A Label-Free Assay for Aminoacylation of tRNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of tRNAs
2.2. Isolation of E. coli aaRS Enzymes
2.3. Saturation Level of Aminoacylation of tRNA
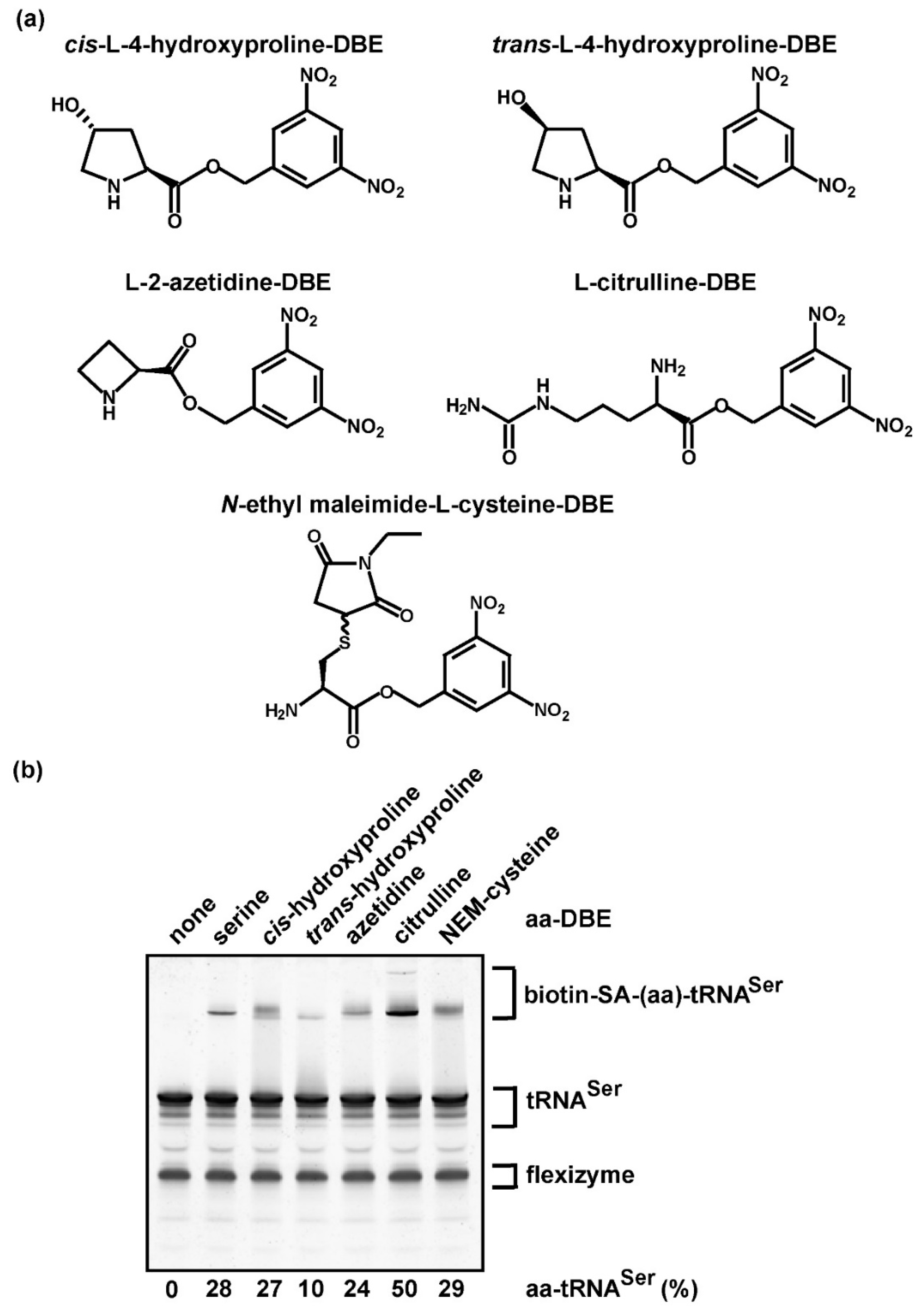
2.4. Flexizyme-Catalyzed Aminoacylation of E. coli tRNASer with Non-proteinogenic Amino Acids
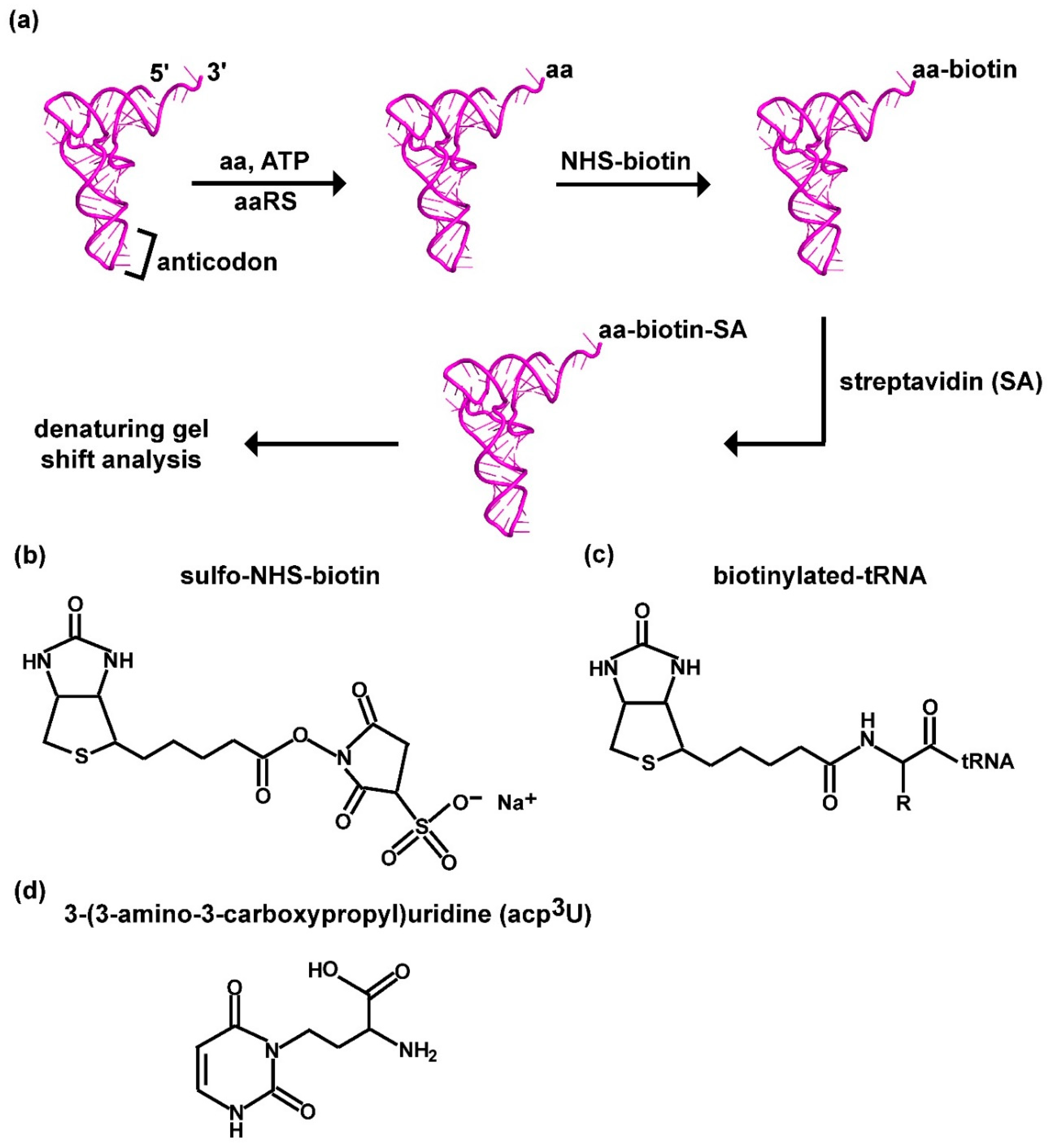
2.5. Biotin-SA Conjugation
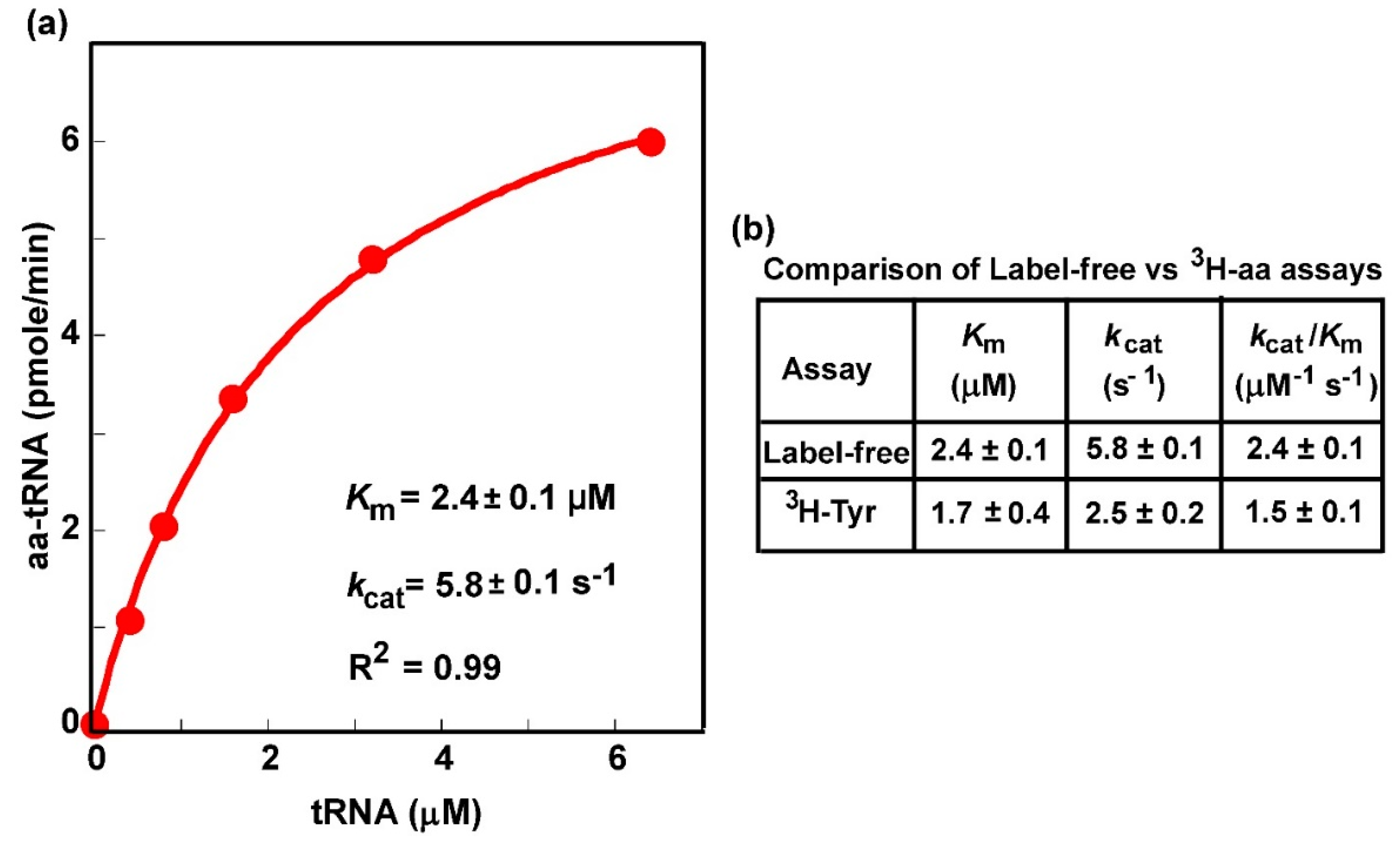
2.6. Determination of Kinetic Parameters.
3. Results
3.1. A Label-Free Assay for Aminoacylation of tRNA by Biotin-SA Conjugation
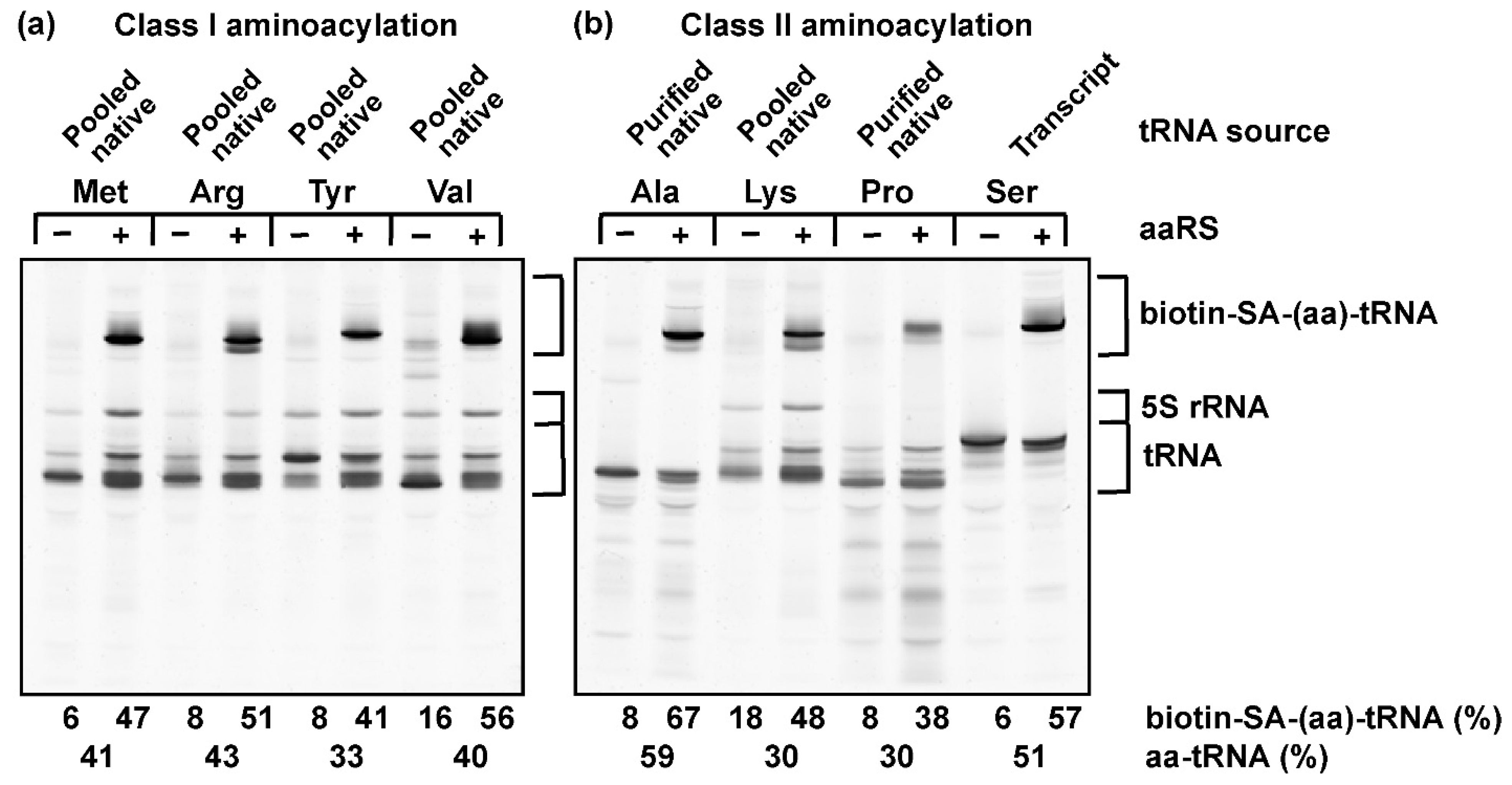
3.2. The Label-Free Assay Is Applicable to a Wide Range of Amino Acids and tRNAs
3.3. Stoichiometry of Quantifying Aminoacylation by the Label-Free Assay
3.4. The Label-Free Assay Is Applicable to Non-Proteinogenic Amino Acids
3.5. The Label-Free Assay Determines Kinetic Parameters of Aminoacylation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ibba, M.; Thomann, H.U.; Hong, K.W.; Sherman, J.M.; Weygand-Durasevic, I.; Sever, S.; Stange-Thomann, N.; Praetorius, M.; Söll, D. Substrate selection by aminoacyl-tRNA synthetases. Nucleic Acids Symp. Ser. 1995, 26, 40–42. [Google Scholar]
- Bremer, H.; Dennis, P.P. Modulation of Chemical Composition and Other Parameters of the Cell at Different Exponential Growth Rates. EcoSal Plus 2008, 3. [Google Scholar] [CrossRef]
- Dai, X.; Zhu, M.; Warren, M.; Balakrishnan, R.; Patsalo, V.; Okano, H.; Williamson, J.R.; Fredrick, K.; Wang, Y.; Hwa, T. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2016, 2, 16231. [Google Scholar] [CrossRef]
