Resolvases, Dissolvases, and Helicases in Homologous Recombination: Clearing the Road for Chromosome Segregation
Abstract
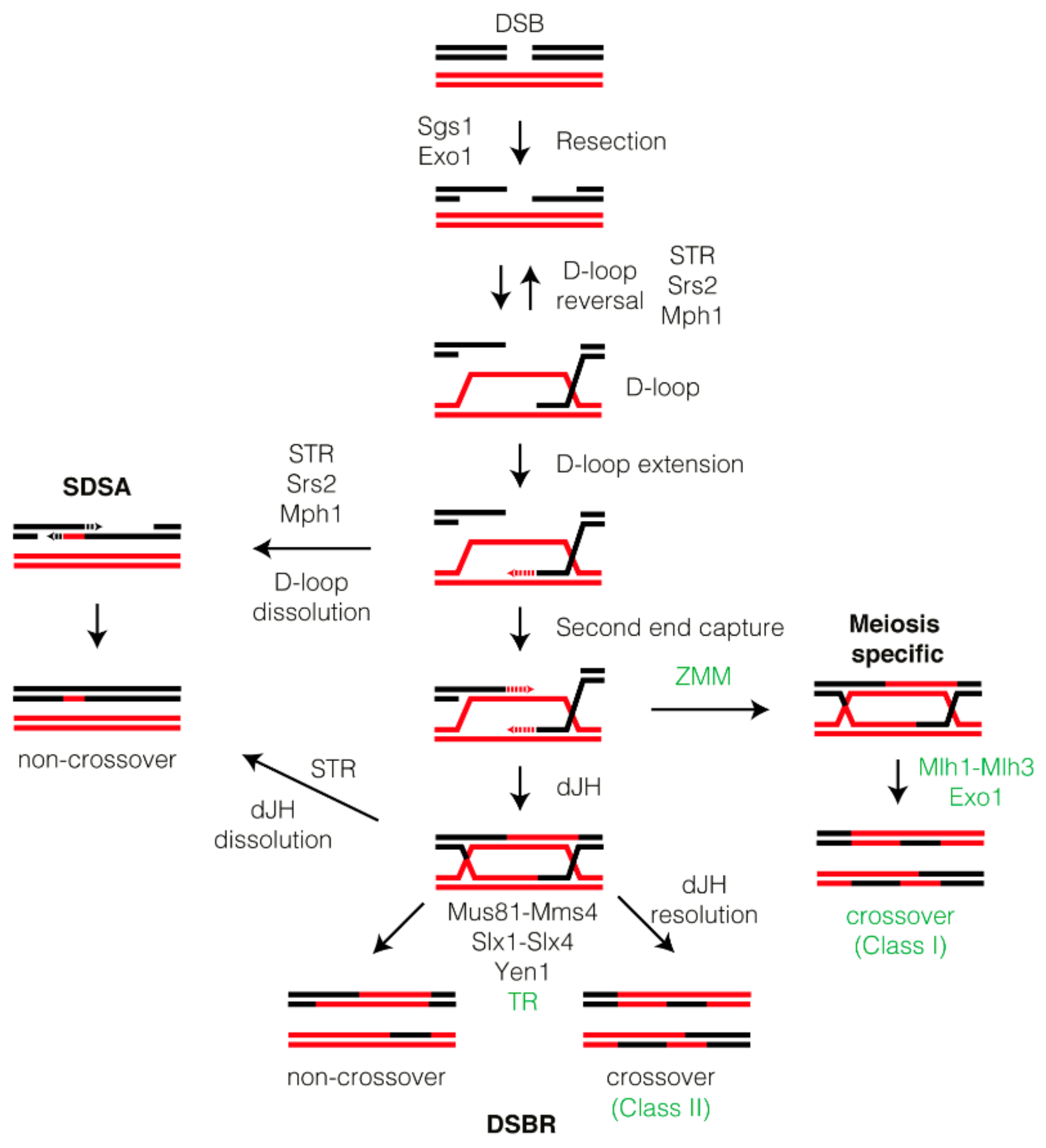
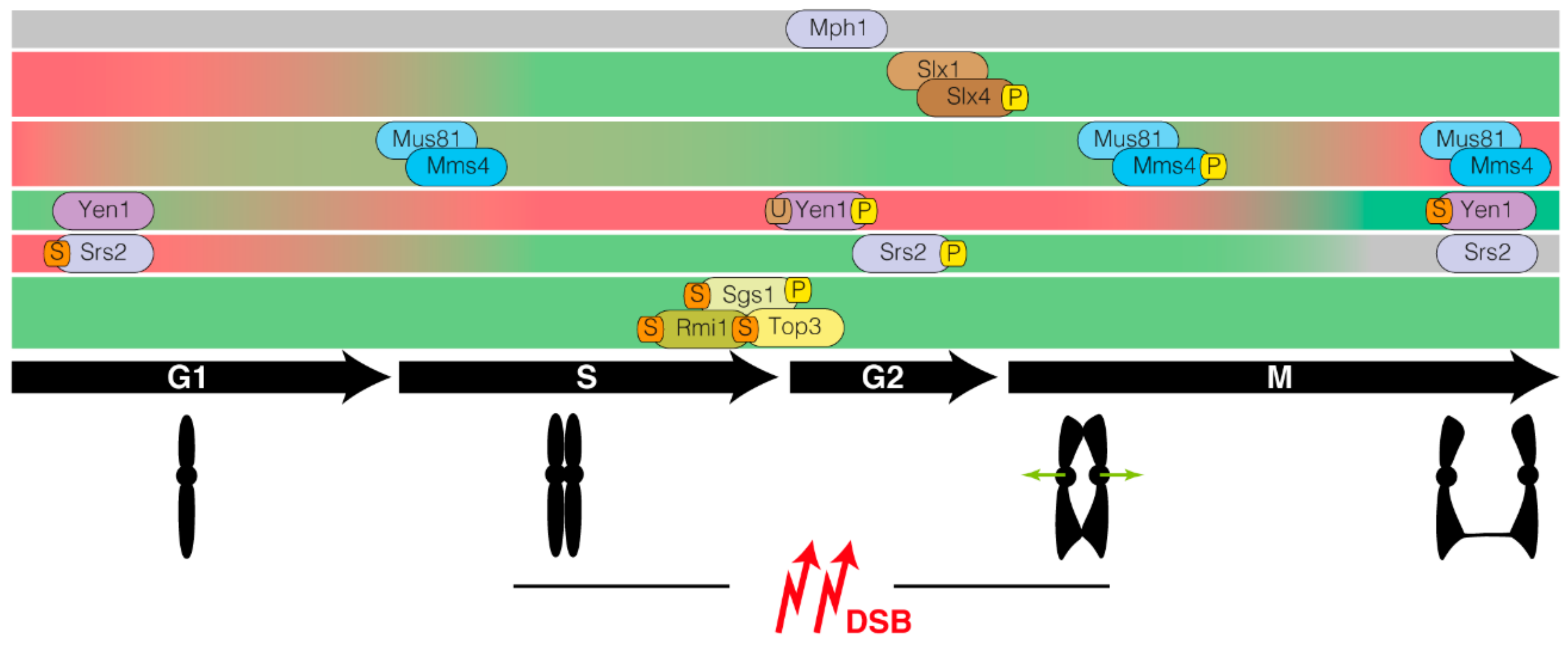
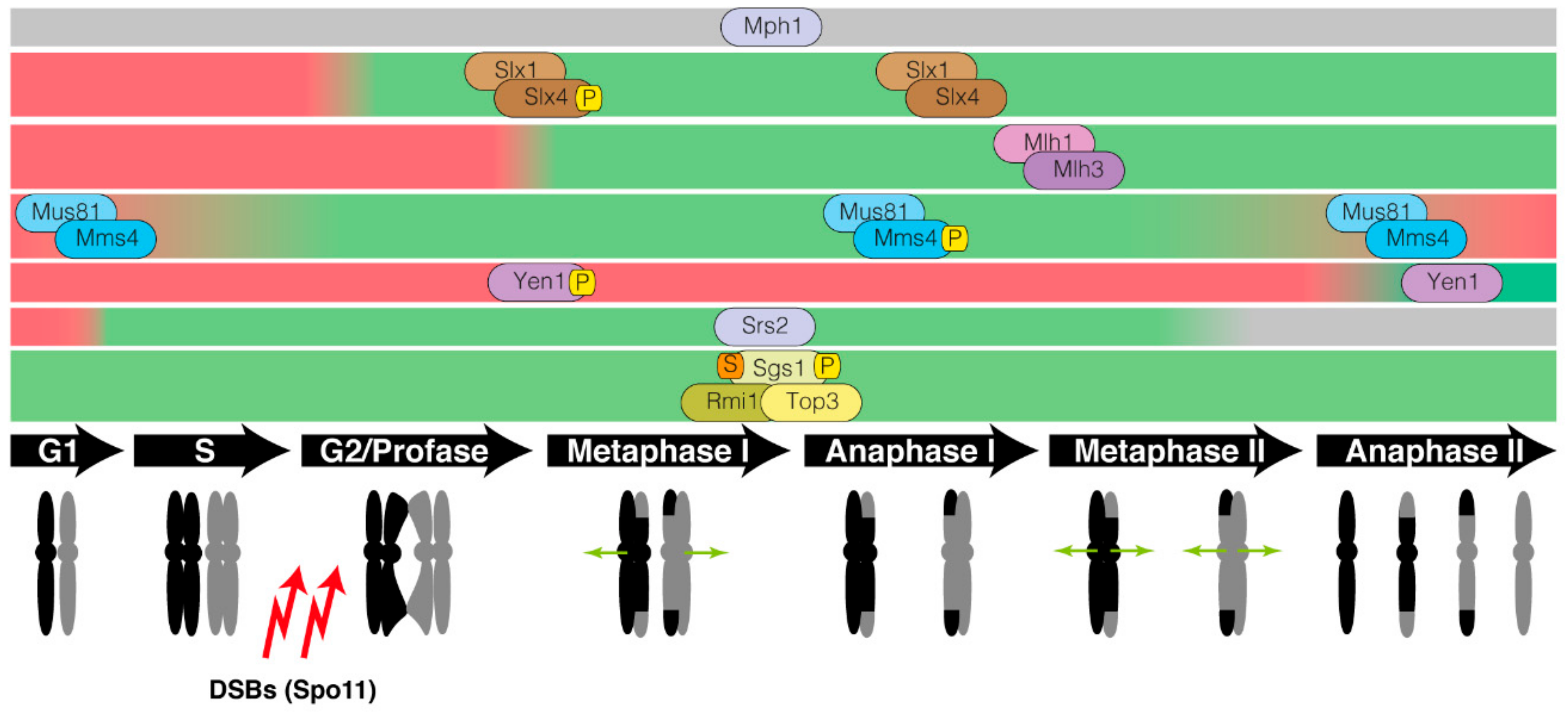
1. Introduction
2. Processing of Recombination Intermediates by Helicases
2.1. The Srs2 Helicase-Translocase
2.2. The Mph1 Helicase Contributes to D-Loop Dismantling
2.3. The Dissolvase Role of the Multifaceted STR Complex
3. Nucleolytic Resolution of Holliday Junctions
3.1. The Mus81/Mms4 Complex: A Crucial Actor in Joint Molecule Resolution
3.2. Yen1: The Last Chance to Resolve DNA Linkages
3.3. Slx1-Slx4: A Scaffold for Other Structure-Selective Nucleases
3.4. Mismatch Repair-Independent Role for Mlh1-Mlh3/Exo1 in the Resolution of Recombination Intermediates
4. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Garcia, V.; Phelps, S.E.; Gray, S.; Neale, M.J. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 2011, 479, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Chung, W.H.; Shim, E.Y.; Lee, S.E.; Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 2008, 134, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Biswas, H.; Goto, G.; Wang, W.; Sung, P.; Sugimoto, K. Ddc2ATRIP promotes Mec1ATR activation at RPA-ssDNA tracts. PLoS Genet. 2019, 15, e1008294. [Google Scholar] [CrossRef] [PubMed]
- Herruzo, E.; Ontoso, D.; Gonzalez-Arranz, S.; Cavero, S.; Lechuga, A.; San-Segundo, P.A. The Pch2 AAA+ ATPase promotes phosphorylation of the Hop1 meiotic checkpoint adaptor in response to synaptonemal complex defects. Nucleic Acids Res. 2016, 44, 7722–7741. [Google Scholar] [CrossRef]
- Ontoso, D.; Acosta, I.; van Leeuwen, F.; Freire, R.; San-Segundo, P.A. Dot1-dependent histone H3K79 methylation promotes activation of the Mek1 meiotic checkpoint effector kinase by regulating the Hop1 adaptor. PLoS Genet. 2013, 9, e1003262. [Google Scholar] [CrossRef]
- Refolio, E.; Cavero, S.; Marcon, E.; Freire, R.; San-Segundo, P.A. The Ddc2/ATRIP checkpoint protein monitors meiotic recombination intermediates. J. Cell Sci. 2011, 124, 2488–2500. [Google Scholar] [CrossRef]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef]
- Bishop, D.K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 1994, 79, 1081–1092. [Google Scholar] [CrossRef]
- Lao, J.P.; Cloud, V.; Huang, C.C.; Grubb, J.; Thacker, D.; Lee, C.Y.; Dresser, M.E.; Hunter, N.; Bishop, D.K. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013, 9, e1003978. [Google Scholar] [CrossRef]
- Cloud, V.; Chan, Y.L.; Grubb, J.; Budke, B.; Bishop, D.K. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 2012, 337, 1222–1225. [Google Scholar] [CrossRef]
- Lee, J.Y.; Terakawa, T.; Qi, Z.; Steinfeld, J.B.; Redding, S.; Kwon, Y.; Gaines, W.A.; Zhao, W.; Sung, P.; Greene, E.C. Base triplet stepping by the Rad51/RecA family of recombinases. Science 2015, 349, 977–981. [Google Scholar] [CrossRef] [PubMed]
- Steinfeld, J.B.; Belan, O.; Kwon, Y.; Terakawa, T.; Al-Zain, A.; Smith, M.J.; Crickard, J.B.; Qi, Z.; Zhao, W.; Rothstein, R.; et al. Defining the influence of Rad51 and Dmc1 lineage-specific amino acids on genetic recombination. Genes Dev. 2019, 33, 1191–1207. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Shah, S.S.; Wright, W.D.; Gore, S.K.; Koszul, R.; Heyer, W.D. Dynamic processing of displacement loops during recombinational DNA repair. Mol. Cell 2019, 73, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Ehmsen, K.T.; Heyer, W.D. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 2008, 36, 2182–2195. [Google Scholar] [CrossRef]
- Gaskell, L.J.; Osman, F.; Gilbert, R.J.; Whitby, M.C. Mus81 cleavage of Holliday junctions: A failsafe for processing meiotic recombination intermediates? EMBO J. 2007, 26, 1891–1901. [Google Scholar] [CrossRef]
- Hollingsworth, N.M.; Brill, S.J. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes Dev. 2004, 18, 117–125. [Google Scholar] [CrossRef]
- Whitby, M.C. Making crossovers during meiosis. Biochem. Soc. Trans. 2005, 33, 1451–1455. [Google Scholar] [CrossRef]
- Haber, J.E. A life investigating pathways that repair broken chromosomes. Annu. Rev. Genet. 2016, 50, 1–28. [Google Scholar] [CrossRef]
- Hunter, N. Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect Biol. 2015, 7. [Google Scholar] [CrossRef]
- Keeney, S.; Lange, J.; Mohibullah, N. Self-organization of meiotic recombination initiation: General principles and molecular pathways. Annu. Rev. Genet. 2014, 48, 187–214. [Google Scholar] [CrossRef]
