Transcriptome Analysis Reveals Functional Diversity in Salivary Glands of Plant Virus Vector, Graminella nigrifrons
Abstract
:1. Introduction
2. Experimental Procedures
2.1. Insects
2.2. Total RNA Isolation and Library Preparation
2.3. Transcriptome Assembly and Differential Expression Analysis
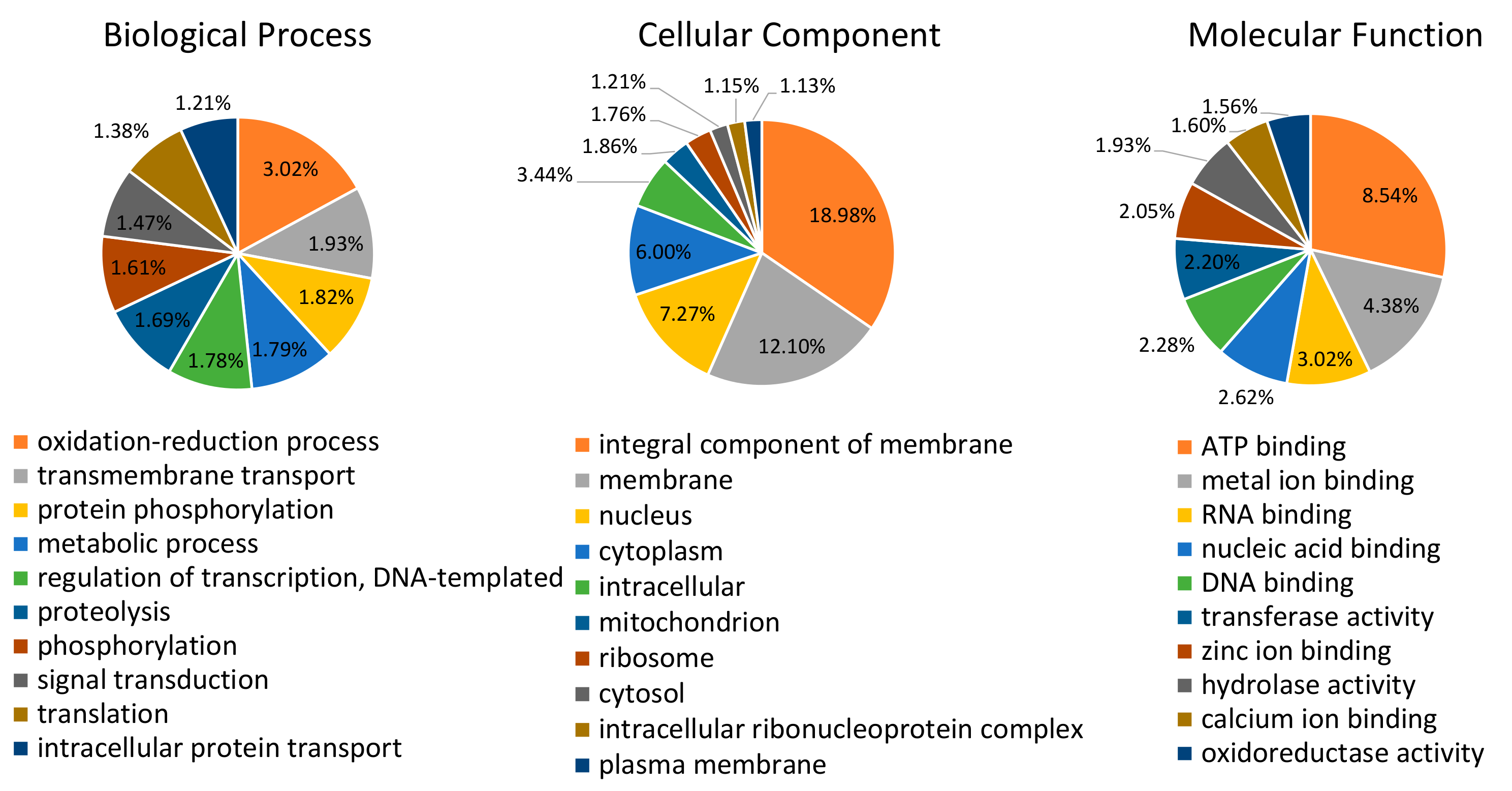
2.4. Functional Annotation of the Salivary Gland Transcriptome
2.5. Real Time Quantitative PCR (RT-qPCR) for Expression Analysis in Maize-Fed vs. Starved Insects
3. Results
3.1. Deep Sequencing and Assembly of the Salivary Gland and Carcass Transcriptomes
3.2. Characterization of the Salivary Gland Transcriptome and its Predicted Proteome
3.3. Differential Expression of Transcripts Between the Salivary Glands and Carcass
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Bernal, J.S.; Medina, R.F. Agriculture sows pests: How crop domestication, host shifts, and agricultural intensification can create insect pests from herbivores. Curr. Opin. Insect Sci. 2018, 26, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, P.R.; Raven, P.H. Butterflies and plants: A study in coevolution. Evolution 1964, 18, 586–608. [Google Scholar] [CrossRef]
- Zhu-Salzman, K.; Bi, J.-L.; Liu, T.-X. Molecular strategies of plant defense and insect counter-defense. Insect Sci. 2005, 12, 3–15. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Van der Hoorn, R.A.L.; Terauchi, R.; Kamoun, S. Emerging Concepts in Effector Biology of Plant-Associated Organisms. MPMI 2009, 22, 115–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogenhout, S.A.; Bos, J.I. Effector proteins that modulate plant–insect interactions. Curr. Opin. Plant Biol. 2011, 14, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Low, P.S.; Merida, J.R. The oxidative burst in plant defense: Function and signal transduction. Physiol. Plant. 1996, 96, 533–542. [Google Scholar] [CrossRef]
- Will, T.; Van Bel, A.J.E. Physical and chemical interactions between aphids and plants. J. Exp. Bot. 2006, 57, 729–737. [Google Scholar] [CrossRef] [Green Version]
- Will, T.; Tjallingii, W.F.; Thönnessen, A.; Van Bel, A.J.E. Molecular sabotage of plant defense by aphid saliva. PNAS 2007, 104, 10536–10541. [Google Scholar] [CrossRef] [Green Version]
