Campylobacter jejuni Cas9 Modulates the Transcriptome in Caco-2 Intestinal Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Eukaryotic Cell Maintenance
2.3. Transwell Cellular Assay
2.4. RNA and Microarray Handling for Transcriptomics of C. jejuni Infected Human Cells
2.5. Statistical and Functional Analysis of Microarray Data
2.6. Biological Interpretation of Transcriptome Datasets
2.7. Pathway Analysis
3. Results
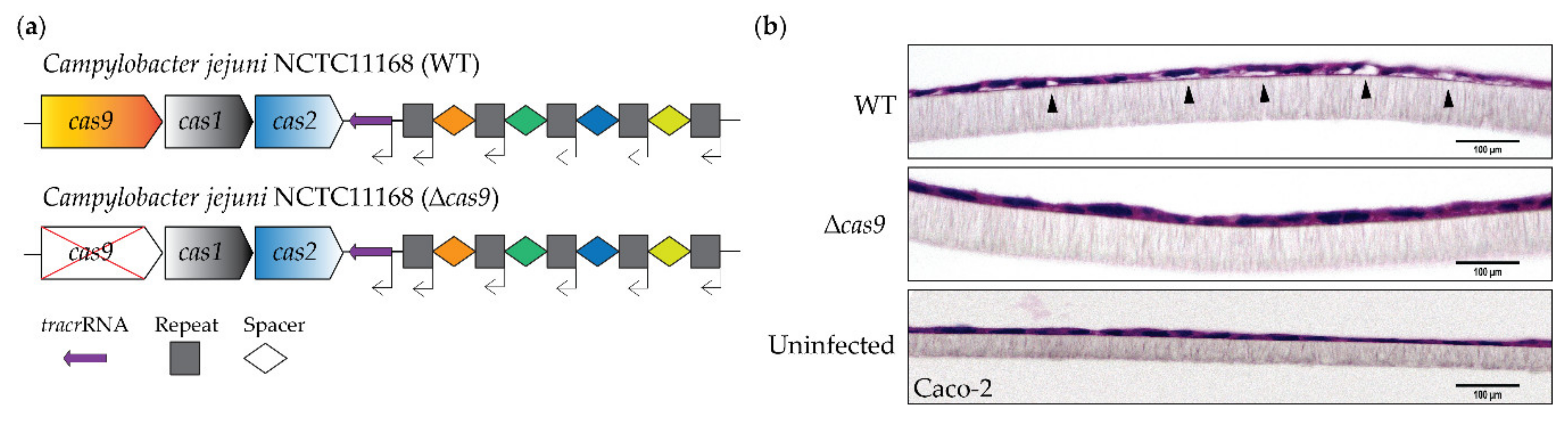
3.1. CjeCas9 Triggers Caco-2 Cell Damage
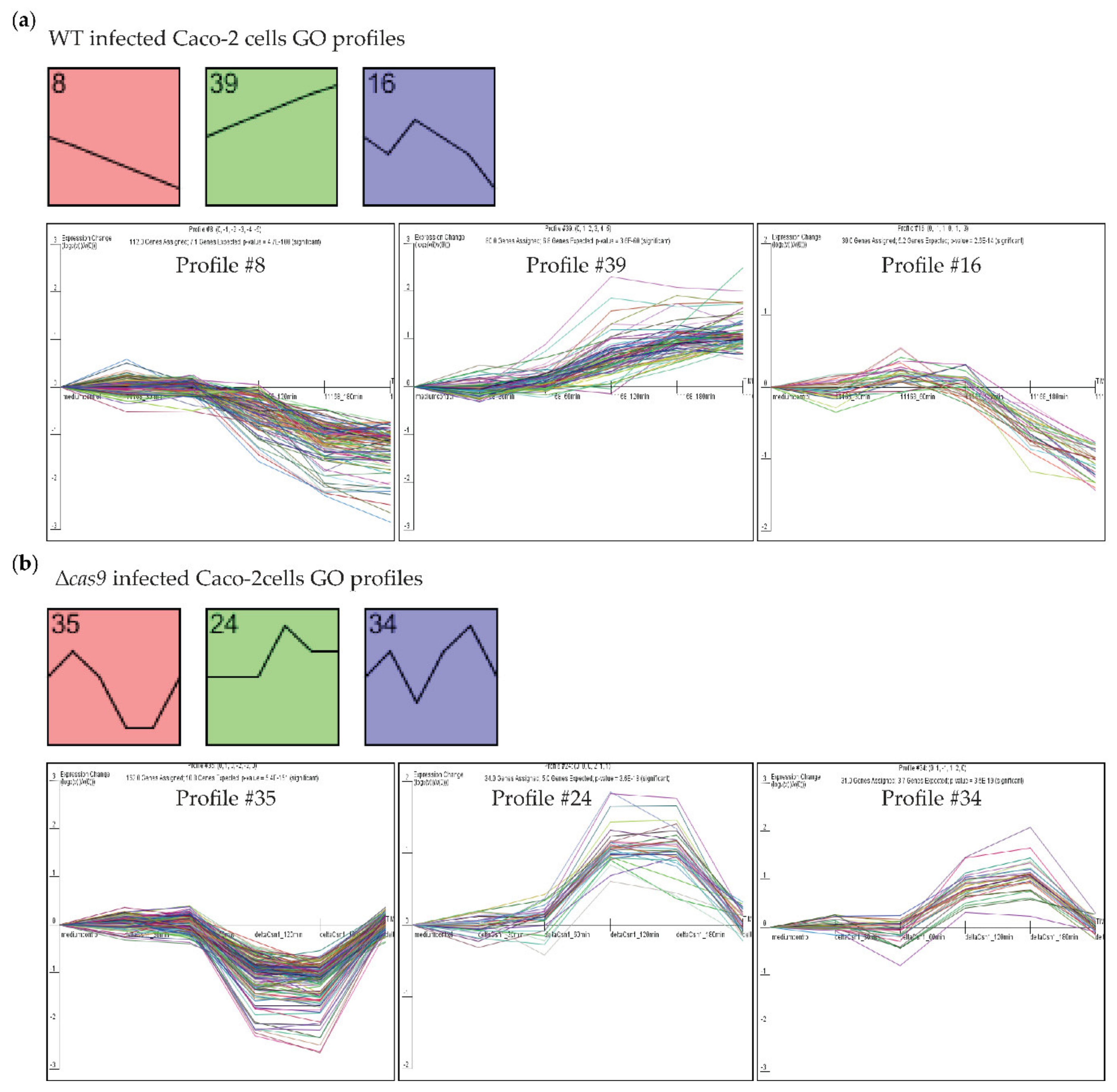
3.2. CjeCas9 Modulates Transcriptomes of Caco-2 Cells during the Early Stages of Infection
3.3. Challenges with WT C. jejuni Strains Alter Gene Expression Profiles of Caco-2 Cells over Time and Are Associated with Cell Damage
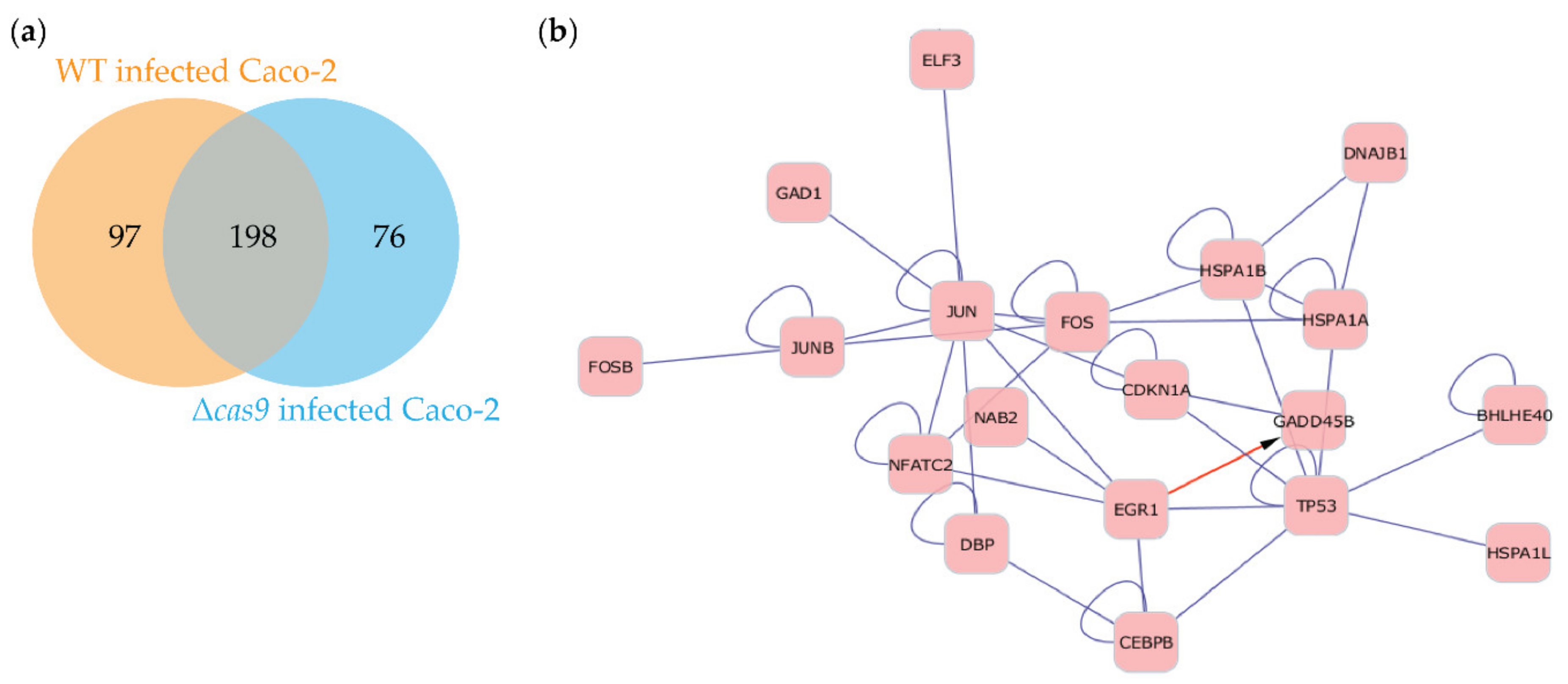
3.4. Challenging Caco-2 Cells with a Wild-Type CjeCas9-Producing C. jejuni Strain is Associated with the Induction of Cell Death and Pro-Inflammatory Signaling Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Young, K.T.; Davis, L.M.; Dirita, V.J. Campylobacter jejuni: Mmolecular biology and pathogenesis. Nat. Rev. Microbiol. 2007, 5, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Kalischuk, L.D.; Inglis, G.D.; Buret, A.G. Campylobacter jejuni induces transcellular translocation of commensal bacteria via lipid rafts. Gut Pathog. 2009, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Kalischuk, L.D.; Leggett, F.; Inglis, G.D. Campylobacter jejuni induces transcytosis of commensal bacteria across the intestinal epithelium through M-like cells. Gut Pathog. 2010, 2, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalischuk, L.D.; Inglis, G.D.; Buret, A.G. Strain-dependent induction of epithelial cell oncosis by Campylobacter jejuni is correlated with invasion ability and is independent of cytolethal distending toxin. Microbiology 2007, 153, 2952–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wassenaar, T.M. Toxin production by Campylobacter spp. Clin. Microbiol. Rev. 1997, 10, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Bolton, D.J. Campylobacter virulence and survival factors. Food Microbiol. 2015, 48, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Dasti, J.I.; Tareen, A.M.; Lugert, R.; Zautner, A.E.; Groß, U. Campylobacter jejuni: A brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. 2010, 300, 205–211. [Google Scholar] [CrossRef]