- Piazza, A.; Heyer, W.D. Moving forward one step back at a time: Reversibility during homologous recombination. Curr Genet. 2019, 65, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Prado, F. Homologous recombination: To fork and beyond. Genes 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Ranjha, L.; Howard, S.M.; Cejka, P. Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma 2018, 127, 187–214. [Google Scholar] [CrossRef] [PubMed]
- Falquet, B.; Rass, U. Structure-specific endonucleases and the resolution of chromosome underreplication. Genes 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Forsburg, S.L. Regulation of structure-specific endonucleases in replication stress. Genes 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Klein, H.L. Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef]
- Aboussekhra, A.; Chanet, R.; Zgaga, Z.; Cassier-Chauvat, C.; Heude, M.; Fabre, F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989, 17, 7211–7219. [Google Scholar] [CrossRef]
- Colavito, S.; Macris-Kiss, M.; Seong, C.; Gleeson, O.; Greene, E.C.; Klein, H.L.; Krejci, L.; Sung, P. Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 2009, 37, 6754–6764. [Google Scholar] [CrossRef]
- Kaniecki, K.; De Tullio, L.; Gibb, B.; Kwon, Y.; Sung, P.; Greene, E.C. Dissociation of Rad51 presynaptic complexes and heteroduplex DNA joints by tandem assemblies of Srs2. Cell Rep. 2017, 21, 3166–3177. [Google Scholar] [CrossRef]
- Liu, J.; Ede, C.; Wright, W.D.; Gore, S.K.; Jenkins, S.S.; Freudenthal, B.D.; Todd Washington, M.; Veaute, X.; Heyer, W.D. Srs2 promotes synthesis-dependent strand annealing by disrupting DNA polymerase δ-extending D-loops. eLife 2017, 6. [Google Scholar] [CrossRef]
- Qiu, Y.; Antony, E.; Doganay, S.; Koh, H.R.; Lohman, T.M.; Myong, S. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat. Commun. 2013, 4, 2281. [Google Scholar] [CrossRef] [PubMed]
- Krejci, L.; Van Komen, S.; Li, Y.; Villemain, J.; Reddy, M.S.; Klein, H.; Ellenberger, T.; Sung, P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 2003, 423, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Veaute, X.; Jeusset, J.; Soustelle, C.; Kowalczykowski, S.C.; Le Cam, E.; Fabre, F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 2003, 423, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Rong, L.; Palladino, F.; Aguilera, A.; Klein, H.L. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 1991, 127, 75–85. [Google Scholar]
- Gangloff, S.; Soustelle, C.; Fabre, F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000, 25, 192–194. [Google Scholar] [CrossRef]
- Klein, H.L. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Δ with other DNA repair genes in Saccharomyces cerevisiae. Genetics 2001, 157, 557–565. [Google Scholar]
- Antony, E.; Tomko, E.J.; Xiao, Q.; Krejci, L.; Lohman, T.M.; Ellenberger, T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell 2009, 35, 105–115. [Google Scholar] [CrossRef]
- Lytle, A.K.; Origanti, S.S.; Qiu, Y.; VonGermeten, J.; Myong, S.; Antony, E. Context-dependent remodeling of Rad51-DNA complexes by Srs2 is mediated by a specific protein-protein interaction. J. Mol. Biol. 2014, 426, 1883–1897. [Google Scholar] [CrossRef]
- Sung, P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997, 11, 1111–1121. [Google Scholar] [CrossRef]
- Liu, J.; Renault, L.; Veaute, X.; Fabre, F.; Stahlberg, H.; Heyer, W.D. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 2011, 479, 245–248. [Google Scholar] [CrossRef]
- De Tullio, L.; Kaniecki, K.; Kwon, Y.; Crickard, J.B.; Sung, P.; Greene, E.C. Yeast Srs2 helicase promotes redistribution of single-stranded DNA-Bound RPA and Rad52 in homologous recombination regulation. Cell Rep. 2017, 21, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Chavdarova, M.; Marini, V.; Sisakova, A.; Sedlackova, H.; Vigasova, D.; Brill, S.J.; Lisby, M.; Krejci, L. Srs2 promotes Mus81-Mms4-mediated resolution of recombination intermediates. Nucleic Acids Res. 2015, 43, 3626–3642. [Google Scholar] [CrossRef] [PubMed]
- Ira, G.; Malkova, A.; Liberi, G.; Foiani, M.; Haber, J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 2003, 115, 401–411. [Google Scholar] [CrossRef]
- Papouli, E.; Chen, S.; Davies, A.A.; Huttner, D.; Krejci, L.; Sung, P.; Ulrich, H.D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 2005, 19, 123–133. [Google Scholar] [CrossRef]
- Pfander, B.; Moldovan, G.L.; Sacher, M.; Hoege, C.; Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005, 436, 428–433. [Google Scholar] [CrossRef]
- Armstrong, A.A.; Mohideen, F.; Lima, C.D. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 2012, 483, 59–63. [Google Scholar] [CrossRef]
- Miura, T.; Shibata, T.; Kusano, K. Putative antirecombinase Srs2 DNA helicase promotes noncrossover homologous recombination avoiding loss of heterozygosity. Proc. Natl. Acad. Sci. USA 2013, 110, 16067–16072. [Google Scholar] [CrossRef]