- Ognjenović, J.; Simonović, M. Human aminoacyl-tRNA synthetases in diseases of the nervous system. RNA Biol. 2017, 15, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Kashina, A. Protein arginylation, a global biological regulator that targets actin cytoskeleton and the muscle. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2014, 297, 1630–1636. [Google Scholar] [CrossRef] [PubMed]
- Avcilar-Kucukgoze, I.; Gamper, H.; Polte, C.; Ignatova, Z.; Kraetzner, R.; Shtutman, M.; Hou, Y.M.; Dong, D.W.; Kashina, A. TRNA(Arg)-Derived Fragments Can Serve as Arginine Donors for Protein Arginylation. Cell Chem. Biol. 2020, 27, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Hoben, P.; Söll, D. [8] Glutaminyl-tRNA synthetase of Escherichia coli. Methods Enzymol. 1985, 113, 55–59. [Google Scholar] [CrossRef]
- Zhang, C.M.; Christian, T.; Newberry, K.J.; Perona, J.J.; Hou, Y.M. Zinc-mediated amino acid discrimination in cysteinyl-tRNA synthetase. J. Mol. Biol. 2003, 327, 911–917. [Google Scholar] [CrossRef]
- Zhang, C.M.; Perona, J.J.; Hou, Y.M. Amino Acid Discrimination by a Highly Differentiated Metal Center of an Aminoacyl-tRNA Synthetas. Biochemistry 2003, 42, 10931–10937. [Google Scholar] [CrossRef]
- Ibba, M.; Hong, K.W.; Sherman, J.M.; Sever, S.; Soll, D. Interactions between tRNA identity nucleotides and their recognition sites in glutaminyl-tRNA synthetase determine the cognate amino acid affinity of the enzyme. Proc. Natl. Acad. Sci. USA 1996, 93, 6953–6958. [Google Scholar] [CrossRef]