- Burnham, P.M.; Hendrixson, D.R. Campylobacter jejuni: Collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol. 2018, 16, 551–565. [Google Scholar] [CrossRef]
- Bhavsar, S.; Kapadnis, B. Virulence factors of Campylobacter. Int. J. Microbiol. 2006, 3, 1–7. [Google Scholar]
- Guerry, P. Campylobacter flagella: Not just for motility. Trends Microbiol. 2007, 15, 456–461. [Google Scholar] [CrossRef]
- Louwen, R.; Heikema, A.; Van Belkum, A.; Ott, A.; Gilbert, M.; Ang, W.; Endtz, H.P.; Bergman, M.P.; Nieuwenhuis, E.E. The sialylated lipooligosaccharide outer core in Campylobacter jejuni is an important determinant for epithelial cell invasion. Infect. Immun. 2008, 76, 4431–4438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louwen, R.; Nieuwenhuis, E.E.S.; van Marrewijk, L.; Horst-Kreft, D.; de Ruiter, L.; Heikema, A.P.; van Wamel, W.J.B.; Wagenaar, J.A.; Endtz, H.P.; Samsom, J.; et al. Campylobacter jejuni translocation across intestinal epithelial cells is facilitated by ganglioside-like lipooligosaccharide structures. Infect. Immun. 2012, 80, 3307–3318. [Google Scholar] [CrossRef] [Green Version]
- Watson, R.O.; Galán, J.E. Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 2008, 4, e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, C.; Mohanraju, P.; Stubbs, A.; Dugar, G.; Hoogstrate, Y.; Kremers, G.-J.; van Cappellen, W.A.; Horst-Kreft, D.; Laffeber, C.; Lebbink, J.H.G.; et al. Guide-free Cas9 from pathogenic Campylobacter jejuni bacteria causes severe damage to DNA. Sci. Adv. 2020, 6, eaaz4849. [Google Scholar] [CrossRef]
- Chumduri, C.; Gurumurthy, R.K.; Zietlow, R.; Meyer, T.F. Subversion of host genome integrity by bacterial pathogens. Nat. Rev. Mol. Cell Biol. 2016, 17, 659–673. [Google Scholar] [CrossRef]
- Lara-Tejero, M.; Galán, J.E. A Bacterial Toxin That Controls Cell Cycle Progression as a Deoxyribonuclease I-Like Protein. Science 2000, 290, 354–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, R.B.; Hassane, D.C.; Cottle, D.L.; Pickett, C.L. Interactions of Campylobacter jejuni cytolethal distending toxin subunits CdtA and CdtC with HeLa cells. Infect. Immun. 2003, 71, 4883–4890. [Google Scholar] [CrossRef] [Green Version]
