Comparative Review of the Responses of Listeria monocytogenes and Escherichia coli to Low pH Stress
Abstract
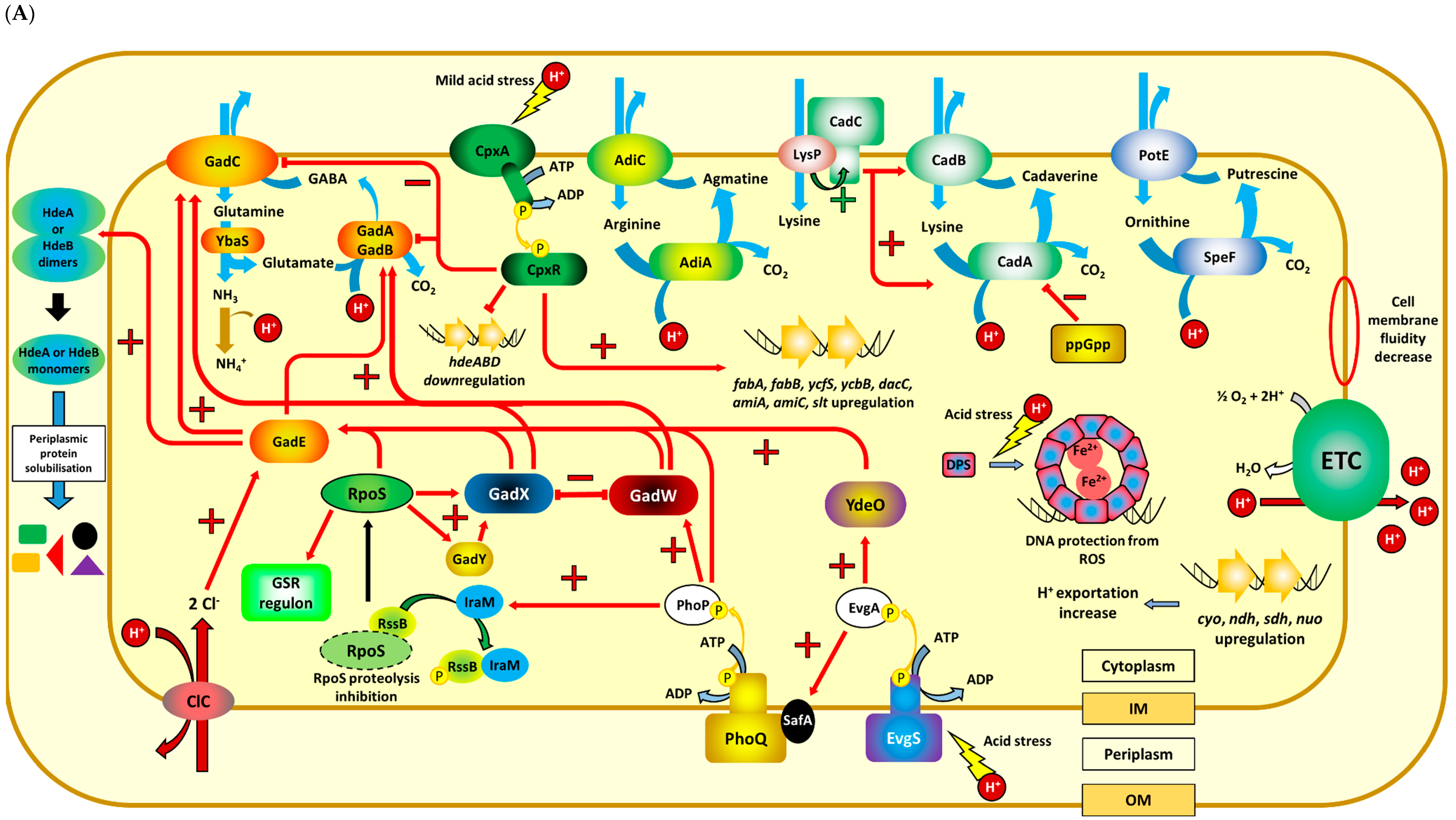
:1. Introduction
2. Maintaining Intracellular pH under Acidic Conditions
3. Protective and Repair Mechanisms against Acid Stress
4. Sensing and Regulatory Processes during Acid Stress
5. Short-Chain Organic Acid Stress
6. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lund, P.A.; De Biase, D.; Liran, O.; Scheler, O.; Mira, N.P.; Cetecioglu, Z.; Fernández, E.N.; Bover-Cid, S.; Hall, R.; Sauer, M.; et al. Understanding How Microorganisms Respond to Acid pH Is Central to Their Control and Successful Exploitation. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Ishii, S.; Ksoll, W.B.; Hicks, R.E.; Sadowsky, M.J. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 2006, 72, 612–621. [Google Scholar] [CrossRef] [Green Version]
- Chiang, S.M.; Dong, T.; Edge, T.A.; Schellhorn, H.E. Phenotypic diversity caused by differential RpoS activity among environmental Escherichia coli isolates. Appl. Environ. Microbiol. 2011, 77, 7915–7923. [Google Scholar] [CrossRef] [Green Version]
- Byappanahalli, M.N.; Yan, T.; Hamilton, M.J.; Ishii, S.; Fujioka, R.S.; Whitman, R.L.; Sadowsky, M.J. The population structure of Escherichia coli isolated from subtropical and temperate soils. Sci. Total Environ. 2012, 417–418, 273–279. [Google Scholar] [CrossRef] [Green Version]
- Brennan, F.P.; Grant, J.; Botting, C.H.; O’Flaherty, V.; Richards, K.G.; Abram, F. Insights into the low-temperature adaptation and nutritional flexibility of a soil-persistent Escherichia coli. FEMS Microbiol. Ecol. 2013, 84, 75–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NicAogáin, K.; O’Byrne, C.P. The Role of Stress and Stress Adaptations in Determining the Fate of the Bacterial Pathogen Listeria monocytogenes in the Food Chain. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorey, A.; Marinho, C.; Piveteau, P.; O’Byrne, C. Role and regulation of the stress activated sigma factor sigma B (σB) in the saprophytic and host-associated life stages of Listeria monocytogenes. Adv. Appl. Microbiol. 2019, 106, 1–48. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, D.N.; Arcari, T.; O’Byrne, C.P. The σB-Mediated General Stress Response of Listeria monocytogenes: Life and Death Decision Making in a Pathogen. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C. Surviving the Acid Test: Responses of Gram-Positive Bacteria to Low pH. Microbiol. Mol. Biol. Rev. 2003, 67, 429–453. [Google Scholar] [CrossRef] [Green Version]
- Kanjee, U.; Houry, W.A. Mechanisms of Acid Resistance in Escherichia coli. Annu. Rev. Microbiol. 2013, 67, 65–81. [Google Scholar] [CrossRef] [Green Version]
- Foster, J.W. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat. Rev. Microbiol. 2004, 2, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Feehily, C.; Karatzas, K.A.G. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 2013, 114, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.K.; Kassam, T.; Singh, B.; Elliott, J.F. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 1992, 174, 5820–5826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Biase, D.; Tramonti, A.; Bossa, F.; Visca, P. The response to stationary-phase stress conditions in Escherichia coli: Role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 1999, 32, 1198–1211. [Google Scholar] [CrossRef] [Green Version]
- Cotter, P.D.; Gahan, C.G.M.; Hill, C. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 2001, 40, 465–475. [Google Scholar] [CrossRef]
- Ryan, S.; Begley, M.; Gahan, C.G.M.; Hill, C. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: Regulation and role in acid tolerance. Environ. Microbiol. 2009, 11, 432–445. [Google Scholar] [CrossRef]
- Chen, J.; Cheng, C.; Xia, Y.; Zhao, H.; Fang, C.; Shan, Y.; Wu, B.; Fang, W. Lmo0036, an ornithine and putrescine carbamoyltransferase in Listeria monocytogenes, participates in arginine deiminase and agmatine deiminase pathways and mediates acid tolerance. Microbiology 2011, 157, 3150–3161. [Google Scholar] [CrossRef] [Green Version]
- Meng, S.Y.; Bennett, G.N. Nucleotide sequence of the Escherichia coli cad operon: A system for neutralization of low extracellular pH. J. Bacteriol. 1992, 174, 2659–2669. [Google Scholar] [CrossRef] [Green Version]
- Kashiwagi, K.; Suzuki, T.; Suzuki, F.; Furuchi, T.; Kobayashi, H.; Igarashi, K. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J. Biol. Chem. 1991, 266, 20922–20927. [Google Scholar]
- Xiao, Z.; Xu, P. Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 2007, 33, 127–140. [Google Scholar] [CrossRef]