- Zeng, Q.Y.; Peng, G.X.; Li, G.; Zhou, J.B.; Zheng, W.Q.; Xue, M.Q.; Wang, E.; Zhou, X.L. The G3-U70-independent tRNA recognition by human mitochondrial alanyl-tRNA synthetase. Nucleic Acids Res. 2019, 47, 3072–3085. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.L.; Ruan, Z.R.; Huang, Q.; Tan, M.; Wang, E. Translational fidelity maintenance preventing Ser mis-incorporation at Thr codon in protein from eukaryote. Nucleic Acids Res. 2012, 41, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.L.; Fang, Z.P.; Ruan, Z.R.; Wang, M.; Liu, R.J.; Tan, M.; Anella, F.M.; Wang, E. Aminoacylation and translational quality control strategy employed by leucyl-tRNA synthetase from a human pathogen with genetic code ambiguity. Nucleic Acids Res. 2013, 41, 9825–9838. [Google Scholar] [CrossRef] [PubMed]
- LeDoux, S.; Uhlenbeck, O.C. [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 2008, 44, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, T.; Liu, C.; Morinaga, H.; Kim, S.; Hou, Y.M. Pyrophosphorolysis of CCA Addition: Implication for Fidelity. J. Mol. Biol. 2011, 414, 28–43. [Google Scholar] [CrossRef]
- Zhang, C.M.; Hou, Y.M. Domain−Domain Communication for tRNA Aminoacylation: The Importance of Covalent Connectivity. Biochemistry 2005, 44, 7240–7249. [Google Scholar] [CrossRef]
- Liu, C.; Gamper, H.; Shtivelband, S.; Hauenstein, S.; Perona, J.J.; Hou, Y.M. Kinetic Quality Control of Anticodon Recognition by a Eukaryotic Aminoacyl-tRNA Synthetase. J. Mol. Biol. 2007, 367, 1063–1078. [Google Scholar] [CrossRef][Green Version]
- Liu, C.; Gamper, H.; Liu, H.; Cooperman, B.S.; Hou, Y.M. Potential for interdependent development of tRNA determinants for aminoacylation and ribosome decoding. Nat. Commun. 2011, 2, 1–8. [Google Scholar] [CrossRef]
- Liu, C.; Sanders, J.M.; Pascal, J.M.; Hou, Y.M. Adaptation to tRNA acceptor stem structure by flexible adjustment in the catalytic domain of class I tRNA synthetases. RNA 2011, 18, 213–221. [Google Scholar] [CrossRef]