- Saponaro, M.; Callahan, D.; Zheng, X.; Krejci, L.; Haber, J.E.; Klein, H.L.; Liberi, G. Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet. 2010, 6, e1000858. [Google Scholar] [CrossRef]
- Palladino, F.; Klein, H.L. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 1992, 132, 23–37. [Google Scholar]
- Sasanuma, H.; Furihata, Y.; Shinohara, M.; Shinohara, A. Remodeling of the Rad51 DNA strand-exchange protein by the Srs2 helicase. Genetics 2013, 194, 859–872. [Google Scholar] [CrossRef]
- Hunt, L.J.; Ahmed, E.A.; Kaur, H.; Ahuja, J.S.; Hulme, L.; Chou, T.C.; Lichten, M.; Goldman, A.S.H. S. cerevisiae Srs2 helicase ensures normal recombination intermediate metabolism during meiosis and prevents accumulation of Rad51 aggregates. Chromosoma 2019. [Google Scholar] [CrossRef] [PubMed]
- Sasanuma, H.; Sakurai, H.S.M.; Furihata, Y.; Challa, K.; Palmer, L.; Gasser, S.M.; Shinohara, M.; Shinohara, A. Srs2 helicase prevents the formation of toxic DNA damage during late prophase I of yeast meiosis. Chromosoma 2019. [Google Scholar] [CrossRef] [PubMed]
- Crickard, J.B.; Kaniecki, K.; Kwon, Y.; Sung, P.; Greene, E.C. Meiosis-specific recombinase Dmc1 is a potent inhibitor of the Srs2 antirecombinase. Proc. Natl. Acad. Sci. USA 2018, 115, E10041–E10048. [Google Scholar] [CrossRef] [PubMed]
- Wild, P.; Susperregui, A.; Piazza, I.; Dorig, C.; Oke, A.; Arter, M.; Yamaguchi, M.; Hilditch, A.T.; Vuina, K.; Chan, K.C.; et al. Network rewiring of homologous recombination enzymes during mitotic proliferation and meiosis. Mol. Cell 2019, 75, 859–874. [Google Scholar] [CrossRef]
- Scheller, J.; Schurer, A.; Rudolph, C.; Hettwer, S.; Kramer, W. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 2000, 155, 1069–1081. [Google Scholar]
- Schurer, K.A.; Rudolph, C.; Ulrich, H.D.; Kramer, W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from Homologous recombination, but not from postreplicative repair. Genetics 2004, 166, 1673–1686. [Google Scholar] [CrossRef]
- Prakash, R.; Krejci, L.; Van Komen, S.; Anke Schurer, K.; Kramer, W.; Sung, P. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 2005, 280, 7854–7860. [Google Scholar] [CrossRef]
- Banerjee, S.; Smith, S.; Oum, J.H.; Liaw, H.J.; Hwang, J.Y.; Sikdar, N.; Motegi, A.; Lee, S.E.; Myung, K. Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J. Cell Biol. 2008, 181, 1083–1093. [Google Scholar] [CrossRef]
- Mitchel, K.; Lehner, K.; Jinks-Robertson, S. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet. 2013, 9, e1003340. [Google Scholar] [CrossRef]
- Sebesta, M.; Burkovics, P.; Haracska, L.; Krejci, L. Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair 2011, 10, 567–576. [Google Scholar] [CrossRef]
- Mazon, G.; Symington, L.S. Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination intermediates. Mol. Cell 2013, 52, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Lafuente-Barquero, J.; Luke-Glaser, S.; Graf, M.; Silva, S.; Gomez-Gonzalez, B.; Lockhart, A.; Lisby, M.; Aguilera, A.; Luke, B. The Smc5/6 complex regulates the yeast Mph1 helicase at RNA-DNA hybrid-mediated DNA damage. PLoS Genet. 2017, 13, e1007136. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Choi, K.; Szakal, B.; Arenz, J.; Duan, X.; Ye, H.; Branzei, D.; Zhao, X. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl. Acad. Sci. USA 2009, 106, 21252–21257. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Altmannova, V.; Luke-Glaser, S.; Henriksen, P.; Gallina, I.; Yang, X.; Choudhary, C.; Luke, B.; Krejci, L.; Lisby, M. Mte1 interacts with Mph1 and promotes crossover recombination and telomere maintenance. Genes Dev. 2016, 30, 700–717. [Google Scholar] [CrossRef]
- Xue, X.; Papusha, A.; Choi, K.; Bonner, J.N.; Kumar, S.; Niu, H.; Kaur, H.; Zheng, X.F.; Donnianni, R.A.; Lu, L.; et al. Differential regulation of the anti-crossover and replication fork regression activities of Mph1 by Mte1. Genes Dev. 2016, 30, 687–699. [Google Scholar] [CrossRef]
- Yimit, A.; Kim, T.; Anand, R.P.; Meister, S.; Ou, J.; Haber, J.E.; Zhang, Z.; Brown, G.W. MTE1 Functions with MPH1 in double-strand break repair. Genetics 2016, 203, 147–157. [Google Scholar] [CrossRef]
- Joshi, N.; Brown, M.S.; Bishop, D.K.; Borner, G.V. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol. Cell 2015, 57, 797–811. [Google Scholar] [CrossRef]
- Bernstein, K.A.; Shor, E.; Sunjevaric, I.; Fumasoni, M.; Burgess, R.C.; Foiani, M.; Branzei, D.; Rothstein, R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 2009, 28, 915–925. [Google Scholar] [CrossRef]
- Gangloff, S.; McDonald, J.P.; Bendixen, C.; Arthur, L.; Rothstein, R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A Potential eukaryotic reverse gyrase. Mol. Cell Biol. 1994, 14, 8391–8398. [Google Scholar] [CrossRef]
