PML Regulates the Epidermal Differentiation Complex and Skin Morphogenesis during Mouse Embryogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Antibodies
2.3. Isolation and Culturing of Epidermal Keratinocytes
2.4. Immunohistochemistry (IHC) Analysis
2.5. RNA-Seq Analysis
2.6. qPCR Analysis
3. Results
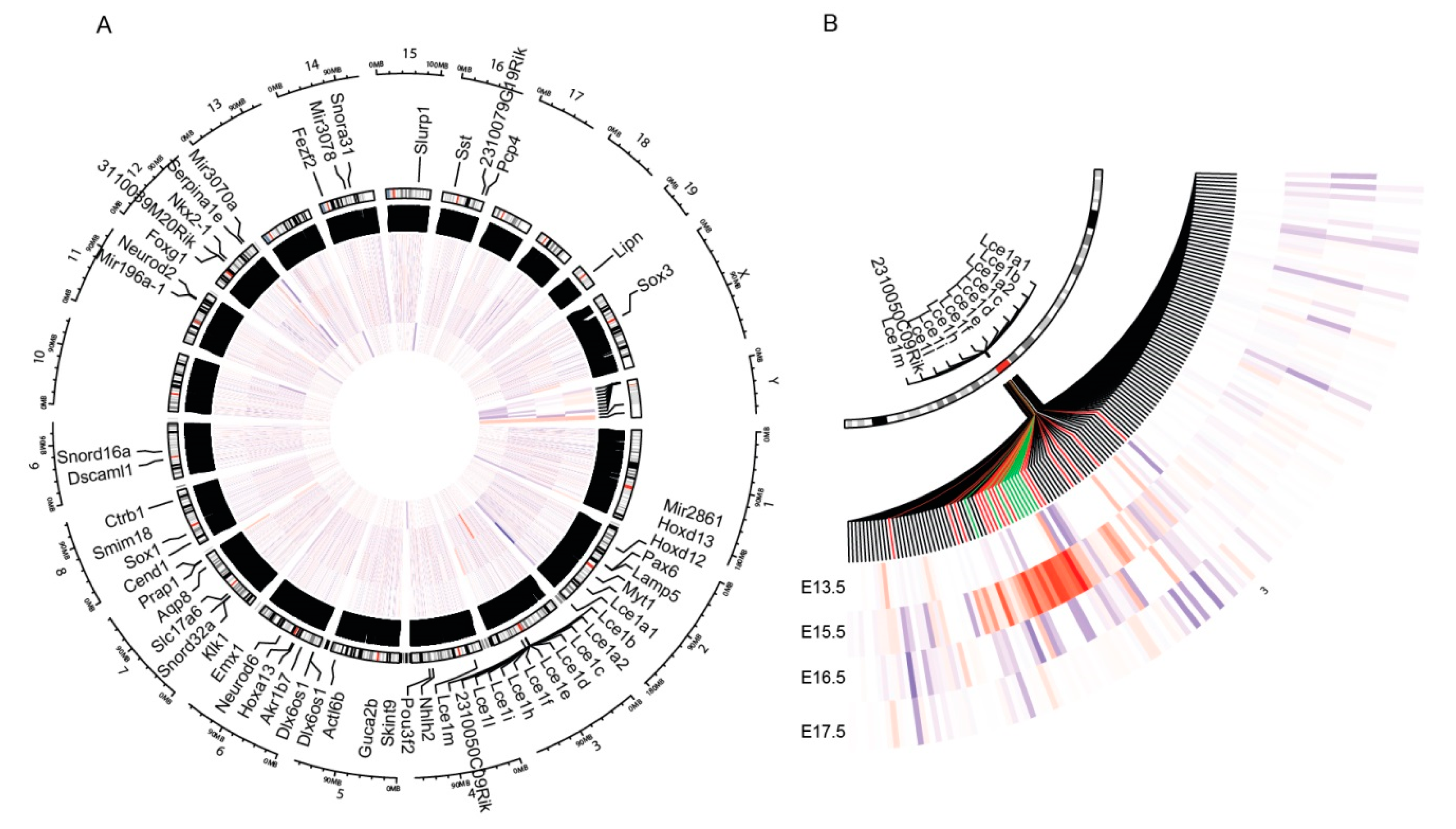
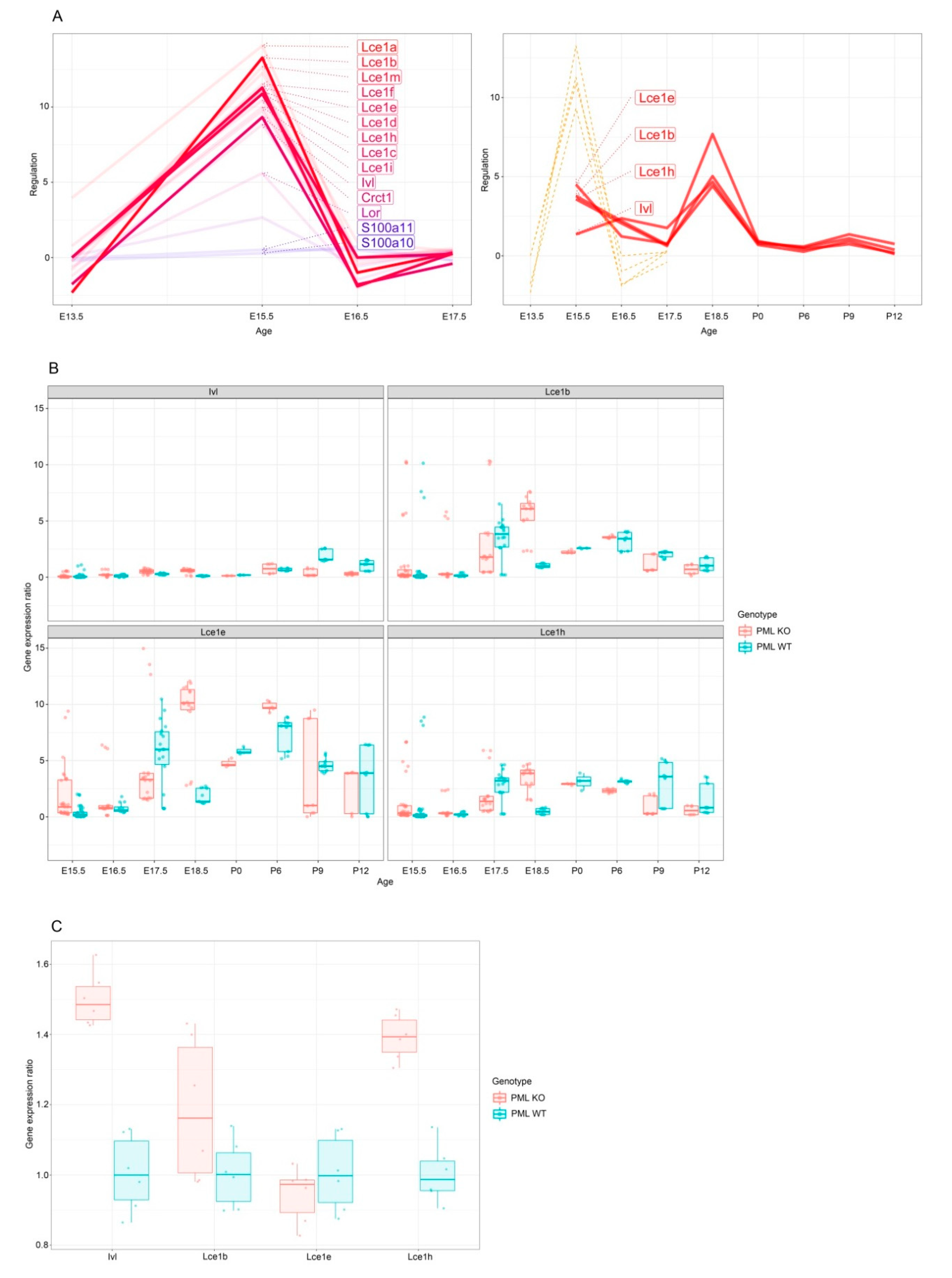
3.1. PML Regulates EDC Genes during Embryogenesis
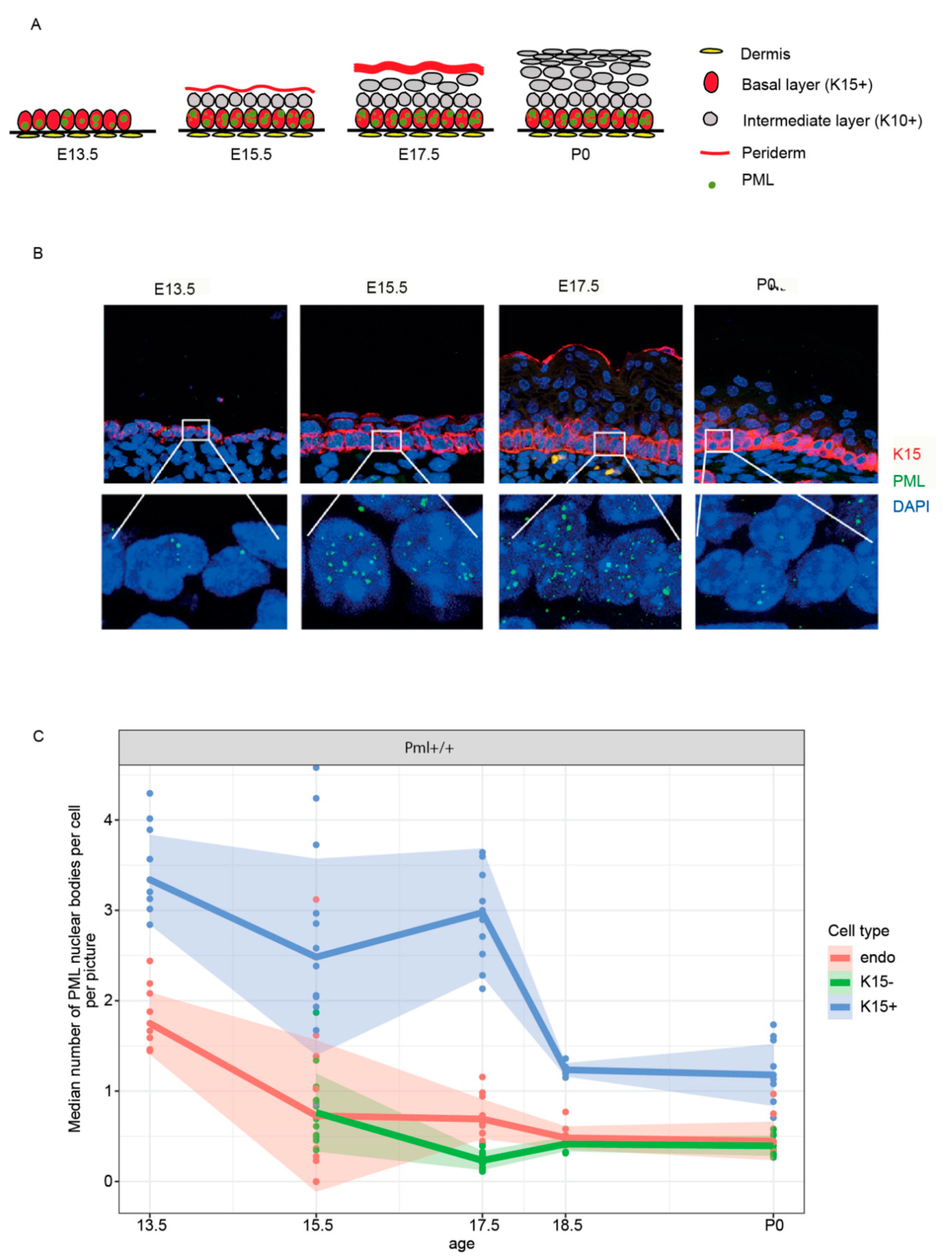
3.2. PML Expression Is Elevated in Basal Keratinocytes during Embryogenesis
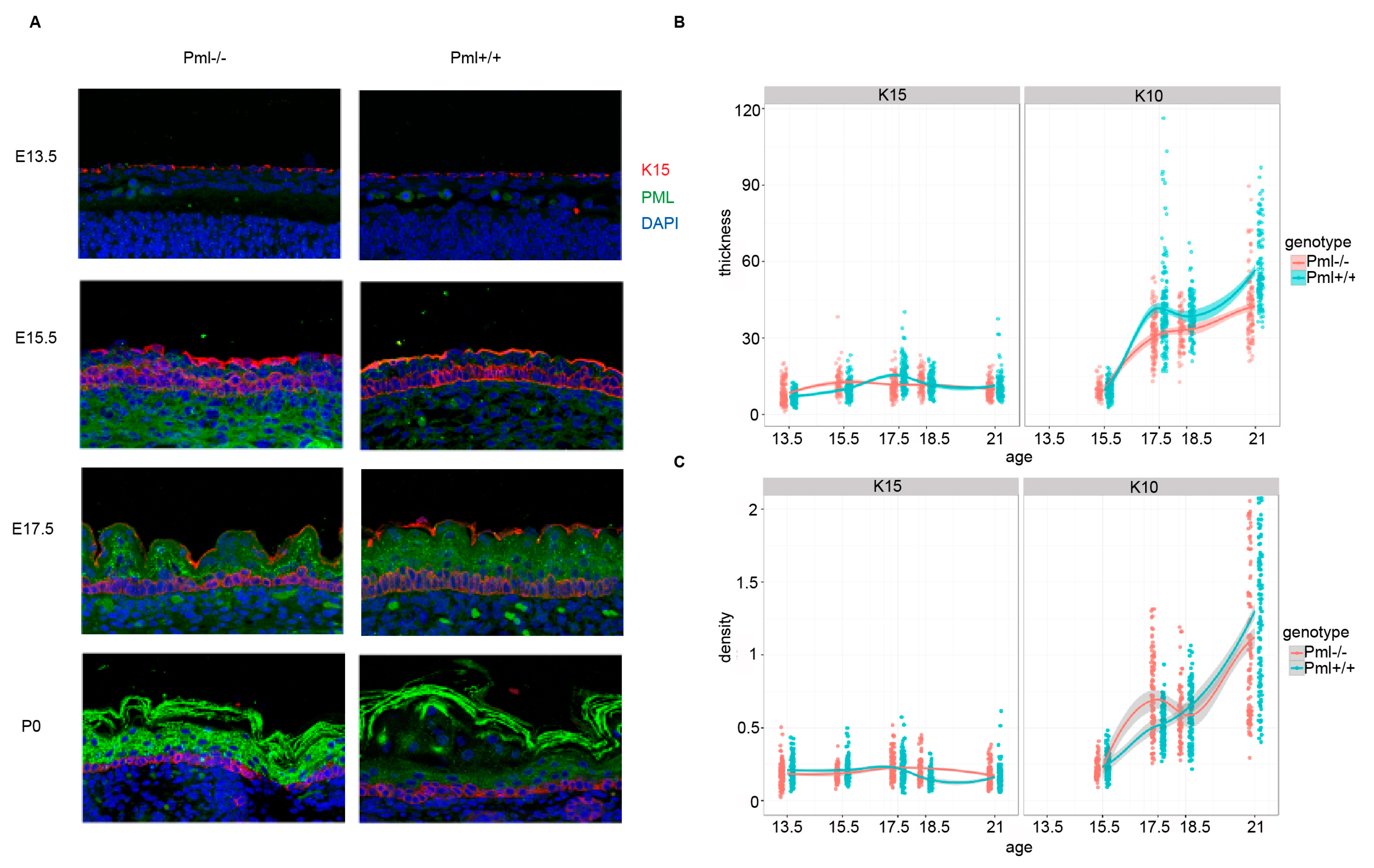
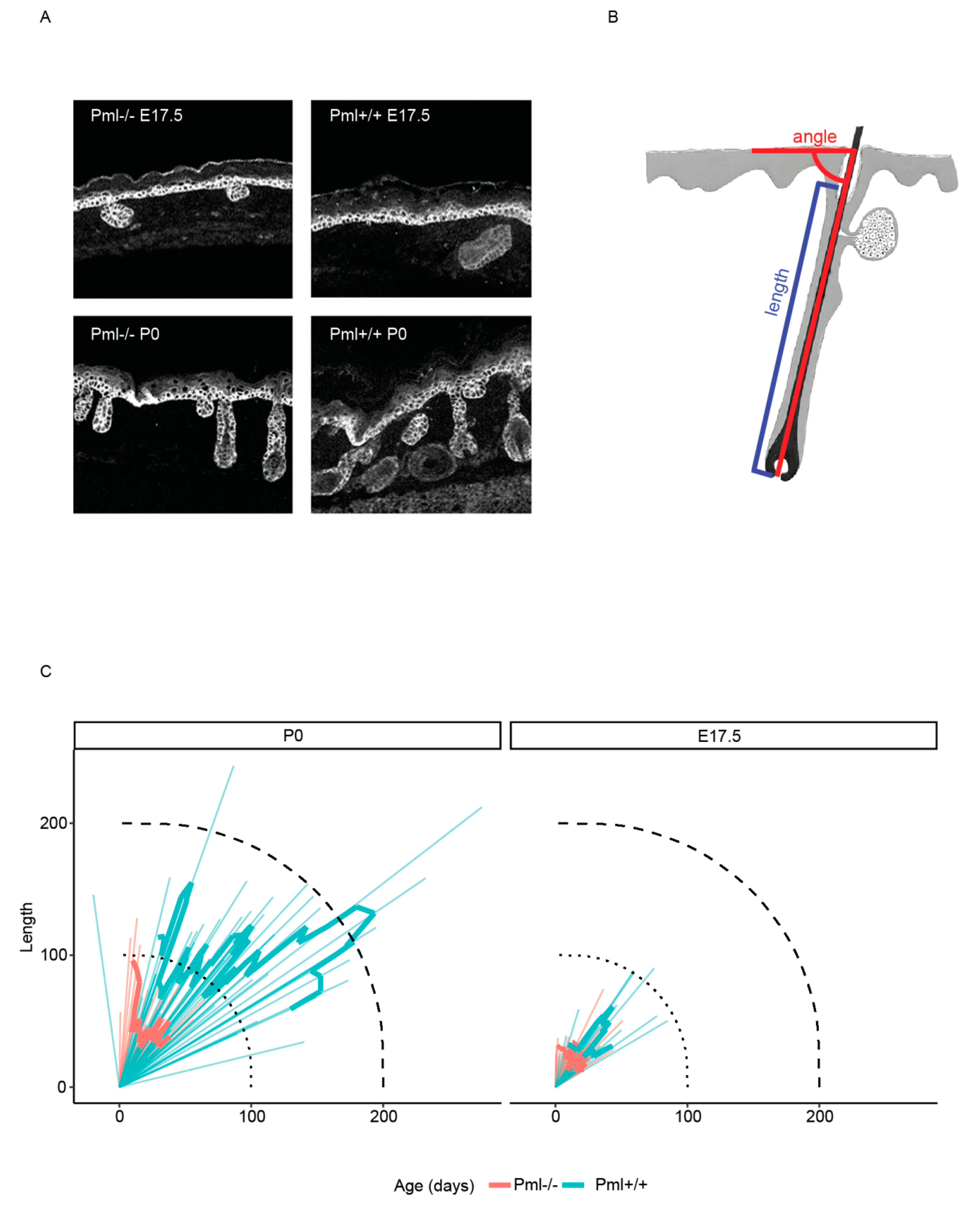
3.3. PML Depletion Affects Epidermal Thickness and Hair Follicle Size during Embryogenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, Z.G.; Ruggero, D.; Ronchetti, S.; Zhong, S.; Gaboli, M.; Rivi, R.; Pandolfi, P.P. PML is essential for multiple apoptotic pathways. Nat. Genet. 1998, 20, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Dellaire, G.; Ching, R.W.; Ahmed, K.; Jalali, F.; Tse, K.C.; Bristow, R.G.; Bazett-Jones, D.P. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J. Cell Biol. 2006, 175, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Boe, S.O.; Haave, M.; Jul-Larsen, A.; Grudic, A.; Bjerkvig, R.; Lonning, P.E. Promyelocytic leukemia nuclear bodies are predetermined processing sites for damaged DNA. J. Cell Sci. 2006, 119 Pt 16, 3284–3295. [Google Scholar] [CrossRef] [Green Version]