- Pickett, C.L.; Pesci, E.C.; Cottle, D.L.; Russell, G.; Erdem, A.N.; Zeytin, H. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect. Immun. 1996, 64, 2070–2078. [Google Scholar] [CrossRef] [Green Version]
- Mortensen, N.P.; Schiellerup, P.; Boisen, N.; Klein, B.M.; Locht, H.; Abuoun, M.; Newell, D.; Krogfelt, K.A. The role of Campylobacter jejuni cytolethal distending toxin in gastroenteritis: toxin detection, antibody production, and clinical outcome. Apmis 2011, 119, 626–634. [Google Scholar] [CrossRef]
- Nielsen, H.; Persson, S.; Olsen, K.E.P.; Ejlertsen, T.; Kristensen, B.; Schønheyder, H.C. Bacteraemia with Campylobacter jejuni: no association with the virulence genes iam, cdtB, capA or virB. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 357–358. [Google Scholar] [CrossRef]
- Louwen, R.; Horst-Kreft, D.; De Boer, A.G.; Van Der Graaf, L.; de Knegt, G.; Hamersma, M.; Heikema, A.P.; Timms, A.R.; Jacobs, B.C.; Wagenaar, J.A. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain–Barré syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 207–226. [Google Scholar] [CrossRef] [PubMed]
- Wine, E.; Chan, V.L.; Sherman, P.M. Campylobacter jejuni Mediated Disruption of Polarized Epithelial Monolayers is Cell-Type Specific, Time Dependent, and Correlates With Bacterial Invasion. Pediatric Res. 2008, 64, 599–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkhill, J.; Wren, B.W.; Mungall, K.; Ketley, J.M.; Churcher, C.; Basham, D.; Chillingworth, T.; Davies, R.M.; Feltwell, T.; Holroyd, S.; et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000, 403, 665–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidalgo, I.J.; Raub, T.J.; Borchardt, R.T. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 1989, 96, 736–749. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.; Kools, H.; de Groot, P.J.; Gavai, A.K.; Basnet, R.K.; Cheng, F.; Wu, J.; Wang, X.; Lommen, A.; Hooiveld, G.J.E.J.; et al. MADMAX—Management and analysis database for multiple ~omics experiments. J. Integr. Bioinform. 2011, 8, 160. [Google Scholar] [CrossRef]