- Bocquet, N.; Bizard, A.H.; Abdulrahman, W.; Larsen, N.B.; Faty, M.; Cavadini, S.; Bunker, R.D.; Kowalczykowski, S.C.; Cejka, P.; Hickson, I.D.; et al. Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIα and RMI1. Nat. Struct. Mol. Biol. 2014, 21, 261–268. [Google Scholar] [CrossRef]
- Cejka, P.; Plank, J.L.; Dombrowski, C.C.; Kowalczykowski, S.C. Decatenation of DNA by the S. cerevisiae Sgs1-Top3-Rmi1 and RPA complex: A mechanism for disentangling chromosomes. Mol. Cell 2012, 47, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Mimitou, E.P.; Symington, L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 2008, 455, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Kasaciunaite, K.; Fettes, F.; Levikova, M.; Daldrop, P.; Anand, R.; Cejka, P.; Seidel, R. Competing interaction partners modulate the activity of Sgs1 helicase during DNA end resection. EMBO J. 2019, 38, e101516. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Chung, W.H.; Zhu, Z.; Kwon, Y.; Zhao, W.; Chi, P.; Prakash, R.; Seong, C.; Liu, D.; Lu, L.; et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 2010, 467, 108–111. [Google Scholar] [CrossRef]
- Mimitou, E.P.; Yamada, S.; Keeney, S. A global view of meiotic double-strand break end resection. Science 2017, 355, 40–45. [Google Scholar] [CrossRef] [PubMed]
- De Muyt, A.; Jessop, L.; Kolar, E.; Sourirajan, A.; Chen, J.; Dayani, Y.; Lichten, M. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 2012, 46, 43–53. [Google Scholar] [CrossRef]
- Zakharyevich, K.; Tang, S.; Ma, Y.; Hunter, N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 2012, 149, 334–347. [Google Scholar] [CrossRef]
- Fasching, C.L.; Cejka, P.; Kowalczykowski, S.C.; Heyer, W.D. Top3-Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol. Cell 2015, 57, 595–606. [Google Scholar] [CrossRef]
- Haber, J.E. TOPping off meiosis. Mol. Cell 2015, 57, 1142. [Google Scholar] [CrossRef]
- Crickard, J.B.; Xue, C.; Wang, W.; Kwon, Y.; Sung, P.; Greene, E.C. The RecQ helicase Sgs1 drives ATP-dependent disruption of Rad51 filaments. Nucleic Acids Res. 2019, 47, 4694–4706. [Google Scholar] [CrossRef]
- Jessop, L.; Lichten, M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell 2008, 31, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Jessop, L.; Rockmill, B.; Roeder, G.S.; Lichten, M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2006, 2, e155. [Google Scholar] [CrossRef] [PubMed]
- Marsolier-Kergoat, M.C.; Khan, M.M.; Schott, J.; Zhu, X.; Llorente, B. Mechanistic view and genetic control of DNA recombination during meiosis. Mol. Cell 2018, 70, 9–20.e26. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.D.; Lao, J.P.; Hwang, P.Y.; Taylor, A.F.; Smith, G.R.; Hunter, N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 2007, 130, 259–272. [Google Scholar] [CrossRef]
- Kaur, H.; De Muyt, A.; Lichten, M. Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol. Cell 2015, 57, 583–594. [Google Scholar] [CrossRef]
- Tang, S.; Wu, M.K.Y.; Zhang, R.; Hunter, N. Pervasive and essential roles of the Top3-Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol. Cell 2015, 57, 607–621. [Google Scholar] [CrossRef]
- Kaur, H.; Krishnaprasad, G.N.; Lichten, M. Unresolved recombination intermediates cause a RAD9-dependent cell cycle arrest in Saccharomyces cerevisiae. Genetics 2019, 213, 805–818. [Google Scholar] [CrossRef]
- Ampatzidou, E.; Irmisch, A.; O’Connell, M.J.; Murray, J.M. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell Biol. 2006, 26, 9387–9401. [Google Scholar] [CrossRef]
- Bermudez-Lopez, M.; Ceschia, A.; de Piccoli, G.; Colomina, N.; Pasero, P.; Aragon, L.; Torres-Rosell, J. The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res. 2010, 38, 6502–6512. [Google Scholar] [CrossRef]
- Branzei, D.; Sollier, J.; Liberi, G.; Zhao, X.; Maeda, D.; Seki, M.; Enomoto, T.; Ohta, K.; Foiani, M. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 2006, 127, 509–522. [Google Scholar] [CrossRef]
- Sollier, J.; Driscoll, R.; Castellucci, F.; Foiani, M.; Jackson, S.P.; Branzei, D. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol. Biol. Cell 2009, 20, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Bermudez-Lopez, M.; Villoria, M.T.; Esteras, M.; Jarmuz, A.; Torres-Rosell, J.; Clemente-Blanco, A.; Aragon, L. Sgs1’s roles in DNA end resection, HJ dissolution, and crossover suppression require a two-step SUMO regulation dependent on Smc5/6. Genes Dev. 2016, 30, 1339–1356. [Google Scholar] [CrossRef] [PubMed]