- Ferro, I.; Liebeton, K.; Ignatova, Z. Growth-Rate Dependent Regulation of tRNA Level and Charging in Bacillus licheniformis. J. Mol. Biol. 2017, 429, 3102–3112. [Google Scholar] [CrossRef]
- Hou, Y.M.; Westhof, E.; Giege, R. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 6776–6780. [Google Scholar] [CrossRef] [PubMed]
- Netzer, N.; Goodenbour, J.M.; Davide, A.; Dittmar, K.A.; Jones, R.B.; Schneider, J.R.; Boone, D.; Eves, E.M.; Rosner, M.R.; Gibbs, J.S.; et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 2009, 462, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Zaborske, J.; Pan, T. Genome-wide analysis of aminoacylation (charging) levels of tRNA using microarrays. J. Vis. Exp. 2010, e2007. [Google Scholar] [CrossRef] [PubMed]
- Avcilar-Kucukgoze, I.; Bartholomäus, A.; Varela, J.A.C.; Kaml, R.F.X.; Neubauer, P.; Budisa, N.; Ignatova, Z. Discharging tRNAs: A tug of war between translation and detoxification inEscherichia coli. Nucleic Acids Res. 2016, 44, 8324–8334. [Google Scholar] [CrossRef]
- Zheng, G.; Qin, Y.; Clark, W.C.; Dai, Q.; Yi, C.; He, C.; Lambowitz, A.M.; Pan, T. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 2015, 12, 835–837. [Google Scholar] [CrossRef]
- Cozen, A.E.; Quartley, E.; Holmes, A.D.; Hrabeta-Robinson, E.; Phizicky, E.M.; Lowe, T.M. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 2015, 12, 879–884. [Google Scholar] [CrossRef]
- Murakami, H.; Ohta, A.; Ashigai, H.; Suga, H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat. Methods 2006, 3, 357–359. [Google Scholar] [CrossRef]