- Heber, S.; Sick, B. Quality assessment of Affymetrix GeneChip data. Omics J. Integr. Biol. 2006, 10, 358–368. [Google Scholar] [CrossRef] [Green Version]
- Dai, M.; Wang, P.; Boyd, A.D.; Kostov, G.; Athey, B.; Jones, E.G.; Bunney, W.E.; Myers, R.M.; Speed, T.P.; Akil, H.; et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef] [Green Version]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3. [Google Scholar] [CrossRef]
- Sartor, M.A.; Tomlinson, C.R.; Wesselkamper, S.C.; Sivaganesan, S.; Leikauf, G.D.; Medvedovic, M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinform. 2006, 7, 538. [Google Scholar] [CrossRef] [Green Version]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.K.; Braynen, W.; Keshav, K.; Pavlidis, P. ErmineJ: Tool for functional analysis of gene expression data sets. BMC Bioinform. 2005, 6, 269. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Baarlen, P.; Troost, F.; van der Meer, C.; Hooiveld, G.; Boekschoten, M.; Brummer, R.J.M.; Kleerebezem, M. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 4562–4569. [Google Scholar] [CrossRef] [Green Version]
- Sampson, T.R.; Saroj, S.D.; Llewellyn, A.C.; Tzeng, Y.L.; Weiss, D.S. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 2013. [Google Scholar] [CrossRef] [Green Version]
- Louwen, R.; Staals, R.H.; Endtz, H.P.; van Baarlen, P.; van der Oost, J. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol. Mol. Biol. Rev. 2014, 78, 74–88. [Google Scholar] [CrossRef] [Green Version]
- Breitling, R.; Armengaud, P.; Amtmann, A.; Herzyk, P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004, 573, 83–92. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Friis, L.M.; Keelan, M.; Taylor, D.E. Campylobacter jejuni drives MyD88-independent interleukin-6 secretion via Toll-like receptor 2. Infect. Immun. 2009, 77, 1553–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aschner, J.L.; Aschner, M. Nutritional aspects of manganese homeostasis. Mol. Asp. Med. 2005, 26, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Kerstan, D.; Quamme, G.A. Intestinal Absorption of Magnesium BT—Calcium in Internal Medicine; Springer: London, UK, 2002; pp. 171–183. [Google Scholar] [CrossRef]