- Bonner, J.N.; Choi, K.; Xue, X.; Torres, N.P.; Szakal, B.; Wei, L.; Wan, B.; Arter, M.; Matos, J.; Sung, P.; et al. Smc5/6 mediated sumoylation of the Sgs1-Top3-Rmi1 complex promotes removal of recombination intermediates. Cell Rep. 2016, 16, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Farmer, S.; San-Segundo, P.A.; Aragon, L. The Smc5-Smc6 complex is required to remove chromosome junctions in meiosis. PLoS ONE 2011, 6, e20948. [Google Scholar] [CrossRef]
- Lilienthal, I.; Kanno, T.; Sjogren, C. Inhibition of the Smc5/6 complex during meiosis perturbs joint molecule formation and resolution without significantly changing crossover or non-crossover levels. PLoS Genet. 2013, 9, e1003898. [Google Scholar] [CrossRef]
- Xaver, M.; Huang, L.; Chen, D.; Klein, F. Smc5/6-Mms21 prevents and eliminates inappropriate recombination intermediates in meiosis. PLoS Genet. 2013, 9, e1004067. [Google Scholar] [CrossRef]
- Hegnauer, A.M.; Hustedt, N.; Shimada, K.; Pike, B.L.; Vogel, M.; Amsler, P.; Rubin, S.M.; van Leeuwen, F.; Guenole, A.; van Attikum, H.; et al. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 2012, 31, 3768–3783. [Google Scholar] [CrossRef]
- Villoria, M.T.; Gutierrez-Escribano, P.; Alonso-Rodriguez, E.; Ramos, F.; Merino, E.; Campos, A.; Montoya, A.; Kramer, H.; Aragon, L.; Clemente-Blanco, A. PP4 phosphatase cooperates in recombinational DNA repair by enhancing double-strand break end resection. Nucleic Acids Res. 2019, 47, 10706–10727. [Google Scholar] [CrossRef]
- Interthal, H.; Heyer, W.D. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000, 263, 812–827. [Google Scholar] [CrossRef]
- Kaliraman, V.; Mullen, J.R.; Fricke, W.M.; Bastin-Shanower, S.A.; Brill, S.J. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001, 15, 2730–2740. [Google Scholar] [CrossRef]
- Bastin-Shanower, S.A.; Fricke, W.M.; Mullen, J.R.; Brill, S.J. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell Biol. 2003, 23, 3487–3496. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; McDonald, N.; West, S.C. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008, 77, 259–287. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.K.; Wright, W.D.; Ehmsen, K.T.; Evans, J.E.; Stahlberg, H.; Heyer, W.D. Mus81-Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair. Mol. Cell Biol. 2012, 32, 3065–3080. [Google Scholar] [CrossRef] [PubMed]
- Dayani, Y.; Simchen, G.; Lichten, M. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 2011, 7, e1002083. [Google Scholar] [CrossRef]
- Saugar, I.; Vazquez, M.V.; Gallo-Fernandez, M.; Ortiz-Bazan, M.A.; Segurado, M.; Calzada, A.; Tercero, J.A. Temporal regulation of the Mus81-Mms4 endonuclease ensures cell survival under conditions of DNA damage. Nucleic Acids Res. 2013, 41, 8943–8958. [Google Scholar] [CrossRef]
- Garcia-Luis, J.; Machin, F. Mus81-Mms4 and Yen1 resolve a novel anaphase bridge formed by noncanonical Holliday junctions. Nat. Commun. 2014, 5, 5652. [Google Scholar] [CrossRef]
- Gallo-Fernandez, M.; Saugar, I.; Ortiz-Bazan, M.A.; Vazquez, M.V.; Tercero, J.A. Cell cycle-dependent regulation of the nuclease activity of Mus81-Eme1/Mms4. Nucleic Acids Res. 2012, 40, 8325–8335. [Google Scholar] [CrossRef]
- Matos, J.; Blanco, M.G.; Maslen, S.; Skehel, J.M.; West, S.C. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 2011, 147, 158–172. [Google Scholar] [CrossRef]
- Matos, J.; Blanco, M.G.; West, S.C. Cell-cycle kinases coordinate the resolution of recombination intermediates with chromosome segregation. Cell Rep. 2013, 4, 76–86. [Google Scholar] [CrossRef]
- Princz, L.N.; Wild, P.; Bittmann, J.; Aguado, F.J.; Blanco, M.G.; Matos, J.; Pfander, B. Dbf4-dependent kinase and the Rtt107 scaffold promote Mus81-Mms4 resolvase activation during mitosis. EMBO J. 2017, 36, 664–678. [Google Scholar] [CrossRef]
- Szakal, B.; Branzei, D. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J. 2013, 32, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Mayle, R.; Campbell, I.M.; Beck, C.R.; Yu, Y.; Wilson, M.; Shaw, C.A.; Bjergbaek, L.; Lupski, J.R.; Ira, G. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science 2015, 349, 742–747. [Google Scholar] [CrossRef] [PubMed]
- Froget, B.; Blaisonneau, J.; Lambert, S.; Baldacci, G. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Mol. Biol. Cell 2008, 19, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Budzowska, M.; Davies, S.L.; van Drunen, E.; Onizawa, H.; Beverloo, H.B.; Maas, A.; Essers, J.; Hickson, I.D.; Kanaar, R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007, 14, 1096–1104. [Google Scholar] [CrossRef] [PubMed]
- Kai, M.; Boddy, M.N.; Russell, P.; Wang, T.S. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 2005, 19, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Matulova, P.; Marini, V.; Burgess, R.C.; Sisakova, A.; Kwon, Y.; Rothstein, R.; Sung, P.; Krejci, L. Cooperativity of Mus81.Mms4 with Rad54 in the resolution of recombination and replication intermediates. J. Biol. Chem. 2009, 284, 7733–7745. [Google Scholar] [CrossRef]