- Murakami, H.; Kourouklis, D.; Suga, H. Using a Solid-Phase Ribozyme Aminoacylation System to Reprogram the Genetic Code. Chem. Biol. 2003, 10, 1077–1084. [Google Scholar] [CrossRef]
- Zhang, C.-M.; Perona, J.J.; Ryu, K.; Francklyn, C.; Hou, Y.-M. Distinct Kinetic Mechanisms of the Two Classes of Aminoacyl-tRNA Synthetases. J. Mol. Biol. 2006, 361, 300–311. [Google Scholar] [CrossRef]
- Shitivelband, S.; Hou, Y.-M. Breaking the Stereo Barrier of Amino Acid Attachment to tRNA by a Single Nucleotide. J. Mol. Biol. 2005, 348, 513–521. [Google Scholar] [CrossRef]
- Köhrer, C.; Rajbhandary, U.L. The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases. Methods 2008, 44, 129–138. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eriani, G.; Delarue, M.; Poch, O.; Gangloff, J.; Moras, D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 1990, 347, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Taiji, M.; Yokoyama, S.; Miyazawa, T. Transacylation rates of (aminoacyl)adenosine moiety at the 3’-terminus of aminoacyl transfer ribonucleic acid. Biochemistry 1983, 22, 3220–3225. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.L.; Schmeing, T.M.; Moore, P.B.; Steitz, T.A. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. USA 2002, 99, 11670–11675. [Google Scholar] [CrossRef]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crecy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 2046, D303–D307. [Google Scholar] [CrossRef]
- Friedman, S.; Li, H.J.; Nakanishi, K.; Van Lear, G. 3-(3-amino-3-carboxy-n-propyl)uridine. The structure of the nucleoside in Escherichia coli transfer ribonucleic acid that reacts with phenoxyacetoxysuccinimide. Biochemistry 1974, 13, 2932–2937. [Google Scholar] [CrossRef]