- McCarthy, J.T.; Kumar, R. Divalent cation metabolism: Magnesium. Atlas Dis. Kidney 1999, 1, 1–4. [Google Scholar]
- Lisher, J.; Giedroc, D. Manganese acquisition and homeostasis at the host-pathogen interface. Front. Cell. Infect. Microbiol. 2013, 3, 91. [Google Scholar] [CrossRef] [Green Version]
- Konkel, M.E.; Mead, D.J.; Hayes, S.F.; Cieplak, W. Translocation of Campylobacter jejuni across human polarized epithelial cell monolayer cultures. J. Infect. Dis. 1992, 166, 308–315. [Google Scholar] [CrossRef]
- Louwen, R.; Hays, J.P. Is there an unrecognised role for Campylobacter infections in (chronic) inflammatory diseases? World 2013, 4, 002. [Google Scholar]
- Van Spreeuwel, J.P.; Duursma, G.C.; Meijer, C.J.; Bax, R.; Rosekrans, P.C.; Lindeman, J. Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut 1985, 26, 945–951. [Google Scholar] [CrossRef] [Green Version]
- Wooldridge, K.G.; Ketley, J.M. Campylobacter-host cell interactions. Trends Microbiol. 1997, 5, 96–102. [Google Scholar] [CrossRef]
- Siegl, C.; Rudel, T. Modulation of p53 during bacterial infections. Nat. Rev. Microbiol. 2015, 13, 741–748. [Google Scholar] [CrossRef]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [Green Version]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Schultz, L.B.; Chehab, N.H.; Malikzay, A.; Halazonetis, T.D. P53 Binding Protein 1 (53bp1) Is an Early Participant in the Cellular Response to DNA Double-Strand Breaks. J. Cell Biol. 2000, 151, 1381–1390. [Google Scholar] [CrossRef] [Green Version]
- Enache, O.M.; Rendo, V.; Abdusamad, M.; Lam, D.; Davison, D.; Pal, S.; Currimjee, N.; Hess, J.; Pantel, S.; Nag, A.; et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 2020, 52, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Haapaniemi, E.; Botla, S.; Persson, J.; Schmierer, B.; Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018. [Google Scholar] [CrossRef] [Green Version]
- Ihry, R.J.; Worringer, K.A.; Salick, M.R.; Frias, E.; Ho, D.; Theriault, K.; Kommineni, S.; Chen, J.; Sondey, M.; Ye, C.; et al. P53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mellits, K.H.; Mullen, J.; Wand, M.; Armbruster, G.; Patel, A.; Connerton, P.L.; Skelly, M.; Connerton, I.F. Activation of the transcription factor NF-$κ$B by Campylobacter jejuni. Microbiology 2002, 148, 2753–2763. [Google Scholar] [CrossRef] [Green Version]