- Sebesta, M.; Urulangodi, M.; Stefanovie, B.; Szakal, B.; Pacesa, M.; Lisby, M.; Branzei, D.; Krejci, L. Esc2 promotes Mus81 complex-activity via its SUMO-like and DNA binding domains. Nucleic Acids Res. 2017, 45, 215–230. [Google Scholar] [CrossRef]
- Saugar, I.; Jimenez-Martin, A.; Tercero, J.A. Subnuclear relocalization of structure-specific endonucleases in response to DNA damage. Cell Rep. 2017, 20, 1553–1562. [Google Scholar] [CrossRef]
- De los Santos, T.; Loidl, J.; Larkin, B.; Hollingsworth, N.M. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 2001, 159, 1511–1525. [Google Scholar]
- De los Santos, T.; Hunter, N.; Lee, C.; Larkin, B.; Loidl, J.; Hollingsworth, N.M. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 2003, 164, 81–94. [Google Scholar]
- Argueso, J.L.; Wanat, J.; Gemici, Z.; Alani, E. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 2004, 168, 1805–1816. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.D.; Lao, J.P.; Taylor, A.F.; Smith, G.R.; Hunter, N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell 2008, 31, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Sourirajan, A.; Lichten, M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008, 22, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Kimura, S.; Ishibashi, T.; Mori, Y.; Hashimoto, J.; Sakaguchi, K. OsSEND-1: A new RAD2 nuclease family member in higher plants. Plant Mol. Biol. 2003, 51, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, G.; Kanai, Y.; Takata, K.; Takeuchi, R.; Shimanouchi, K.; Ruike, T.; Furukawa, T.; Kimura, S.; Sakaguchi, K. DmGEN, a novel RAD2 family endo-exonuclease from Drosophila melanogaster. Nucleic Acids Res. 2004, 32, 6251–6259. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ip, S.C.; Rass, U.; Blanco, M.G.; Flynn, H.R.; Skehel, J.M.; West, S.C. Identification of Holliday junction resolvases from humans and yeast. Nature 2008, 456, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Blanco, M.G.; Matos, J.; Rass, U.; Ip, S.C.; West, S.C. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair 2010, 9, 394–402. [Google Scholar] [CrossRef]
- Ho, C.K.; Mazon, G.; Lam, A.F.; Symington, L.S. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell 2010, 40, 988–1000. [Google Scholar] [CrossRef]
- Tay, Y.D.; Wu, L. Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J. Biol. Chem. 2010, 285, 11427–11432. [Google Scholar] [CrossRef]
- Schwartz, E.K.; Heyer, W.D. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 2011, 120, 109–127. [Google Scholar] [CrossRef]
- Pardo, B.; Aguilera, A. Complex chromosomal rearrangements mediated by break-induced replication involve structure-selective endonucleases. PLoS Genet. 2012, 8, e1002979. [Google Scholar] [CrossRef] [PubMed]
- Ashton, T.M.; Mankouri, H.W.; Heidenblut, A.; McHugh, P.J.; Hickson, I.D. Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol. Cell Biol. 2011, 31, 1921–1933. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Galvan, S.; Tous, C.; Blanco, M.G.; Schwartz, E.K.; Ehmsen, K.T.; West, S.C.; Heyer, W.D.; Aguilera, A. Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol. Cell Biol. 2012, 32, 1592–1603. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef]
- Blanco, M.G.; Matos, J.; West, S.C. Dual control of Yen1 nuclease activity and cellular localization by Cdk and Cdc14 prevents genome instability. Mol. Cell 2014, 54, 94–106. [Google Scholar] [CrossRef]
- Eissler, C.L.; Mazon, G.; Powers, B.L.; Savinov, S.N.; Symington, L.S.; Hall, M.C. The Cdk/cDc14 module controls activation of the Yen1 holliday junction resolvase to promote genome stability. Mol. Cell 2014, 54, 80–93. [Google Scholar] [CrossRef]
- Garcia-Luis, J.; Clemente-Blanco, A.; Aragon, L.; Machin, F. Cdc14 targets the Holliday junction resolvase Yen1 to the nucleus in early anaphase. Cell Cycle 2014, 13, 1392–1399. [Google Scholar] [CrossRef]
- Talhaoui, I.; Bernal, M.; Mullen, J.R.; Dorison, H.; Palancade, B.; Brill, S.J.; Mazon, G. Slx5-Slx8 ubiquitin ligase targets active pools of the Yen1 nuclease to limit crossover formation. Nat. Commun. 2018, 9, 5016. [Google Scholar] [CrossRef]
- Agmon, N.; Yovel, M.; Harari, Y.; Liefshitz, B.; Kupiec, M. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011, 39, 7009–7019. [Google Scholar] [CrossRef]
- Arter, M.; Hurtado-Nieves, V.; Oke, A.; Zhuge, T.; Wettstein, R.; Fung, J.C.; Blanco, M.G.; Matos, J. Regulated crossing-over requires inactivation of Yen1/GEN1 resolvase during meiotic prophase I. Dev. Cell 2018, 45, 785–800. [Google Scholar] [CrossRef]