- Castanie-Cornet, M.-P.; Foster, J.W. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 2001, 147, 709–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, P.; Ma, D.; Chen, Y.; Guo, Y.; Chen, G.-Q.; Deng, H.; Shi, Y. L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 2013, 23, 635–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karatzas, K.-A.G.; Suur, L.; O’Byrne, C.P. Characterization of the Intracellular Glutamate Decarboxylase System: Analysis of Its Function, Transcription, and Role in the Acid Resistance of Various Strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2012, 78, 3571–3579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, S.; Begley, M.; Hill, C.; Gahan, C.G.M. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 2010, 109, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ryan, S.; Gahan, C.G.M.; Hill, C. Presence of GadD1 Glutamate Decarboxylase in Selected Listeria monocytogenes Strains Is Associated with an Ability To Grow at Low pH. Appl. Environ. Microbiol. 2005, 71, 2832–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karatzas, K.-A.G.; Brennan, O.; Heavin, S.; Morrissey, J.; O’Byrne, C.P. Intracellular Accumulation of High Levels of γ-Aminobutyrate by Listeria monocytogenes 10403S in Response to Low pH: Uncoupling of γ-Aminobutyrate Synthesis from Efflux in a Chemically Defined Medium. Appl. Environ. Microbiol. 2010, 76, 3529–3537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuhaus, K.; Satorhelyi, P.; Schauer, K.; Scherer, S.; Fuchs, T.M. Acid shock of Listeria monocytogenes at low environmental temperatures induces prfA, epithelial cell invasion, and lethality towards Caenorhabditis elegans. BMC Genom. 2013, 14, 285. [Google Scholar] [CrossRef] [Green Version]
- Wemekamp-Kamphuis, H.H.; Wouters, J.A.; de Leeuw, P.P.L.A.; Hain, T.; Chakraborty, T.; Abee, T. Identification of Sigma Factor σB-Controlled Genes and Their Impact on Acid Stress, High Hydrostatic Pressure, and Freeze Survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 2004, 70, 3457–3466. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Lee, I.S.; Frey, J.; Slonczewski, J.L.; Foster, J.W. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 1995, 177, 4097–4104. [Google Scholar] [CrossRef] [Green Version]
- Castanie-Cornet, M.-P.; Penfound, T.A.; Smith, D.; Elliott, J.F.; Foster, J.W. Control of Acid Resistance in Escherichia coli. J. Bacteriol. 1999, 181, 3525–3535. [Google Scholar] [CrossRef] [Green Version]
- Gong, S.; Richard, H.; Foster, J.W. YjdE (AdiC) Is the Arginine:Agmatine Antiporter Essential for Arginine-Dependent Acid Resistance in Escherichia coli. J. Bacteriol. 2003, 185, 4402–4409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, R.; Williams, C.; Miller, C. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J. Bacteriol. 2003, 185, 6556–6561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.; Chen, J.; Shan, Y.; Fang, C.; Liu, Y.; Xia, Y.; Song, H.; Fang, W. Listeria monocytogenes ArcA contributes to acid tolerance. J. Med Microbiol. 2013, 62, 813–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotter, P.; Gahan, C.; Hill, C. Analysis of the role of the Listeria monocytogenes F0F1-ATPase operon in the acid tolerance response. Int. J. Food Microbiol. 2000, 60, 137–146. [Google Scholar] [CrossRef]
- Cheng, C.; Chen, J.; Fang, C.; Xia, Y.; Shan, Y.; Liu, Y.; Wen, G.; Song, H.; Fang, W. Listeria monocytogenes aguA1, but Not aguA2, Encodes a Functional Agmatine Deiminase. J. Biol. Chem. 2013, 288, 26606–26615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, C.A.; Knuckley, B. Mechanistic studies of the agmatine deiminase from Listeria monocytogenes. Biochem. J. 2016, 473, 1553–1561. [Google Scholar] [CrossRef] [Green Version]
- Kanjee, U.; Gutsche, I.; Alexopoulos, E.; Zhao, B.; El Bakkouri, M.; Thibault, G.; Liu, K.; Ramachandran, S.; Snider, J.; Pai, E.F.; et al. Linkage between the bacterial acid stress and stringent responses: The structure of the inducible lysine decarboxylase. EMBO J. 2011, 30, 931–944. [Google Scholar] [CrossRef] [Green Version]
- Soksawatmaekhin, W.; Kuraishi, A.; Sakata, K.; Kashiwagi, K.; Igarashi, K. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol. Microbiol. 2004, 51, 1401–1412. [Google Scholar] [CrossRef]
- Kanjee, U.; Gutsche, I.; Ramachandran, S.; Houry, W.A. The enzymatic activities of the Escherichia coli basic aliphatic amino acid decarboxylases exhibit a pH zone of inhibition. Biochemistry 2011, 50, 9388–9398. [Google Scholar] [CrossRef]
- Bowman, J.P.; Lee Chang, K.J.; Pinfold, T.; Ross, T. Transcriptomic and Phenotypic Responses of Listeria monocytogenes Strains Possessing Different Growth Efficiencies under Acidic Conditions. Appl. Environ. Microbiol. 2010, 76, 4836–4850. [Google Scholar] [CrossRef] [Green Version]
- Stasiewicz, M.J.; Wiedmann, M.; Bergholz, T.M. The transcriptional response of Listeria monocytogenes during adaptation to growth on lactate and diacetate includes synergistic changes that increase fermentative Acetoin production. Appl. Environ. Microbiol. 2011, 77, 5294–5306. [Google Scholar] [CrossRef] [Green Version]
- Madeo, M.; O’Riordan, N.; Fuchs, T.; Utratna, M.; Karatzas, K.; O’Byrne, C. Thiamine plays a critical role in the acid tolerance of Listeria monocytogenes. FEMS Microbiol. Lett. 2011, 326, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, H.; Foster, J.W. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 2004, 186, 6032–6041. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Fukamachi, T.; Saito, H.; Kobayashi, H. Respiration and the F1Fo-ATPase Enhance Survival under Acidic Conditions in Escherichia coli. PLoS ONE 2012, 7, e52577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurer, L.M.; Yohannes, E.; Bondurant, S.S.; Radmacher, M.; Slonczewski, J.L. pH Regulates Genes for Flagellar Motility, Catabolism, and Oxidative Stress in Escherichia coli K-12. J. Bacteriol. 2005, 187, 304–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, R.; Iverson, T.M.; Accardi, A.; Miller, C. A biological role for prokaryotic ClC chloride channels. Nature 2002, 419, 715–718. [Google Scholar] [CrossRef]