- Al-Sayeqh, A.F.; Loughlin, M.F.; Dillon, E.; Mellits, K.H.; Connerton, I.F. Campylobacter jejuni activates NF-κB independently of TLR2, TLR4, Nod1 and Nod2 receptors. Microb. Pathog. 2010, 49, 294–304. [Google Scholar] [CrossRef]
- Wang, W.; Mani, A.M.; Wu, Z.-H. DNA damage-induced nuclear factor-kappa B activation and its roles in cancer progression. J. Cancer Metastasis Treat. 2017, 3, 45. [Google Scholar] [CrossRef]
- Elkon, R.; Rashi-Elkeles, S.; Lerenthal, Y.; Linhart, C.; Tenne, T.; Amariglio, N.; Rechavi, G.; Shamir, R.; Shiloh, Y. Dissection of a DNA-damage-induced transcriptional network using a combination of microarrays, RNA interference and computational promoter analysis. Genome Biol. 2005, 6, R43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lippert, E.; Karrasch, T.; Sun, X.; Allard, B.; Herfarth, H.H.; Threadgill, D.; Jobin, C. Gnotobiotic IL-10−/−; NF-κBEGFP Mice Develop Rapid and Severe Colitis Following Campylobacter jejuni Infection. PLoS ONE 2009, 4, e7413. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations |

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saha, C.; Horst-Kreft, D.; Kross, I.; van der Spek, P.J.; Louwen, R.; van Baarlen, P. Campylobacter jejuni Cas9 Modulates the Transcriptome in Caco-2 Intestinal Epithelial Cells. Genes 2020, 11, 1193. https://doi.org/10.3390/genes11101193
Saha C, Horst-Kreft D, Kross I, van der Spek PJ, Louwen R, van Baarlen P. Campylobacter jejuni Cas9 Modulates the Transcriptome in Caco-2 Intestinal Epithelial Cells. Genes. 2020; 11(10):1193. https://doi.org/10.3390/genes11101193
Chicago/Turabian StyleSaha, Chinmoy, Deborah Horst-Kreft, Inez Kross, Peter J. van der Spek, Rogier Louwen, and Peter van Baarlen. 2020. "Campylobacter jejuni Cas9 Modulates the Transcriptome in Caco-2 Intestinal Epithelial Cells" Genes 11, no. 10: 1193. https://doi.org/10.3390/genes11101193
APA StyleSaha, C., Horst-Kreft, D., Kross, I., van der Spek, P. J., Louwen, R., & van Baarlen, P. (2020). Campylobacter jejuni Cas9 Modulates the Transcriptome in Caco-2 Intestinal Epithelial Cells. Genes, 11(10), 1193. https://doi.org/10.3390/genes11101193