- Aasland, R.; Gibson, T.J.; Stewart, A.F. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends. Biochem. Sci. 1995, 20, 56–59. [Google Scholar] [CrossRef]
- Fricke, W.M.; Brill, S.J. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003, 17, 1768–1778. [Google Scholar] [CrossRef] [PubMed]
- Gaur, V.; Wyatt, H.D.M.; Komorowska, W.; Szczepanowski, R.H.; de Sanctis, D.; Gorecka, K.M.; West, S.C.; Nowotny, M. Structural and mechanistic analysis of the Slx1-Slx4 endonuclease. Cell Rep. 2015, 10, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Mullen, J.R.; Kaliraman, V.; Ibrahim, S.S.; Brill, S.J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 2001, 157, 103–118. [Google Scholar]
- Fabre, F.; Chan, A.; Heyer, W.D.; Gangloff, S. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 2002, 99, 16887–16892. [Google Scholar] [CrossRef]
- Coulon, S.; Gaillard, P.H.L.; Chahwan, C.; McDonald, W.H.; Yates, J.R., 3rd; Russell, P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol. Biol. Cell 2004, 15, 71–80. [Google Scholar] [CrossRef]
- Ito, T.; Chiba, T.; Ozawa, R.; Yoshida, M.; Hattori, M.; Sakaki, Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 2001, 98, 4569–4574. [Google Scholar] [CrossRef]
- Lyndaker, A.M.; Goldfarb, T.; Alani, E. Mutants defective in Rad1-Rad10-Slx4 exhibit a unique pattern of viability during mating-type switching in Saccharomyces cerevisiae. Genetics 2008, 179, 1807–1821. [Google Scholar] [CrossRef]
- Mazon, G.; Lam, A.F.; Ho, C.K.; Kupiec, M.; Symington, L.S. The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nat. Struct. Mol. Biol. 2012, 19, 964–971. [Google Scholar] [CrossRef]
- Roberts, T.M.; Kobor, M.S.; Bastin-Shanower, S.A.; Ii, M.; Horte, S.A.; Gin, J.W.; Emili, A.; Rine, J.; Brill, S.J.; Brown, G.W. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol. Biol. Cell 2006, 17, 539–548. [Google Scholar] [CrossRef]
- Dibitetto, D.; Ferrari, M.; Rawal, C.C.; Balint, A.; Kim, T.; Zhang, Z.; Smolka, M.B.; Brown, G.W.; Marini, F.; Pellicioli, A. Slx4 and Rtt107 control checkpoint signalling and DNA resection at double-strand breaks. Nucleic Acids Res. 2016, 44, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Flott, S.; Alabert, C.; Toh, G.W.; Toth, R.; Sugawara, N.; Campbell, D.G.; Haber, J.E.; Pasero, P.; Rouse, J. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol. Cell Biol. 2007, 27, 6433–6445. [Google Scholar] [CrossRef] [PubMed]
- Flott, S.; Rouse, J. Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem. J. 2005, 391, 325–333. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Princz, L.N.; Gritenaite, D.; Pfander, B. The Slx4-Dpb11 scaffold complex: Coordinating the response to replication fork stalling in S-phase and the subsequent mitosis. Cell Cycle 2015, 14, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Gritenaite, D.; Princz, L.N.; Szakal, B.; Bantele, S.C.; Wendeler, L.; Schilbach, S.; Habermann, B.H.; Matos, J.; Lisby, M.; Branzei, D.; et al. A cell cycle-regulated Slx4-Dpb11 complex promotes the resolution of DNA repair intermediates linked to stalled replication. Genes Dev. 2014, 28, 1604–1619. [Google Scholar] [CrossRef] [PubMed]
- Ohouo, P.Y.; Bastos de Oliveira, F.M.; Liu, Y.; Ma, C.J.; Smolka, M.B. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature 2013, 493, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Ohouo, P.Y.; Bastos de Oliveira, F.M.; Almeida, B.S.; Smolka, M.B. DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol. Cell 2010, 39, 300–306. [Google Scholar] [CrossRef]
- Higashide, M.; Shinohara, M. Budding yeast SLX4 contributes to the appropriate distribution of crossovers and meiotic double-strand break formation on bivalents during meiosis. G3 2016, 6, 2033–2042. [Google Scholar] [CrossRef]
- Prolla, T.A.; Christie, D.M.; Liskay, R.M. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol. Cell Biol. 1994, 14, 407–415. [Google Scholar] [CrossRef][Green Version]
- Flores-Rozas, H.; Kolodner, R.D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. USA 1998, 95, 12404–12409. [Google Scholar] [CrossRef]
- Hunter, N.; Borts, R.H. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 1997, 11, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Harfe, B.D.; Minesinger, B.K.; Jinks-Robertson, S. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol. 2000, 10, 145–148. [Google Scholar] [CrossRef]
- Wang, T.F.; Kleckner, N.; Hunter, N. Functional specificity of MutL homologs in yeast: Evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 1999, 96, 13914–13919. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, N.M.; Ponte, L.; Halsey, C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995, 9, 1728–1739. [Google Scholar] [CrossRef]