- Accardi, A.; Miller, C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl- channels. Nature 2004, 427, 803–807. [Google Scholar] [CrossRef]
- Gut, H.; Pennacchietti, E.; John, R.A.; Bossa, F.; Capitani, G.; De Biase, D.; Grütter, M.G. Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB. EMBO J. 2006, 25, 2643–2651. [Google Scholar] [CrossRef]
- Brown, J.L.; Ross, T.; McMeekin, T.A.; Nichols, P.D. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance. Int. J. Food Microbiol. 1997, 37, 163–173. [Google Scholar] [CrossRef]
- Fozo, E.M.; Quivey, R.G. Shifts in the Membrane Fatty Acid Profile of Streptococcus mutans Enhance Survival in Acidic Environments. Appl. Environ. Microbiol. 2004, 70, 929–936. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.H.; Kim, S.; Kim, H.G.; Lee, J.; Lee, I.S.; Park, Y.K. The formation of cyclopropane fatty acids in Salmonella enterica serovar Typhimurium. Microbiology 2005, 151, 209–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.-Y.; Cronan, J.E. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 1999, 33, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Shabala, L.; Ross, T. Cyclopropane fatty acids improve Escherichia coli survival in acidified minimal media by reducing membrane permeability to H+ and enhanced ability to extrude H+. Res. Microbiol. 2008, 159, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.W.; Cronan, J.E. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 1997, 61, 429–441. [Google Scholar] [CrossRef]
- Hari, S.B.; Grant, R.A.; Sauer, R.T. Structural and Functional Analysis of E. coli Cyclopropane Fatty Acid Synthase. Structure 2018, 26, 1251–1258. [Google Scholar] [CrossRef] [Green Version]
- Poger, D.; Mark, A.E. A ring to rule them all: The effect of cyclopropane Fatty acids on the fluidity of lipid bilayers. J. Phys. Chem. B 2015, 119, 5487–5495. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, B.; Zhang, X.; Yang, Y.; Hardwidge, P.R.; Ren, W.; Zhu, G. The role of bacterial cell envelope structures in acid stress resistance in E. coli. Appl. Microbiol. Biotechnol. 2020, 104, 2911–2921. [Google Scholar] [CrossRef]
- Giotis, E.S.; McDowell, D.A.; Blair, I.S.; Wilkinson, B.J. Role of Branched-Chain Fatty Acids in pH Stress Tolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 2007, 73, 997–1001. [Google Scholar] [CrossRef] [Green Version]
- Mastronicolis, S.K.; Berberi, A.; Diakogiannis, I.; Petrova, E.; Kiaki, I.; Baltzi, T.; Xenikakis, P. Alteration of the phospho- or neutral lipid content and fatty acid composition in Listeria monocytogenes due to acid adaptation mechanisms for hydrochloric, acetic and lactic acids at pH 5.5 or benzoic acid at neutral pH. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2010, 98, 307–316. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhao, Z.; Tong, W.; Ding, Y.; Liu, B.; Shi, Y.; Wang, J.; Sun, S.; Liu, M.; Wang, Y.; et al. An acid-tolerance response system protecting exponentially growing Escherichia coli. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Nikaido, H.; Vaara, M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985, 49, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajiwala, K.S.; Burley, S.K. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria 1 1Edited by P. E. Wright. J. Mol. Biol. 2000, 295, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Kern, R.; Malki, A.; Abdallah, J.; Tagourti, J.; Richarme, G. Escherichia coli HdeB Is an Acid Stress Chaperone. J. Bacteriol. 2007, 189, 603–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malki, A.; Le, H.-T.; Milles, S.; Kern, R.; Caldas, T.; Abdallah, J.; Richarme, G. Solubilization of Protein Aggregates by the Acid Stress Chaperones HdeA and HdeB. J. Biol. Chem. 2008, 283, 13679–13687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stull, F.; Hipp, H.; Stockbridge, R.B.; Bardwell, J.C.A. In vivo chloride concentrations surge to proteotoxic levels during acid stress. Nat. Chem. Biol. 2018, 14, 1051–1058. [Google Scholar] [CrossRef]
- Foit, L.; George, J.S.; Zhang, B.W.; Brooks, C.L.; Bardwell, J.C.A. Chaperone activation by unfolding. Proc. Natl. Acad. Sci. USA 2013, 110, E1254–E1262. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.-C.; Yang, C.; Ding, J.; Niu, X.; Hu, Y.; Jin, C. Characterizations of the Interactions between Escherichia coli Periplasmic Chaperone HdeA and Its Native Substrates during Acid Stress. Biochemistry 2017, 56, 5748–5757. [Google Scholar] [CrossRef]
- Dahl, J.-U.; Koldewey, P.; Salmon, L.; Horowitz, S.; Bardwell, J.C.A.; Jakob, U. HdeB Functions as an Acid-protective Chaperone in Bacteria. J. Biol. Chem. 2015, 290, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Mujacic, M.; Baneyx, F. Chaperone Hsp31 contributes to acid resistance in stationary-phase Escherichia coli. Appl. Environ. Microbiol. 2007, 73, 1014–1018. [Google Scholar] [CrossRef] [Green Version]
- Hanawa, T.; Fukuda, M.; Kawakamis, H.; Hirano, H.; Kamiya, S.; Yamamoto, T. The Listeria monocytogenes DnaK chaperone is required for stress tolerance and efficient phagocytosis with macrophages. Cell Stress Chaperones 1999, 4, 118–128. [Google Scholar] [CrossRef] [Green Version]