- Dietrich, C.H. Chapter 15—Auchenorrhyncha: (Cicadas, Spittlebugs, Leafhoppers, Treehoppers, and Planthoppers). In Encyclopedia of Insects, 2nd ed.; Resh, V.H., Cardé, R.T., Eds.; Academic Press: San Diego, CA, USA, 2009; pp. 56–64. ISBN 978-0-12-374144-8. [Google Scholar]
- Huang, H.-J.; Lu, J.-B.; Li, Q.; Bao, Y.-Y.; Zhang, C.-X. Combined transcriptomic/proteomic analysis of salivary gland and secreted saliva in three planthopper species. J. Proteom. 2018, 172, 25–35. [Google Scholar] [CrossRef]
- Carolan, J.C.; Caragea, D.; Reardon, K.T.; Mutti, N.S.; Dittmer, N.; Pappan, K.; Cui, F.; Castaneto, M.; Poulain, J.; Dossat, C.; et al. Predicted Effector Molecules in the Salivary Secretome of the Pea Aphid (Acyrthosiphon pisum): A Dual Transcriptomic/Proteomic Approach. J. Proteome Res. 2011, 10, 1505–1518. [Google Scholar] [CrossRef]
- Stoner, W.N.; Gustin, R.D. Biology of Graminella nigrifrons (Homoptera: Cicadellidae), a Vector of Corn (Maize) Stunt Virus. Ann. Entomol. Soc. Am. 1967, 60, 496–505. [Google Scholar] [CrossRef]
- Arocha-Rosete, Y.; Kent, P.; Agrawal, V.; Hunt, D.; Hamilton, A.; Bertaccini, A.; Scott, J.; Crosby, W.; Michelutti, R. Identification of Graminella nigrifrons as a potential vector for phytoplasmas affecting Prunus and Pyrus species in Canada. Can. J. Plant. Path. 2011, 33, 465–474. [Google Scholar] [CrossRef]
- Granados, R.; Maramorosch, K.; Everett, T.; Pirone, T. Transmission of Corn Stunt Virus by A New Leafhopper Vector Graminella Nigrifrons (Forbes). Contrib. Boyce Thompson Inst. 1966, 23, 275–280. [Google Scholar]
- Nault, L.R.; Gordon, D.T.; Robertson, D.C.; Bradfute, O.E. Host range of maize chlorotic dwarf virus. Plant. Dis. Report. 1976, 60, 374–377. [Google Scholar]
- Redinbaugh, M.G.; Seifers, D.L.; Meulia, T.; Abt, J.J.; Anderson, R.J.; Styer, W.E.; Ackerman, J.; Salomon, R.; Houghton, W.; Creamer, R.; et al. Maize fine streak virus, a New Leafhopper-Transmitted Rhabdovirus. Phytopathology 2002, 92, 1167–1174. [Google Scholar] [CrossRef]
- Todd, J.C.; Ammar, E.-D.; Redinbaugh, M.G.; Hoy, C.; Hogenhout, S.A. Plant Host Range and Leafhopper Transmission of Maize fine streak virus. Phytopathology 2010, 100, 1138–1145. [Google Scholar] [CrossRef] [Green Version]
- Simon, A. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 December 2018).