- Ross-Macdonald, P.; Roeder, G.S. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 1994, 79, 1069–1080. [Google Scholar] [CrossRef]
- Manhart, C.M.; Ni, X.; White, M.A.; Ortega, J.; Surtees, J.A.; Alani, E. The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans. PLoS Biol. 2017, 15, e2001164. [Google Scholar] [CrossRef]
- Ranjha, L.; Anand, R.; Cejka, P. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J. Biol. Chem. 2014, 289, 5674–5686. [Google Scholar] [CrossRef]
- Rogacheva, M.V.; Manhart, C.M.; Chen, C.; Guarne, A.; Surtees, J.; Alani, E. Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J. Biol. Chem. 2014, 289, 5664–5673. [Google Scholar] [CrossRef]
- Nishant, K.T.; Plys, A.J.; Alani, E. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 2008, 179, 747–755. [Google Scholar] [CrossRef]
- Sonntag Brown, M.; Lim, E.; Chen, C.; Nishant, K.T.; Alani, E. Genetic analysis of mlh3 mutations reveals interactions between crossover promoting factors during meiosis in baker’s yeast. G3 2013, 3, 9–22. [Google Scholar] [CrossRef][Green Version]
- Claeys Bouuaert, C.; Keeney, S. Distinct DNA-binding surfaces in the ATPase and linker domains of MutLγ determine its substrate specificities and exert separable functions in meiotic recombination and mismatch repair. PLoS Genet. 2017, 13, e1006722. [Google Scholar] [CrossRef] [PubMed]
- Zakharyevich, K.; Ma, Y.; Tang, S.; Hwang, P.Y.; Boiteux, S.; Hunter, N. Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double Holliday junctions. Mol. Cell 2010, 40, 1001–1015. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lach, F.P.; Desetty, R.; Hanenberg, H.; Auerbach, A.D.; Smogorzewska, A. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 2011, 43, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Stoepker, C.; Hain, K.; Schuster, B.; Hilhorst-Hofstee, Y.; Rooimans, M.A.; Steltenpool, J.; Oostra, A.B.; Eirich, K.; Korthof, E.T.; Nieuwint, A.W.; et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 2011, 43, 138–141. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M., Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 2013, 13, 542–558. [Google Scholar] [CrossRef]
- Ellis, N.A.; Groden, J.; Ye, T.Z.; Straughen, J.; Lennon, D.J.; Ciocci, S.; Proytcheva, M.; German, J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 1995, 83, 655–666. [Google Scholar] [CrossRef]
- Larsen, N.B.; Hickson, I.D. RecQ Helicases: Conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 2013, 767, 161–184. [Google Scholar] [CrossRef]
- Ji, G.; Long, Y.; Zhou, Y.; Huang, C.; Gu, A.; Wang, X. Common variants in mismatch repair genes associated with increased risk of sperm DNA damage and male infertility. BMC Med. 2012, 10, 49. [Google Scholar] [CrossRef]
- MacGregor, T.P.; Carter, R.; Gillies, R.S.; Findlay, J.M.; Kartsonaki, C.; Castro-Giner, F.; Sahgal, N.; Wang, L.M.; Chetty, R.; Maynard, N.D.; et al. Translational study identifies XPF and MUS81 as predictive biomarkers for oxaliplatin-based peri-operative chemotherapy in patients with esophageal adenocarcinoma. Sci. Rep. 2018, 8, 7265. [Google Scholar] [CrossRef]
- Schlacher, K. PARPi focus the spotlight on replication fork protection in cancer. Nat. Cell Biol. 2017, 19, 1309–1310. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, W.; Shen, Q.; Zhang, P.; Wang, L.; Tao, R.; Li, H.; Ma, X.; Zeng, X.; Cheong, J.H.; et al. The DNA endonuclease Mus81 regulates ZEB1 expression and serves as a target of BET4 inhibitors in gastric cancer. Mol. Cancer Ther. 2019, 18, 1439–1450. [Google Scholar] [CrossRef] [PubMed]
- Zhong, A.; Zheng, H.; Zhang, H.; Sun, J.; Shen, J.; Deng, M.; Chen, M.; Lu, R.; Guo, L. MUS81 inhibition increases the sensitivity to therapy effect in epithelial ovarian cancer via regulating CyclinB pathway. J. Cancer 2019, 10, 2276–2287. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
San-Segundo, P.A.; Clemente-Blanco, A. Resolvases, Dissolvases, and Helicases in Homologous Recombination: Clearing the Road for Chromosome Segregation. Genes 2020, 11, 71. https://doi.org/10.3390/genes11010071
San-Segundo PA, Clemente-Blanco A. Resolvases, Dissolvases, and Helicases in Homologous Recombination: Clearing the Road for Chromosome Segregation. Genes. 2020; 11(1):71. https://doi.org/10.3390/genes11010071
Chicago/Turabian StyleSan-Segundo, Pedro A., and Andrés Clemente-Blanco. 2020. "Resolvases, Dissolvases, and Helicases in Homologous Recombination: Clearing the Road for Chromosome Segregation" Genes 11, no. 1: 71. https://doi.org/10.3390/genes11010071
APA StyleSan-Segundo, P. A., & Clemente-Blanco, A. (2020). Resolvases, Dissolvases, and Helicases in Homologous Recombination: Clearing the Road for Chromosome Segregation. Genes, 11(1), 71. https://doi.org/10.3390/genes11010071