- Chiancone, E.; Ceci, P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim. et Biophys. Acta (BBA) Gen. Subj. 2010, 1800, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Baumler, D.J.; Kaspar, C.W. Contribution of dps to Acid Stress Tolerance and Oxidative Stress Tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 2000, 66, 3911–3916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, S.; Finkel, S.E. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 2004, 186, 4192–4198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, K.; Hung, K.; Baumler, D.J.; Byrd, J.J.; Kaspar, C.W. Acid stress damage of DNA is prevented by Dps binding in Escherichia coli O157:H7. BMC Microbiol. 2008, 8, 181. [Google Scholar] [CrossRef] [Green Version]
- Altuvia, S.; Almirón, M.; Huisman, G.; Kolter, R.; Storz, G. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol. 1994, 13, 265–272. [Google Scholar] [CrossRef]
- Grant, R.A.; Filman, D.J.; Finkel, S.E.; Kolter, R.; Hogle, J.M. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 1998, 5, 294–303. [Google Scholar] [CrossRef]
- Calhoun, L.N.; Kwon, Y.M. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: A review: Escherichia coli Dps protein. J. Appl. Microbiol. 2011, 110, 375–386. [Google Scholar] [CrossRef]
- Bellapadrona, G.; Ardini, M.; Ceci, P.; Stefanini, S.; Chiancone, E. Dps proteins prevent Fenton-mediated oxidative damage by trapping hydroxyl radicals within the protein shell. Free Radic. Biol. Med. 2010, 48, 292–297. [Google Scholar] [CrossRef]
- Karas, V.O.; Westerlaken, I.; Meyer, A.S. The DNA-Binding Protein from Starved Cells (Dps) Utilizes Dual Functions To Defend Cells against Multiple Stresses. J. Bacteriol. 2015, 197, 3206–3215. [Google Scholar] [CrossRef] [Green Version]
- Hébraud, M.; Guzzo, J. The main cold shock protein of Listeria monocytogenes belongs to the family of ferritin-like proteins. FEMS Microbiol. Lett. 2000, 190, 29–34. [Google Scholar] [CrossRef]
- Olsen, K.N.; Larsen, M.H.; Gahan, C.G.M.; Kallipolitis, B.; Wolf, X.A.; Rea, R.; Hill, C.; Ingmer, H. The Dps-like protein Fri of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiology 2005, 151, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Dussurget, O.; Dumas, E.; Archambaud, C.; Chafsey, I.; Chambon, C.; Hébraud, M.; Cossart, P. Listeria monocytogenes ferritin protects against multiple stresses and is required for virulence. FEMS Microbiol. Lett. 2005, 250, 253–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krawczyk-Balska, A.; Lipiak, M. Critical Role of a Ferritin-Like Protein in the Control of Listeria monocytogenes Cell Envelope Structure and Stability under β-lactam Pressure. PLoS ONE 2013, 8, e77808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milecka, D.; Samluk, A.; Wasiak, K.; Krawczyk-Balska, A. An essential role of a ferritin-like protein in acid stress tolerance of Listeria monocytogenes. Arch. Microbiol. 2015, 197, 347–351. [Google Scholar] [CrossRef] [Green Version]
- Polidoro, M.; De Biase, D.; Montagnini, B.; Guarrera, L.; Cavallo, S.; Valenti, P.; Stefanini, S.; Chiancone, E. The expression of the dodecameric ferritin in Listeria spp. is induced by iron limitation and stationary growth phase. Gene 2002, 296, 121–128. [Google Scholar] [CrossRef]
- Butala, M.; Zgur-Bertok, D.; Busby, S.J.W. The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 2009, 66, 82–93. [Google Scholar] [CrossRef]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef] [Green Version]
- Sousa, F.J.R.; Lima, L.M.T.R.; Pacheco, A.B.F.; Oliveira, C.L.P.; Torriani, I.; Almeida, D.F.; Foguel, D.; Silva, J.L.; Mohana-Borges, R. Tetramerization of the LexA repressor in solution: Implications for gene regulation of the E.coli SOS system at acidic pH. J. Mol. Biol. 2006, 359, 1059–1074. [Google Scholar] [CrossRef]
- Van der Veen, S.; van Schalkwijk, S.; Molenaar, D.; de Vos, W.M.; Abee, T.; Wells-Bennik, M.H.J. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology (Read. Engl.) 2010, 156, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Wiedmann, M.; Arvik, T.J.; Hurley, R.J.; Boor, K.J. General Stress Transcription Factor ςB and Its Role in Acid Tolerance and Virulence ofListeria monocytogenes. J. Bacteriol. 1998, 180, 3650–3656. [Google Scholar] [CrossRef] [Green Version]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-Mediated General Stress Response in Escherichia coli. Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, G.T.; Bonocora, R.P.; Schep, A.N.; Beeler, S.M.; Fong, A.J.L.; Shull, L.M.; Batachari, L.E.; Dillon, M.; Evans, C.; Becker, C.J.; et al. Genome-Wide Transcriptional Response to Varying RpoS Levels in Escherichia coli K-12. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eguchi, Y.; Utsumi, R. Alkali metals in addition to acidic pH activate the EvgS histidine kinase sensor in Escherichia coli. J. Bacteriol. 2014, 196, 3140–3149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eguchi, Y.; Utsumi, R. Two-component Systems in Sensing and Adapting to Acid Stress in Escherichia Coli. In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; de Bruijn, F.J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 927–934. ISBN 978-1-119-00481-3. [Google Scholar]
- Sen, H.; Aggarwal, N.; Ishionwu, C.; Hussain, N.; Parmar, C.; Jamshad, M.; Bavro, V.N.; Lund, P.A. Structural and Functional Analysis of the Escherichia coli Acid-Sensing Histidine Kinase EvgS. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.D.; Bell, J.; Clarke, K.; Chandler, R.; Pathak, P.; Xia, Y.; Marshall, R.L.; Weinstock, G.M.; Loman, N.J.; Winn, P.J.; et al. Characterization of mutations in the PAS domain of the EvgS sensor kinase selected by laboratory evolution for acid resistance in Escherichia coli. Mol. Microbiol. 2014, 93, 911–927. [Google Scholar] [CrossRef] [Green Version]