- Ohashi, Z.; Maeda, M.; McCloskey, J.A.; Nishimura, S. 3-(3-Amino-3-carboxypropyl)uridine. Novel modified nucleoside isolated from Escherichia coli phenylalanine transfer ribonucleic acid. Biochemistry 1974, 13, 2620–2625. [Google Scholar] [CrossRef]
- Fei, J.; Wang, J.; Sternberg, S.H.; MacDougall, D.D.; Elvekrog, M.M.; Pulukkunat, D.K.; Englander, M.T.; Gonzalez, R.L. A Highly Purified, Fluorescently Labeled In Vitro Translation System for Single-Molecule Studies of Protein Synthesis. Methods Enzymol. 2010, 472, 221–259. [Google Scholar] [CrossRef]
- Jühling, F.; Mörl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Pütz, J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef]
- Masuda, I.; Matsubara, R.; Christian, T.; Rojas, E.R.; Yadavalli, S.S.; Zhang, L.; Goulian, M.; Foster, L.J.; Huang, K.C.; Hou, Y.M. TRNA Methylation Is a Global Determinant of Bacterial Multi-drug Resistance. Cell Syst. 2019, 8, 302–314. [Google Scholar] [CrossRef]
- Katoh, T.; Wohlgemuth, I.; Nagano, M.; Rodnina, M.V.; Suga, H. Essential structural elements in tRNAPro for EF-P-mediated alleviation of translation stalling. Nat. Commun. 2016, 7, 11657. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Lajoie, M.J.; Englert, M.; Söll, D. Rewriting the Genetic Code. Annu. Rev. Microbiol. 2017, 71, 557–577. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Murakami, H.; Suga, H.; Ferré-D’Amaré, A.R. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature 2008, 454, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Fujino, T.; Kondo, T.; Suga, H.; Murakami, H. Exploring the Minimal RNA Substrate of Flexizymes. ChemBioChem 2019, 20, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Po, P.; Delaney, E.; Gamper, H.; Szantai-Kis, D.M.; Speight, L.; Tu, L.; Kosolapov, A.; Petersson, E.J.; Hou, Y.M.; Deutsch, C. Effect of Nascent Peptide Steric Bulk on Elongation Kinetics in the Ribosome Exit Tunnel. J. Mol. Biol. 2017, 429, 1873–1888. [Google Scholar] [CrossRef]
- Masuda, I.; Igarashi, T.; Sakaguchi, R.; Nitharwal, R.G.; Takase, R.; Han, K.Y.; Leslie, B.J.; Liu, C.; Gamper, H.; Ha, T.; et al. A genetically encoded fluorescent tRNA is active in live-cell protein synthesis. Nucleic Acids Res. 2016, 45, 4081–4093. [Google Scholar] [CrossRef]
- Niehues, S.; Bussmann, J.; Steffes, G.; Erdmann, I.; Köhrer, C.; Sun, L.; Wagner, M.; Schäfer, K.; Wang, G.; Koerdt, S.N.; et al. Impaired protein translation in Drosophila models for Charcot–Marie–Tooth neuropathy caused by mutant tRNA synthetases. Nat. Commun. 2015, 6, 7520. [Google Scholar] [CrossRef]
- Bheda, P.; Swatkoski, S.; Fiedler, K.L.; Boeke, J.D.; Cotter, R.J.; Wolberger, C. Biotinylation of lysine method identifies acetylated histone H3 lysine 79 in Saccharomyces cerevisiae as a substrate for Sir2. Proc. Natl. Acad. Sci. USA 2012, 109, E916–E925. [Google Scholar] [CrossRef]
- Picard, M.; Zhang, J.; Hancock, S.; Derbeneva, O.; Golhar, R.; Golik, P.; O’Hearn, S.; Levy, S.; Potluri, P.; Lvova, M.; et al. Progressive increase in mtDNA 3243A > G heteroplasmy causes abrupt transcriptional reprogramming. Proc. Natl. Acad. Sci. USA 2014, 111, E4033–E4042. [Google Scholar] [CrossRef]
- Börner, G.V.; Zeviani, M.; Tiranti, V.; Carrara, F.; Hoffmann, S.; Gerbitz, K.D.; Lochmüller, H.; Pongratz, D.; Klopstock, T.; Melberg, A.; et al. Decreased aminoacylation of mutant tRNAs in MELAS but not in MERRF patients. Hum. Mol. Genet. 2000, 9, 467–475. [Google Scholar] [CrossRef]