- Zhong, S.; Hu, P.; Ye, T.Z.; Stan, R.; Ellis, N.A.; Pandolfi, P.P. A role for PML and the nuclear body in genomic stability. Oncogene 1999, 18, 7941–7947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sloan, E.; Orr, A.; Everett, R.D. MORC3, a Component of PML Nuclear Bodies, Has a Role in Restricting Herpes Simplex Virus 1 and Human Cytomegalovirus. J. Virol. 2016, 90, 8621–8633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geoffroy, M.C.; Chelbi-Alix, M.K. Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2011, 31, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Palibrk, V.; Suganthan, R.; Scheffler, K.; Wang, W.; Bjoras, M.; Boe, S.O. PML regulates neuroprotective innate immunity and neuroblast commitment in a hypoxic-ischemic encephalopathy model. Cell Death Dis. 2016, 7, e2320. [Google Scholar] [CrossRef]
- Regad, T.; Bellodi, C.; Nicotera, P.; Salomoni, P. The tumor suppressor Pml regulates cell fate in the developing neocortex. Nat. Neurosci. 2009, 12, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ferguson, B.J.; Khaled, W.T.; Tevendale, M.; Stingl, J.; Poli, V.; Rich, T.; Salomoni, P.; Watson, C.J. PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool. Proc. Natl. Acad. Sci. USA 2009, 106, 4725–4730. [Google Scholar] [CrossRef] [Green Version]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef]
- Dellaire, G.; Farrall, R.; Bickmore, W.A. The Nuclear Protein Database (NPD): Sub-nuclear localisation and functional annotation of the nuclear proteome. Nucleic Acids Res. 2003, 31, 328–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Song, Y.; Tian, B.; Qian, J.; Dong, Y.; Liu, J.; Liu, B.; Sun, Z. Functional proteomic analysis of promyelocytic leukaemia nuclear bodies in irradiation-induced MCF-7 cells. J. Biochem. 2010, 148, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Shiels, C.; Sasieni, P.; Wu, P.J.; Islam, S.A.; Freemont, P.S.; Sheer, D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004, 164, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Ching, R.W.; Dellaire, G.; Eskiw, C.H.; Bazett-Jones, D.P. PML bodies: A meeting place for genomic loci? J. Cell Sci. 2005, 118 Pt 5, 847–854. [Google Scholar] [CrossRef] [Green Version]
- Salsman, J.; Stathakis, A.; Parker, E.; Chung, D.; Anthes, L.E.; Koskowich, K.L.; Lahsaee, S.; Gaston, D.; Kukurba, K.R.; Smith, K.S.; et al. PML nuclear bodies contribute to the basal expression of the mTOR inhibitor DDIT4. Sci. Rep. 2017, 7, 45038. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Luo, Y. Histone Acetylation and the Regulation of Major Histocompatibility Class II Gene Expression. Adv. Protein Chem. Struct. Biol. 2017, 106, 71–111. [Google Scholar]
- Shiina, T.; Blancher, A.; Inoko, H.; Kulski, J.K. Comparative genomics of the human, macaque and mouse major histocompatibility complex. Immunology 2017, 150, 127–138. [Google Scholar] [CrossRef]
- Shiels, C.; Islam, S.A.; Vatcheva, R.; Sasieni, P.; Sternberg, M.J.; Freemont, P.S.; Sheer, D. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J Cell Sci 2001, 114 Pt 20, 3705–3716. [Google Scholar]
- Ulbricht, T.; Alzrigat, M.; Horch, A.; Reuter, N.; von Mikecz, A.; Steimle, V.; Schmitt, E.; Kramer, O.H.; Stamminger, T.; Hemmerich, P. PML promotes MHC class II gene expression by stabilizing the class II transactivator. J. Cell Biol. 2012, 199, 49–63. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.P.; Bischof, O.; Purbey, P.K.; Notani, D.; Urlaub, H.; Dejean, A.; Galande, S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 2007, 9, 45–56. [Google Scholar] [PubMed] [Green Version]
- Ito, K.; Bernardi, R.; Morotti, A.; Matsuoka, S.; Saglio, G.; Ikeda, Y.; Rosenblatt, J.; Avigan, D.E.; Teruya-Feldstein, J.; Pandolfi, P.P. PML targeting eradicates quiescent leukaemia-initiating cells. Nature 2008, 453, 1072–1078. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Carracedo, A.; Weiss, D.; Arai, F.; Ala, U.; Avigan, D.E.; Schafer, Z.T.; Evans, R.M.; Suda, T.; Lee, C.H.; et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 2012, 18, 1350–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Fu, S.; Zhong, W.; Huang, H. PML overexpression inhibits proliferation and promotes the osteogenic differentiation of human mesenchymal stem cells. Oncol. Rep. 2013, 30, 2785–2794. [Google Scholar] [CrossRef]
- Salsman, J.; Rapkin, L.M.; Margam, N.N.; Duncan, R.; Bazett-Jones, D.P.; Dellaire, G. Myogenic differentiation triggers PML nuclear body loss and DAXX relocalization to chromocentres. Cell Death Dis. 2017, 8, e2724. [Google Scholar] [CrossRef]
- Hadjimichael, C.; Chanoumidou, K.; Nikolaou, C.; Klonizakis, A.; Theodosi, G.-I.; Makatounakis, T.; Papamatheakis, J.; Kretsovali, A. Promyelocytic leukemia protein is an essential regulator of stem cell pluripotency and somatic cell reprogramming. Stem Cell Rep. 2017, 8, 1366–1378. [Google Scholar] [CrossRef] [Green Version]
