genome Biol. 2005, 6, R11. [Google Scholar] [CrossRef] [Green Version]
- Gibson, J.D.; Niehuis, O.; Verrelli, B.C.; Gadau, J. Contrasting patterns of selective constraints in nuclear-encoded genes of the oxidative phosphorylation pathway in holometabolous insects and their possible role in hybrid breakdown in Nasonia. Heredity 2010, 104, 310–317. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, R.; Liu, S.; Donath, A.; Peters, R.S.; Ware, J.; Misof, B.; Niehuis, O.; Pfrender, M.E.; Zhou, X. The molecular evolutionary dynamics of oxidative phosphorylation (OXPHOS) genes in Hymenoptera. BMC Evol. Biol. 2017, 17, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.H.; Jia, J.G.; Murphy, R.W.; Huang, D.W. Rapid evolution of the mitochondrial genome in Chalcidoid wasps (Hymenoptera: Chalcidoidea) driven by parasitic lifestyles. PLoS ONE 2011, 6, e26645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.H.; Wang, N.X.; Murphy, R.W.; Cook, J.; Jia, L.Y.; Huang, D.W. Wolbachia infection and dramatic intraspecific mitochondrial DNA divergence in a fig wasp. Evolution 2012, 66, 1907–1916. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Z. The Association between Wolbachia and the Evolution of OXPHOS Genes in a Fig Wasp (Ceratosolen solmsi). Ph.D. Thesis, University of Chinese Academy of Sciences, Beijing, China, 2014. [Google Scholar]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Junier, T.; Zdobnov, E.M. The Newick utilities: High-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 2010, 26, 1669–1670. [Google Scholar] [CrossRef] [Green Version]
- Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 1998, 15, 568–573. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J. Mol. Evol. 1998, 46, 409–418. [Google Scholar] [CrossRef]
- Zhang, J.; Nielsen, R.; Yang, Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wong, W.S.W.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Little, A.G.; Kocha, K.M.; Lougheed, S.C.; Moyes, C.D. Evolution of the nuclear-encoded cytochrome oxidase subunits in vertebrates. Physiol. Genom. 2010, 42, 76–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, R.; Zhang, H.; Kim, J.W.; Shimoda, L.; Dang, C.V.; Semenza, G.L. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 2007, 129, 111–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porplycia, D.; Lau, G.Y.; McDonald, J.; Chen, Z.; Richards, J.G.; Moyes, C.D. Subfunctionalization of COX4 paralogs in fish. Am. J. Physiol. Regul. Integr. C 2017, 312, 671–680. [Google Scholar] [CrossRef]
- Kimura, M. Evolutionary rate at the molecular level. Nature 1968, 217, 624–626. [Google Scholar] [CrossRef]
- Nei, M. The new mutation theory of phenotypic evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 12235–12242. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z. Computational Molecular Evolution; Oxford University Press: New York, NY, USA, 2006; p. 271. [Google Scholar]
- Rhooms, S.-K.; Murari, A.; Goparaju, N.S.V.; Vilanueva, M.; Owusu-Ansah, E. Insights from Drosophila on mitochondrial complex I. Cell. Mol. Life Sci. 2020, 77, 607–618. [Google Scholar] [CrossRef]
- Carr, H.S.; George, G.N.; Winge, D.R. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 2002, 277, 31237–31242. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Ford, H.C.; Carroll, J.; Douglas, C.; Gonzales, E.; Ding, S.; Fearnley, I.M.; Walker, J.E. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc. Natl. Acad. Sci. USA 2018, 115, 2988–2993. [Google Scholar] [CrossRef] [Green Version]
- Gauba, E.; Chen, H.; Guo, L.; Du, H. Cyclophilin D deficiency attenuates mitochondrial F1FO-ATP synthase dysfunction via OSCP in Alzheimer’s disease. Neurobiol. Dis. 2019, 121, 138–147. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Chen, B.; Zhao, D.J.; Kang, L. Functional modulation of mitochondrial cytochrome c oxidase underlies adaptation to high-altitude hypoxia in a Tibetan migratory locust. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.F.; Broughton, R.E. Heterogeneous natural selection on oxidative phosphorylation genes among fishes with extreme high and low aerobic performance. BMC Evol. Biol. 2015, 15, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.-D.; Jiang, G.-F.; Yan, L.-Y.; Li, R.; Mu, Y.; Deng, W.-A. Positive selection drove the adaptation of mitochondrial genes to the demands of flight and high-altitude environments in grasshoppers. Front. Genet. 2018, 9, 605. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene | lnL (Alter) | lnL (Null) | 2ΔlnL | p Value | Positive Sites |
|---|---|---|---|---|---|
| Ndufb5 | −5095.22 | −5100.57 | 10.69 | 0.001 | 75 G ** |
| Ndufb10 | −3997.50 | −4001.73 | 8.45 | 0.004 | 118 N ** |
| COX11 | −4689.52 | −4691.67 | 4.28 | 0.039 | 92 I *, 171 M ** |
| ATPd | −4605.99 | −4608.06 | 4.13 | 0.042 | 47 S *, 129 Q * |
| ATPOSCP | −4263.97 | −4266.42 | 4.89 | 0.027 | 92 G **, 93 T ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Huang, D.; Xin, Z.; Xiao, J. Evolution of Oxidative Phosphorylation (OXPHOS) Genes Reflecting the Evolutionary and Life Histories of Fig Wasps (Hymenoptera, Chalcidoidea). Genes 2020, 11, 1353. https://doi.org/10.3390/genes11111353
Zhou Y, Huang D, Xin Z, Xiao J. Evolution of Oxidative Phosphorylation (OXPHOS) Genes Reflecting the Evolutionary and Life Histories of Fig Wasps (Hymenoptera, Chalcidoidea). Genes. 2020; 11(11):1353. https://doi.org/10.3390/genes11111353
Chicago/Turabian StyleZhou, Yi, Dawei Huang, Zhaozhe Xin, and Jinhua Xiao. 2020. "Evolution of Oxidative Phosphorylation (OXPHOS) Genes Reflecting the Evolutionary and Life Histories of Fig Wasps (Hymenoptera, Chalcidoidea)" Genes 11, no. 11: 1353. https://doi.org/10.3390/genes11111353
APA StyleZhou, Y., Huang, D., Xin, Z., & Xiao, J. (2020). Evolution of Oxidative Phosphorylation (OXPHOS) Genes Reflecting the Evolutionary and Life Histories of Fig Wasps (Hymenoptera, Chalcidoidea). Genes, 11(11), 1353. https://doi.org/10.3390/genes11111353