- Wingett, S. FastQ Screen. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/ (accessed on 1 December 2018).
- Bushnell, B. BBMap Short-Read. Aligner, and Other Bioinformatics Tools; University of California: Berkeley, CA, USA, 2015. [Google Scholar]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-Seq: Reference generation and analysis with Trinity. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.M.; Kelly, S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016, 26, 1134–1144. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.-H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, H. Predicting Secretory Proteins with SignalP. In Protein Function Prediction; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; pp. 59–73. [Google Scholar]
- TMHMM Server v2.0. 2015. Available online: http://www.cbs.dtu.dk/services/TMHMM (accessed on 1 April 2020).
- Armenteros, J.J.A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Cassone, B.J.; Bai, X.; Redinbaugh, M.G.; Michel, A.P. Transcriptome of the Plant Virus Vector Graminella nigrifrons, and the Molecular Interactions of Maize fine streak rhabdovirus Transmission. PLoS ONE 2012, 7, e40613. [Google Scholar] [CrossRef] [Green Version]
- Tassone, E.E.; Cowden, C.C.; Castle, S.J. De novo transcriptome assemblies of four xylem sap-feeding insects. GigaScience 2017, 6, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Tassone, E.E.; Geib, S.M.; Hall, B.; Fabrick, J.A.; Brent, C.S.; Hull, J.J. De novo construction of an expanded transcriptome assembly for the western tarnished plant bug, Lygus hesperus. GigaScience 2016, 5, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, J.I.B.; Prince, D.; Pitino, M.; Maffei, M.E.; Win, J.; Hogenhout, S.A. A Functional Genomics Approach Identifies Candidate Effectors from the Aphid Species Myzus persicae (Green Peach Aphid). PLoS Genet. 2010, 6, e1001216. [Google Scholar] [CrossRef]
- Villarroel, C.A.; Jonckheere, W.; Alba, J.M.; Glas, J.J.; Dermauw, W.; Haring, M.A.; Leeuwen, T.V.; Schuurink, R.C.; Kant, M.R. Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant. J. 2016, 86, 119–131. [Google Scholar] [CrossRef] [Green Version]
- Hattori, M.; Komatsu, S.; Noda, H.; Matsumoto, Y. Proteome Analysis of Watery Saliva Secreted by Green Rice Leafhopper, Nephotettix cincticeps. PLoS ONE 2015, 10, e0123671. [Google Scholar] [CrossRef]
- Yang, C.-H.; Guo, J.-Y.; Chu, D.; Ding, T.-B.; Wei, K.-K.; Cheng, D.-F.; Wan, F.-H. Secretory laccase 1 in Bemisia tabaci MED is involved in whitefly-plant interaction. Sci. Rep. 2017, 7, 3623. [Google Scholar] [CrossRef]
- Ji, R.; Ye, W.; Chen, H.; Zeng, J.; Li, H.; Yu, H.; Li, J.; Lou, Y. A salivary endo-β-1,4-glucanase acts as an effector that enables the brown planthopper to feed on rice. Plant. Physiol. 2017, 173, 1920–1932. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Yu, H.; Jian, Y.; Zeng, J.; Ji, R.; Chen, H.; Lou, Y. A salivary EF-hand calcium-binding protein of the brown planthopper Nilaparvata lugens functions as an effector for defense responses in rice. Sci. Rep. 2017, 7, 40498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, Y.; Suetsugu, Y.; Nakamura, M.; Hattori, M. Transcriptome analysis of the salivary glands of Nephotettix cincticeps (Uhler). J. Insect Physiol. 2014, 71, 170–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellanos, N.; Martínez, L.C.; Silva, E.H.; Teodoro, A.V.; Serrão, J.E.; Oliveira, E.E. Ultrastructural analysis of salivary glands in a phytophagous stink bug revealed the presence of unexpected muscles. PLoS ONE 2017, 12, e0179478. [Google Scholar] [CrossRef]