- Fay, A.; Garcia, Y.; Margeta, M.; Maharjan, S.; Jürgensen, C.; Briceño, J.; Garcia, M.; Yin, S.; Bassaganyas, L.; McMahon, T.; et al. A Mitochondrial tRNA Mutation Causes Axonal CMT in a Large Venezuelan Family. Ann. Neurol. 2020. [Google Scholar] [CrossRef] [PubMed]
| aa-tRNA | With Labeled Amino Acid | With Biotin-SA at 4 °C | With Biotin-SA at RT |
|---|---|---|---|
| Aminoacylation with the cognate aaRS enzyme | |||
| Met-tRNAMet/CAU | 32 | 41 | 26 |
| Arg-tRNAArg/ICG | 27 | 43 | 28 |
| Tyr-tRNATyr/QUA | 33 | 33 | 27 |
| Val-tRNAVal/cmo5UAC | 23 | 40 | 35 |
| Ala-tRNAAla/GGC | 35 | 59 | 49 |
| Lys-tRNALys/mnm5s2UUU | 16 | 30 | 27 |
| Pro-tRNAPro/GGG | 24 | 30 | 23 |
| Ser-tRNASer/GCU | – | 51 | 32 |
| Aminoacylation with dFx flexizyme | |||
| cis-HxyPro-tRNASer | – | 27 | 25 |
| trans-HxyPro-tRNASer | – | 10 | 10 |
| Azetidine-tRNASer | – | 24 | 20 |
| Citrulline-tRNASer | – | 50 | 38 |
| NEM-Cys-tRNASer | – | 29 | 25 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gamper, H.; Hou, Y.-M. A Label-Free Assay for Aminoacylation of tRNA. Genes 2020, 11, 1173. https://doi.org/10.3390/genes11101173
Gamper H, Hou Y-M. A Label-Free Assay for Aminoacylation of tRNA. Genes. 2020; 11(10):1173. https://doi.org/10.3390/genes11101173
Chicago/Turabian StyleGamper, Howard, and Ya-Ming Hou. 2020. "A Label-Free Assay for Aminoacylation of tRNA" Genes 11, no. 10: 1173. https://doi.org/10.3390/genes11101173
APA StyleGamper, H., & Hou, Y.-M. (2020). A Label-Free Assay for Aminoacylation of tRNA. Genes, 11(10), 1173. https://doi.org/10.3390/genes11101173