- de Guzman Strong, C.; Conlan, S.; Deming, C.B.; Cheng, J.; Sears, K.E.; Segre, J.A. A milieu of regulatory elements in the epidermal differentiation complex syntenic block: Implications for atopic dermatitis and psoriasis. Hum. Mol. Genet. 2010, 19, 1453–1460. [Google Scholar] [CrossRef] [Green Version]
- Martin, N.; Patel, S.; Segre, J.A. Long-range comparison of human and mouse Sprr loci to identify conserved noncoding sequences involved in coordinate regulation. Genome Res. 2004, 14, 2430–2438. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.G.; Delva, L.; Gaboli, M.; Rivi, R.; Giorgio, M.; Cordon-Cardo, C.; Grosveld, F.; Pandolfi, P.P. Role of PML in cell growth and the retinoic acid pathway. Science 1998, 279, 1547–1551. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Fessing, M.Y.; Mardaryev, A.N.; Gdula, M.R.; Sharov, A.A.; Sharova, T.Y.; Rapisarda, V.; Gordon, K.B.; Smorodchenko, A.D.; Poterlowicz, K.; Ferone, G.; et al. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J. Cell Biol. 2011, 194, 825–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardaryev, A.N.; Gdula, M.R.; Yarker, J.L.; Emelianov, V.U.; Poterlowicz, K.; Sharov, A.A.; Sharova, T.Y.; Scarpa, J.A.; Joffe, B.; Solovei, I.; et al. p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development (Camb. Engl.) 2014, 141, 101–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Gene | Forward Primer | Revers Primer |
|---|---|---|