- Margittai, É.; Sitia, R. Oxidative Protein Folding in the Secretory Pathway and Redox Signaling Across Compartments and Cells. Traffic 2011, 12, 1–8. [Google Scholar] [CrossRef]
- Klowden, M.J. Metabolic Systems, Physiological Systems in Insects. Preface to the Third Edition. In Physiological Systems in Insects, 3rd ed.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2013; pp. 305–323. [Google Scholar]
- Zhu, K.Y.; Merzendorfer, H.; Zhang, W.; Zhang, J.; Muthukrishnan, S. Biosynthesis, Turnover, and Functions of Chitin in Insects. Annu. Rev. Entomol. 2016, 61, 177–196. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, J.; Sun, J.; Francis, F.; Chen, J. Transcriptome analysis of the salivary glands of the grain aphid, Sitobion Avenae. Sci. Rep. 2017, 7, 15911. [Google Scholar] [CrossRef] [Green Version]
- Felton, G.W.; Summers, C.B. Antioxidant systems in insects. Arch. Insect Biochem. Physiol. 1995, 29, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Després, L.; David, J.-P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, C.E.; Shan, G.; Ottea, J. Overview of carboxylesterases and their role in the metabolism of insecticides. J. Pestic. Sci. 2005, 30, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Triplehorn, B.W.; Nault, L.R.; Horn, D.J. Feeding Behavior of Graminella nigrifrons (Forbes). Ann. Entomol. Soc. Am. 1984, 77, 102–107. [Google Scholar] [CrossRef]
- Su, Y.-L.; Li, J.-M.; Li, M.; Luan, J.-B.; Ye, X.-D.; Wang, X.-W.; Liu, S.-S. Transcriptomic Analysis of the Salivary Glands of an Invasive Whitefly. PLoS ONE 2012, 7, e39303. [Google Scholar] [CrossRef]
- Stafford-Banks, C.A.; Rotenberg, D.; Johnson, B.R.; Whitfield, A.E.; Ullman, D.E. Analysis of the Salivary Gland Transcriptome of Frankliniella occidentalis. PLoS ONE 2014, 9, e94447. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; An, X.-K.; Liu, Y.-D.; Hou, M.-L. Transcriptomic and Expression Analysis of the Salivary Glands in White-Backed Planthoppers, Sogatella furcifera. PLoS ONE 2016, 11, e0159393. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Dai, H.; Zhang, Y.; Chandrasekar, R.; Luo, L.; Hiromasa, Y.; Sheng, C.; Peng, G.; Chen, S.; Tomich, J.M.; et al. Armet is an effector protein mediating aphid–plant interactions. FASEB J. 2015, 29, 2032–2045. [Google Scholar] [CrossRef]

| Assembly Parameters | Salivary Gland Transcriptome | Reference Transcriptome |
|---|---|---|
| Number of sequences | 69,641 | 136,865 |
| Length of the sequences | 201–10,795 | 201–10,795 |
| Number of bases | 37,960,148 | 81,773,441 |
| Number of transcripts with an Open Reading Frame | 9102 | 22,283 |
| N50 * | 701 | 822 |
| GC content | 0.375 | 0.378 |
| Enriched Molecular Function Gene Ontology | Contig ID | Putative Blast Description |
|---|---|---|