- Masuda, N.; Church, G.M. Regulatory network of acid resistance genes in Escherichia coli: Regulatory network of acid resistance genes in E. coli. Mol. Microbiol. 2003, 48, 699–712. [Google Scholar] [CrossRef]
- Ma, Z.; Masuda, N.; Foster, J.W. Characterization of EvgAS-YdeO-GadE Branched Regulatory Circuit Governing Glutamate-Dependent Acid Resistance in Escherichia coli. J. Bacteriol. 2004, 186, 7378–7389. [Google Scholar] [CrossRef] [Green Version]
- Itou, J.; Eguchi, Y.; Utsumi, R. Molecular Mechanism of Transcriptional Cascade Initiated by the EvgS/EvgA System in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 2009, 73, 870–878. [Google Scholar] [CrossRef]
- Ma, Z.; Gong, S.; Richard, H.; Tucker, D.L.; Conway, T.; Foster, J.W. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 2003, 49, 1309–1320. [Google Scholar] [CrossRef] [Green Version]
- Hommais, F.; Krin, E.; Coppée, J.-Y.; Lacroix, C.; Yeramian, E.; Danchin, A.; Bertin, P. GadE (YhiE): A novel activator involved in the response to acid environment in Escherichia coli. Microbiology 2004, 150, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Tramonti, A.; De Canio, M.; Delany, I.; Scarlato, V.; De Biase, D. Mechanisms of Transcription Activation Exerted by GadX and GadW at the gadA and gadBC Gene Promoters of the Glutamate-Based Acid Resistance System in Escherichia coli. J. Bacteriol. 2006, 188, 8118–8127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayed, A.K.; Odom, C.; Foster, J.W. The Escherichia coli AraC-family regulators GadX and GadW activate gadE, the central activator of glutamate-dependent acid resistance. Microbiology 2007, 153, 2584–2592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hommais, F.; Krin, E.; Laurent-Winter, C.; Soutourina, O.; Malpertuy, A.; Le Caer, J.P.; Danchin, A.; Bertin, P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001, 40, 20–36. [Google Scholar] [CrossRef]
- Krin, E.; Danchin, A.; Soutourina, O. Decrypting the H-NS-dependent regulatory cascade of acid stress resistance in Escherichia coli. BMC Microbiol. 2010, 10, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiensuu, T.; Guerreiro, D.N.; Oliveira, A.H.; O’Byrne, C.; Johansson, J. Flick of a switch: Regulatory mechanisms allowing Listeria monocytogenes to transition from a saprophyte to a killer. Microbiology 2019, 165, 819–833. [Google Scholar] [CrossRef]
- Ribeiro, V.B.; Mujahid, S.; Orsi, R.H.; Bergholz, T.M.; Wiedmann, M.; Boor, K.J.; Destro, M.T. Contributions of σB and PrfA to Listeria monocytogenes salt stress under food relevant conditions. Int. J. Food Microbiol. 2014, 177, 98–108. [Google Scholar] [CrossRef]
- Liu, Y.; Orsi, R.H.; Boor, K.J.; Wiedmann, M.; Guariglia-Oropeza, V. Home Alone: Elimination of All but One Alternative Sigma Factor in Listeria monocytogenes Allows Prediction of New Roles for σB. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Cortes, B.W.; Naditz, A.L.; Anast, J.M.; Schmitz-Esser, S. Transcriptome Sequencing of Listeria monocytogenes Reveals Major Gene Expression Changes in Response to Lactic Acid Stress Exposure but a Less Pronounced Response to Oxidative Stress. Front. Microbiol. 2020, 10, 3110. [Google Scholar] [CrossRef]
- Ferreira, A.; Sue, D.; O’Byrne, C.P.; Boor, K.J. Role of Listeria monocytogenes σBin survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 2003, 69, 2692–2698. [Google Scholar] [CrossRef] [Green Version]
- Kazmierczak, M.J.; Mithoe, S.C.; Boor, K.J.; Wiedmann, M. Listeria monocytogenes σB Regulates Stress Response and Virulence Functions. J. Bacteriol. 2003, 185, 5722–5734. [Google Scholar] [CrossRef] [Green Version]
- Raengpradub, S.; Wiedmann, M.; Boor, K.J. Comparative Analysis of the σB-Dependent Stress Responses in Listeria monocytogenes and Listeria innocua Strains Exposed to Selected Stress Conditions. AEM 2008, 74, 158–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, A.; Gray, M.; Wiedmann, M.; Boor, K.J. Comparative Genomic Analysis of the sigB Operon in Listeria monocytogenes and in Other Gram-Positive Bacteria. Curr. Microbiol. 2004, 48, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Vijay, K.; Brody, M.S.; Fredlund, E.; Price, C.W. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 2000, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Been, M.D.; Tempelaars, M.H.; Schaik, W.V.; Moezelaar, R.; Siezen, R.J.; Abee, T. A novel hybrid kinase is essential for regulating the σB-mediated stress response of Bacillus cereus. Environ. Microbiol. 2010, 12, 730–745. [Google Scholar] [CrossRef]
- Impens, F.; Rolhion, N.; Radoshevich, L.; Bécavin, C.; Duval, M.; Mellin, J.; García del Portillo, F.; Pucciarelli, M.G.; Williams, A.H.; Cossart, P. N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat. Microbiol. 2017, 2, 1–12. [Google Scholar] [CrossRef]
- Gao, X.; Lu, F.; Zhou, L.; Dang, S.; Sun, L.; Li, X.; Wang, J.; Shi, Y. Structure and mechanism of an amino acid antiporter. Science 2009, 324, 1565–1568. [Google Scholar] [CrossRef]
- Capitani, G.; De Biase, D.; Aurizi, C.; Gut, H.; Bossa, F.; Grütter, M.G. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 2003, 22, 4027–4037. [Google Scholar] [CrossRef] [Green Version]