| Ivl2 | TGGTTCAGGGACCTCTCAAG | ATGTTTGGGAAAGCCCTTCT |
| Lcs1h | CCTGCTGTAGCTTGGGTTCT | GAGATCTGTGGGGTCTGTGG |
| Lce1e | CAAATGCCAGATCCCAAAGT | AACCCAAGCTACAGCAGGAA |
| Lce1b | GTTGTTGTGGCTCCAGCTCT | CACCACTGCTACAGCATCCA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Połeć, A.; Rowe, A.D.; Blicher, P.; Suganthan, R.; Bjørås, M.; Bøe, S.O. PML Regulates the Epidermal Differentiation Complex and Skin Morphogenesis during Mouse Embryogenesis. Genes 2020, 11, 1130. https://doi.org/10.3390/genes11101130
Połeć A, Rowe AD, Blicher P, Suganthan R, Bjørås M, Bøe SO. PML Regulates the Epidermal Differentiation Complex and Skin Morphogenesis during Mouse Embryogenesis. Genes. 2020; 11(10):1130. https://doi.org/10.3390/genes11101130
Chicago/Turabian StylePołeć, Anna, Alexander D. Rowe, Pernille Blicher, Rajikala Suganthan, Magnar Bjørås, and Stig Ove Bøe. 2020. "PML Regulates the Epidermal Differentiation Complex and Skin Morphogenesis during Mouse Embryogenesis" Genes 11, no. 10: 1130. https://doi.org/10.3390/genes11101130
APA StylePołeć, A., Rowe, A. D., Blicher, P., Suganthan, R., Bjørås, M., & Bøe, S. O. (2020). PML Regulates the Epidermal Differentiation Complex and Skin Morphogenesis during Mouse Embryogenesis. Genes, 11(10), 1130. https://doi.org/10.3390/genes11101130