| Hydrolase activity, acting on ester bonds | TRINITY_DN26981_c0_g1_i1 | DNA repair protein RAD50-like |
| TRINITY_DN29993_c0_g1_i1 | Tyrosine-protein phosphatase 10D isoform X1 | |
| TRINITY_DN16434_c1_g1_i1 | Acid phosphatase-1 | |
| TRINITY_DN24127_c0_g1_i1 | Phosphatidyl glycerophosphatase and protein-tyrosine phosphatase 1 | |
| TRINITY_DN26134_c0_g3_i1 | Pancreatic lipase-related protein 2-like | |
| Hydrolase activity, acting on glycosyl bonds | TRINITY_DN16637_c1_g1_i1 | Cyclin-dependent kinase 8-like |
| TRINITY_DN26139_c0_g7_i1 | α-N-acetylgalactosaminidase | |
| TRINITY_DN24554_c0_g1_i1 | Cellulose-binding protein | |
| TRINITY_DN21305_c0_g1_i2 | Pre-mRNA branch site protein p14 | |
| Chitin binding | TRINITY_DN18138_c0_g1_i1 | Chitin deacetylase 4 |
| TRINITY_DN17883_c0_g1_i1 | Chitin deacetylase 3 | |
| TRINITY_DN21305_c0_g1_i2 | Pre-mRNA branch site protein p14 | |
| Peptidase activity | TRINITY_DN23314_c0_g1_i2 | Cathepsin L |
| TRINITY_DN25538_c0_g1_i1 | Putative serine protease K12H4.7 | |
| TRINITY_DN25934_c0_g1_i2 | Cathepsin B | |
| TRINITY_DN19249_c1_g1_i1 | Putative GPI-anchor transamidase | |
| TRINITY_DN24056_c0_g1_i1 | Calpain-A-like isoform X6 | |
| TRINITY_DN24709_c0_g1_i1 | Cathepsin L | |
| Cation binding | TRINITY_DN26981_c0_g1_i1 | DNA repair protein RAD50-like |
| TRINITY_DN20367_c0_g1_i1 | Transferrin | |
| TRINITY_DN24056_c0_g1_i1 | Calpain-A-like isoform X6 |
| Contig ID | NCBI Reference Accession | Putative Identification | Log2 Fold Change | Padj | SignalP Prediction | Transmembrane Domain Prediction |
|---|---|---|---|---|---|---|
| Cation binding | ||||||
| TRINITY_DN29516_c0_g1_i1 | G9M8X1.1 | Calcium-binding protein SP84 (NcSP84) | 10.94 | 5E-11 | YES | NO |
| TRINITY_DN35919_c0_g1_i1 | XP_031636025.1 | Annexin B9 isoform X1 | 10.15 | 2E-20 | YES | NO |
| TRINITY_DN25027_c0_g1_i1 | XP_028676409.1 | Calmodulin, putative | 10.14 | 1E-29 | YES | NO |
| TRINITY_DN22216_c0_g1_i1 | BBH63273.1 | Laccase-1 Ϯ | 10.01 | 4E-116 | YES | NO |
| TRINITY_DN37325_c2_g6_i1 | XP_016201745.1 | Calmodulin-like protein 1 | 9.35 | 2E-10 | YES | NO |
| TRINITY_DN37587_c0_g1_i4 | XP_012151393.1 | Prolyl 4-hydroxylase subunit α-2 | 9.23 | 2E-09 | NO | - |
| TRINITY_DN39232_c0_g12_i2 | XP_008555414.1 | Angiotensin-converting enzyme-like isoform X1 | 8.83 | 7E-11 | NO | NO |
| TRINITY_DN33957_c0_g1_i1 | BAJ06131.1 | Laccase 1 isoform S | 8.57 | 3E-151 | YES | NO |
| TRINITY_DN19685_c0_g1_i1 | KFM77473.1 | Calcium-binding protein E63-1 Ϯ | 8.38 | 2E-27 | YES | NO |
| TRINITY_DN29669_c0_g1_i4 | XP_022202851.1 | EF-hand calcium-binding domain-containing protein 4A-like isoform X1 | 8.37 | 4E-05 | NO | - |
| TRINITY_DN38312_c0_g3_i1 | XP_026811683.1 | Carbonic anhydrase 2-like | 8.04 | 4E-14 | YES | NO |
| TRINITY_DN29189_c0_g2_i1 | XP_026272268.1 | Prolyl 4-hydroxylase subunit α-1 | 7.91 | 6E-05 | NO | - |
| TRINITY_DN25200_c0_g1_i1 | XP_019167479.1 | Calcium-binding allergen Ole e 8-like | 7.39 | 5E-05 | YES | NO |
| TRINITY_DN17761_c0_g1_i1 | XP_033736401.1 | Calmodulin-like protein 11 | 5.88 | 5E-04 | YES | NO |
| TRINITY_DN13330_c0_g1_i1 | XP_008484676.1 | Uncharacterized protein K02A2.6-like, partial | 5.33 | 1E-01 | NO | NO |
| Hydrolase activity (Ester bonds) | ||||||