- Andréll, J.; Hicks, M.G.; Palmer, T.; Carpenter, E.P.; Iwata, S.; Maher, M.J. Crystal structure of the acid-induced arginine decarboxylase from Escherichia coli: Reversible decamer assembly controls enzyme activity. Biochemistry 2009, 48, 3915–3927. [Google Scholar] [CrossRef]
- Boeker, E.A.; Snell, E.E. Arginine Decarboxylase from Escherichia coli II. DISSOCIATION AND REASSOCIATION OF SUBUNITS. J. Biol. Chem. 1968, 243, 1678–1684. [Google Scholar]
- Pennacchietti, E.; Lammens, T.M.; Capitani, G.; Franssen, M.C.R.; John, R.A.; Bossa, F.; Biase, D.D. Mutation of His465 Alters the pH-dependent Spectroscopic Properties of Escherichia coli Glutamate Decarboxylase and Broadens the Range of Its Activity toward More Alkaline pH. J. Biol. Chem. 2009, 284, 31587–31596. [Google Scholar] [CrossRef] [Green Version]
- Jun, C.; Joo, J.C.; Lee, J.H.; Kim, Y.H. Thermostabilization of glutamate decarboxylase B from Escherichia coli by structure-guided design of its pH-responsive N-terminal interdomain. J. Biotechnol. 2014, 174, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Lu, P.; Yan, C.; Fan, C.; Yin, P.; Wang, J.; Shi, Y. Structure and mechanism of a glutamate–GABA antiporter. Nature 2012, 483, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhou, L.; Jiao, X.; Lu, F.; Yan, C.; Zeng, X.; Wang, J.; Shi, Y. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 2010, 463, 828–832. [Google Scholar] [CrossRef] [PubMed]
- Ilgü, H.; Jeckelmann, J.-M.; Gapsys, V.; Ucurum, Z.; de Groot, B.L.; Fotiadis, D. Insights into the molecular basis for substrate binding and specificity of the wild-type L-arginine/agmatine antiporter AdiC. Proc. Natl. Acad. Sci. USA 2016, 113, 10358–10363. [Google Scholar] [CrossRef] [Green Version]
- Krammer, E.-M.; Prévost, M. Function and Regulation of Acid Resistance Antiporters. J. Membr. Biol. 2019, 252, 465–481. [Google Scholar] [CrossRef] [Green Version]
- De Biase, D.; Lund, P.A. The Escherichia coli Acid Stress Response and Its Significance for Pathogenesis. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 92, pp. 49–88. ISBN 978-0-12-802249-8. [Google Scholar]
- Eichinger, A.; Haneburger, I.; Koller, C.; Jung, K.; Skerra, A. Crystal structure of the sensory domain of Escherichia coli CadC, a member of the ToxR-like protein family. Protein Sci. 2011, 20, 656–669. [Google Scholar] [CrossRef] [Green Version]
- Haneburger, I.; Eichinger, A.; Skerra, A.; Jung, K. New Insights into the Signaling Mechanism of the pH-responsive, Membrane-integrated Transcriptional Activator CadC of Escherichia coli. J. Biol. Chem. 2011, 286, 10681–10689. [Google Scholar] [CrossRef] [Green Version]
- Tetsch, L.; Koller, C.; Dönhöfer, A.; Jung, K. Detection and function of an intramolecular disulfide bond in the pH-responsive CadC of Escherichia coli. BMC Microbiol. 2011, 11, 74. [Google Scholar] [CrossRef] [Green Version]
- Steffes, C.; Ellis, J.; Wu, J.; Rosen, B.P. The lysP gene encodes the lysine-specific permease. J. Bacteriol. 1992, 174, 3242–3249. [Google Scholar] [CrossRef] [Green Version]
- Neely, M.N.; Dell, C.L.; Olson, E.R. Roles of LysP and CadC in mediating the lysine requirement for acid induction of the Escherichia coli cad operon. J. Bacteriol. 1994, 176, 3278–3285. [Google Scholar] [CrossRef] [Green Version]
- Tetsch, L.; Koller, C.; Haneburger, I.; Jung, K. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol. Microbiol. 2008, 67, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Rauschmeier, M.; Schüppel, V.; Tetsch, L.; Jung, K. New Insights into the Interplay Between the Lysine Transporter LysP and the pH Sensor CadC in Escherichia Coli. J. Mol. Biol. 2014, 426, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Kandiah, E.; Carriel, D.; Perard, J.; Malet, H.; Bacia, M.; Liu, K.; Chan, S.W.S.; Houry, W.A.; Ollagnier de Choudens, S.; Elsen, S.; et al. Structural insights into the Escherichia coli lysine decarboxylases and molecular determinants of interaction with the AAA+ ATPase RavA. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.; Dong, Z.; Han, X.; Sun, J.; Wang, H.; Jiang, L.; Yang, Y.; Ma, T.; Chen, Z.; Yu, J.; et al. Listeria monocytogenes 10403S arginine repressor ArgR finely tunes arginine metabolism regulation under acidic conditions. Front. Microbiol. 2017, 8, 145. [Google Scholar] [CrossRef] [Green Version]
- Cotter, P.D.; Emerson, N.; Gahan, C.G.M.; Hill, C. Identification and Disruption of lisRK, a Genetic Locus Encoding a Two-Component Signal Transduction System Involved in Stress Tolerance and Virulence in Listeria monocytogenes. J. Bacteriol. 1999, 181, 6840–6843. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Smith, M.P.; Chapin, K.C.; Baik, H.S.U.K.; Bennett, G.N.; Foster, J.W. Mechanisms of Acid Resistance in Enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 1996, 62, 3094–3100. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Yang, Y.; Dong, Z.; Wang, X.; Fang, C.; Yang, M.; Sun, J.; Xiao, L.; Fang, W.; Song, H. Listeria monocytogenes varies among strains to maintain intracellular pH homeostasis under stresses by different acids as analyzed by a high-throughput microplate-based fluorometr. Front. Microbiol. 2015, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Salmond, C.V.; Kroll, R.G.; Booth, I.R. The effect of food preservatives on pH homeostasis in Escherichia coli. J. Gen. Microbiol. 1984, 130, 2845–2850. [Google Scholar] [CrossRef] [Green Version]
- Siegumfeldt, H.; Rechinger, K.B.; Jakobsen, M. Use of fluorescence ratio imaging for intracellular pH determination of individual bacterial cells in mixed cultures. Microbiology (Reading) 1999, 145 Pt 7, 1703–1709. [Google Scholar] [CrossRef] [Green Version]
- Booth, I.R.; Cash, P.; O’Byrne, C. Sensing and adapting to acid stress. Antonie van Leeuwenhoek 2002, 81, 33–42. [Google Scholar] [CrossRef]
- Hirshfield, I.N.; Terzulli, S.; O’Byrne, C. Weak Organic Acids: A Panoply of Effects on Bacteria. Sci. Prog. 2003, 86, 245–270. [Google Scholar] [CrossRef] [PubMed]
- Roe, A.J.; McLaggan, D.; Davidson, I.; O’Byrne, C.; Booth, I.R. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 1998, 180, 767–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roe, A.J.; Byrne, C.O.; Mclaggan, D.; Booth, I.R. Inhibition of Escherichia coli growth by acetic acid: A problem with methionine biosynthesis and homocysteine toxicity. Microbiology 2002, 148, 2215–2222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutknecht, J. Salicylates and proton transport through lipid bilayer membranes: A model for salicylate-induced uncoupling and swelling in mitochondria. J. Membr. Biol. 1990, 115, 253–260. [Google Scholar] [CrossRef]
- Lewis, K.; Naroditskaya, V.; Ferrante, A.; Fokina, I. Bacterial resistance to uncouplers. J. Bioenerg. Biomembr. 1994, 26, 639–646. [Google Scholar] [CrossRef]
- Wright, E.; Neethirajan, S.; Warriner, K.; Retterer, S.; Srijanto, B. Single cell swimming dynamics of Listeria monocytogenes using a nanoporous microfluidic platform. Lab. Chip 2014, 14, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Alakomi, H.-L.; Skyttä, E.; Saarela, M.; Mattila-Sandholm, T.; Latva-Kala, K.; Helander, I.M. Lactic Acid Permeabilizes Gram-Negative Bacteria by Disrupting the Outer Membrane. Appl. Environ. Microbiol. 2000, 66, 2001–2005. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.A.; Rhee, M.S. Marked Synergistic Bactericidal Effects and Mode of Action of Medium-Chain Fatty Acids in Combination with Organic Acids against Escherichia coli O157:H7. Appl. Environ. Microbiol. 2013, 79, 6552–6560. [Google Scholar] [CrossRef] [Green Version]
- Ning, Y.; Yan, A.; Yang, K.; Wang, Z.; Li, X.; Jia, Y. Antibacterial activity of phenyllactic acid against Listeria monocytogenes and Escherichia coli by dual mechanisms. Food Chem. 2017, 228, 533–540. [Google Scholar] [CrossRef]
- King, T.; Lucchini, S.; Hinton, J.C.D.; Gobius, K. Transcriptomic Analysis of Escherichia coli O157:H7 and K-12 Cultures Exposed to Inorganic and Organic Acids in Stationary Phase Reveals Acidulant- and Strain-Specific Acid Tolerance Responses. Appl. Environ. Microbiol. 2010, 76, 6514–6528. [Google Scholar] [CrossRef] [Green Version]
- Tessema, G.T.; Møretrø, T.; Snipen, L.; Heir, E.; Holck, A.; Naterstad, K.; Axelsson, L. Microarray-based transcriptome of Listeria monocytogenes adapted to sublethal concentrations of acetic acid, lactic acid, and hydrochloric acid. Can. J. Microbiol. 2012, 58, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-J.; Tsai, T.-Y.; Pan, T.-M. Physiological response and protein expression under acid stress of Escherichia coli O157:H7 TWC01 isolated from Taiwan. J. Agric. Food Chem. 2007, 55, 7182–7191. [Google Scholar] [CrossRef]
- Bowman, J.P.; Hages, E.; Nilsson, R.E.; Kocharunchitt, C.; Ross, T. Investigation of the listeria monocytogenes Scott A acid tolerance response and associated physiological and phenotypic features via whole proteome analysis. J. Proteome Res. 2012, 11, 2409–2426. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C.N.; McElhanon, J.; Lee, A.; Leonhart, R.; Siegele, D.A. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 2001, 183, 2178–2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, M.K.; Rohlin, L.; Kao, K.C.; Liao, J.C. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 2002, 277, 13175–13183. [Google Scholar] [CrossRef] [Green Version]
- Tuite, N.L.; Fraser, K.R.; O’byrne, C.P. Homocysteine toxicity in Escherichia coli is caused by a perturbation of branched-chain amino acid biosynthesis. J. Bacteriol. 2005, 187, 4362–4371. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Zhou, F.; Xing, Q.; Cao, Z.; Liu, J.; Zhu, G. The soluble transhydrogenase UdhA affecting the glutamate-dependent acid resistance system of Escherichia coli under acetate stress. Biol. Open 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.P.; Li, H.; Engmann, M.L.; Bischof, K.M.; Kunka, K.S.; Harris, M.E.; Tancredi, A.C.; Ditmars, F.S.; Basting, P.J.; George, N.S.; et al. Inverted Regulation of Multidrug Efflux Pumps, Acid Resistance, and Porins in Benzoate-Evolved Escherichia coli K-12. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [Green Version]
- Creamer, K.E.; Ditmars, F.S.; Basting, P.J.; Kunka, K.S.; Hamdallah, I.N.; Bush, S.P.; Scott, Z.; He, A.; Penix, S.R.; Gonzales, A.S.; et al. Benzoate- and Salicylate-Tolerant Strains of Escherichia coli K-12 Lose Antibiotic Resistance during Laboratory Evolution. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [Green Version]
- Heavin, S.B.; Brennan, O.M.; Morrissey, J.P.; Byrne, C.P.O. Inhibition of Listeria monocytogenes by acetate, benzoate and sorbate: Weak acid tolerance is not influenced by the glutamate decarboxylase system. Lett. Appl. Microbiol. 2009, 49, 179–185. [Google Scholar] [CrossRef]