| TRINITY_DN38399_c0_g1_i1 | XP_022196409.1 | Protein 5NUC-like | 9.45 | 3E-19 | YES | YES |
| TRINITY_DN35420_c0_g1_i4 | XP_014273713.1 | Alkaline ceramidase 3 | 8.34 | 4E-11 | NO | - |
| TRINITY_DN27013_c0_g1_i1 | XP_026275034.1 | Pancreatic triacylglycerol lipase-like | 12.23 | 5E-18 | YES | NO |
| TRINITY_DN27304_c0_g1_i2 | XP_022834705.1 | Phospholipase A1-like | 9.09 | 7E-14 | YES | NO |
| TRINITY_DN36961_c0_g1_i2 | XP_028657491.1 | Deoxyribonuclease-2-α isoform X2 | 8.73 | 6E-11 | NO | NO |
| TRINITY_DN39210_c0_g1_i1 | XP_008195535.1 | Inactive pancreatic lipase-related protein 1 | 8.33 | 3E-09 | NO | NO |
| TRINITY_DN34634_c0_g2_i2 | XP_021935199.1 | Venom acid phosphatase Acph-1-like | 8.30 | 7E-28 | NO | NO |
| TRINITY_DN40024_c0_g1_i2 | RZC33704.1 | Venom carboxylesterase-6-like | 8.17 | 3E-08 | YES | NO |
| TRINITY_DN38228_c0_g1_i8 | XP_024867821.1 | Phospholipase A1 member A-like isoform X1 | 7.97 | 1E-07 | YES | NO |
| Peptidase activity | ||||||
| TRINITY_DN39232_c0_g8_i1 | XP_029977452.1 | Angiotensin-converting enzyme-like | 11.24 | 7E-17 | NO | NO |
| TRINITY_DN39315_c0_g1_i1 | XP_030371519.1 | Aminopeptidase N-like | 9.80 | 8E-31 | NO | - |
| TRINITY_DN39232_c0_g12_i2 | XP_030751661.1 | Angiotensin-converting enzyme-like isoform X1 | 8.83 | 7E-11 | NO | NO |
| TRINITY_DN33549_c0_g5_i6 | VVC42832.1 | Peptidase S1, PA clan, Serine proteases, trypsin domain | 8.46 | 8E-07 | NO | NO |
| TRINITY_DN41406_c0_g4_i3 | XP_031337350.1 | Lysosomal aspartic protease-like | 8.10 | 2E-07 | YES | NO |
| TRINITY_DN38080_c1_g7_i1 | XP_018910288.1 | Zinc metalloproteinase nas-13-like | 8.04 | 4E-06 | NO | NO |
| TRINITY_DN41406_c0_g4_i1 | VTJ90797.1 | Hypothetical predicted protein | 7.58 | 3E-05 | YES | NO |
| TRINITY_DN32600_c0_g1_i2 | XP_026203206.1 | Pepsin A-like | 6.70 | 5E-03 | YES | NO |
| Others | ||||||
| TRINITY_DN34735_c1_g5_i1 | BAQ94509.1 | NcSP19 Ϯ | 12.29 | 2E-115 | YES | YES |
| TRINITY_DN37500_c0_g1_i1 | BAQ94503.1 | NcSP75 Ϯ | 9.31 | 4E-29 | YES | NO |
| TRINITY_DN24271_c0_g1_i1 | BAQ94508.1 | NcSP22 | 10.77 | 2E-29 | YES | NO |
| TRINITY_DN35988_c0_g2_i1 | WP_103338510.1 | Cellulase family glycosylhydrolase Ϯ | 8.90 | 2E-10 | YES | NO |
| TRINITY_DN34815_c0_g1_i1 | RZF49131.1 | Unknown protein | 10.48 | 4E-12 | NO | - |
| TRINITY_DN27409_c0_g3_i1 | XP_022196219.1 | Uncharacterized protein LOC111053608 | 10.29 | 3E-10 | NO | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajarapu, S.P.; Bansal, R.; Mittapelly, P.; Michel, A. Transcriptome Analysis Reveals Functional Diversity in Salivary Glands of Plant Virus Vector, Graminella nigrifrons. Genes 2020, 11, 1289. https://doi.org/10.3390/genes11111289
Rajarapu SP, Bansal R, Mittapelly P, Michel A. Transcriptome Analysis Reveals Functional Diversity in Salivary Glands of Plant Virus Vector, Graminella nigrifrons. Genes. 2020; 11(11):1289. https://doi.org/10.3390/genes11111289
Chicago/Turabian StyleRajarapu, Swapna Priya, Raman Bansal, Priyanka Mittapelly, and Andrew Michel. 2020. "Transcriptome Analysis Reveals Functional Diversity in Salivary Glands of Plant Virus Vector, Graminella nigrifrons" Genes 11, no. 11: 1289. https://doi.org/10.3390/genes11111289