- Yoshimoto, T.; Higashi, H.; Kanatani, A.; Lin, X.S.; Nagai, H.; Oyama, H.; Kurazono, K.; Tsuru, D. Cloning and sequencing of the 7 alpha-hydroxysteroid dehydrogenase gene from Escherichia coli HB101 and characterization of the expressed enzyme. J. Bacteriol. 1991, 173, 2173–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thanassi, D.G.; Cheng, L.W.; Nikaido, H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 1997, 179, 2512–2518. [Google Scholar] [CrossRef] [Green Version]
- Glaser, P.; Frangeul, L.; Buchrieser, C.; Rusniok, C.; Amend, A.; Baquero, F.; Berche, P.; Bloecker, H.; Brandt, P.; Chakraborty, T.; et al. Comparative genomics of Listeria species. Science 2001, 294, 849–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begley, M.; Hill, C.; Gahan, C.G.M. Identification and disruption of btlA, a locus involved in bile tolerance and general stress resistance in Listeria monocytogenes. FEMS Microbiol. Lett. 2003, 218, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Quillin, S.J.; Schwartz, K.T.; Leber, J.H. The novel Listeria monocytogenes bile sensor BrtA controls expression of the cholic acid efflux pump MdrT. Mol. Microbiol. 2011, 81, 129–142. [Google Scholar] [CrossRef]
- White, S.J.; McClung, D.M.; Wilson, J.G.; Roberts, B.N.; Donaldson, J.R. Influence of pH on bile sensitivity amongst various strains of Listeria monocytogenes under aerobic and anaerobic conditions. J. Med Microbiol. 2015, 64, 1287–1296. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.M.; Hill, C. Bile Stress Response in Listeria monocytogenes LO28: Adaptation, Cross-Protection, and Identification of Genetic Loci Involved in Bile Resistance. Appl. Environ. Microbiol. 2002, 68, 6005–6012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.L.; Ricke, S.C.; Donaldson, J.R. Establishment of Listeria monocytogenes in the Gastrointestinal Tract. Microorganisms 2019, 7, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimann, E.; Schmid, B.; Stephan, R.; Tasara, T. The alternative sigma factor sigma(L) of L. monocytogenes promotes growth under diverse environmental stresses. Foodborne Pathog. Dis. 2009, 6, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.; Sleator, R.D.; Casey, P.G.; Hill, C.; Gahan, C.G.M. Specific Osmolyte Transporters Mediate Bile Tolerance in Listeria monocytogenes. Infect. Immun. 2009, 77, 4895–4904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sleator, R.D.; Wemekamp-Kamphuis, H.H.; Gahan, C.G.M.; Abee, T.; Hill, C. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 2005, 55, 1183–1195. [Google Scholar] [CrossRef] [PubMed]
- Feehily, C.; Finnerty, A.; Casey, P.G.; Hill, C.; Gahan, C.G.M.; O’Byrne, C.P.; Karatzas, K.-A.G. Divergent Evolution of the Activity and Regulation of the Glutamate Decarboxylase Systems in Listeria monocytogenes EGD-e and 10403S: Roles in Virulence and Acid Tolerance. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paudyal, R.; Barnes, R.H.; Karatzas, K.A.G. A novel approach in acidic disinfection through inhibition of acid resistance mechanisms; Maleic acid-mediated inhibition of glutamate decarboxylase activity enhances acid sensitivity of Listeria monocytogenes. Food Microbiol. 2018, 69, 96–104. [Google Scholar] [CrossRef] [PubMed]

| Mechanism/Response | E. coli | Key References | L. monocytogenes | Key References |
|---|---|---|---|---|
| pH homeostasis | ||||
| Proton consuming reactions | GAD system | [13,14,118,123] | GAD system | [15,23,25,26] |
| ADI system | [31,32,119,124,125] | ADI system | [16,17,33] | |
| AgDI system | [17,35,36] | |||
| Acetoin production | [20,41] | |||
| CadA | [18,37,38,128,129] | |||
| SpeF | [19,39] | |||
| Proton extrusion mechanism | ClC | [46,47] | ||
| ETC | [10,45] | |||
| FoF1-ATPase | [44] | FoF1-ATPase | [34] | |
| Protection and repair | ||||
| Membrane composition | CFAs | [52,53,54,55] | BCFAs | [58,59] |
| Chaperones | HdeA, HdeB | [62,63,67,68] | DnaK | [70] |
| Hsp31 | [69] | |||
| Dps | [72,74,76] | Fri | [80,81,82,84] | |
| DNA damage | SOS response | [86,87,88] | SOS response | [89] |
| Sensing and regulatory | ||||
| Two-component systems | EvgAS | [95,96,97,98,99] | LisRK | [137] |
| PhoQ-PhoP | [99] | |||
| CpxRA | [60] | |||
| Sensory hub | Stressosome | [8,105,115] | ||
| Alternative Sigma factors | RpoS | [91,92] | SigB | [8,106] |
| Regulators | GadE ‘circuit’ | [100,101,102,103] | ArgR | [16,135] |
| Response to short-chain organic acid | ||||
| Lactic acid | [152] | [109,153,155] | ||
| Acetic acid | [144,145,152,156,157,158] | [40,41,153] | ||
| Benzoic acid | [160,161] | [162] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arcari, T.; Feger, M.-L.; Guerreiro, D.N.; Wu, J.; O’Byrne, C.P. Comparative Review of the Responses of Listeria monocytogenes and Escherichia coli to Low pH Stress. Genes 2020, 11, 1330. https://doi.org/10.3390/genes11111330
Arcari T, Feger M-L, Guerreiro DN, Wu J, O’Byrne CP. Comparative Review of the Responses of Listeria monocytogenes and Escherichia coli to Low pH Stress. Genes. 2020; 11(11):1330. https://doi.org/10.3390/genes11111330
Chicago/Turabian StyleArcari, Talia, Marie-Lucie Feger, Duarte N. Guerreiro, Jialun Wu, and Conor P. O’Byrne. 2020. "Comparative Review of the Responses of Listeria monocytogenes and Escherichia coli to Low pH Stress" Genes 11, no. 11: 1330. https://doi.org/10.3390/genes11111330
APA StyleArcari, T., Feger, M.-L., Guerreiro, D. N., Wu, J., & O’Byrne, C. P. (2020). Comparative Review of the Responses of Listeria monocytogenes and Escherichia coli to Low pH Stress. Genes, 11(11), 1330. https://doi.org/10.3390/genes11111330





