Identification of Exogenous Nitric Oxide-Responsive miRNAs from Alfalfa (Medicago sativa L.) under Drought Stress by High-Throughput Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Small RNA Library Construction and Sequencing
2.3. Bioinformatic Identification of Conserved and Novel miRNAs
2.4. Differential Expression Analysis of Known and Novel miRNAs
2.5. Prediction of miRNA Targets, Gene Ontology (GO), and KEGG Pathway Analysis
2.6. Validation of miRNA and Target Gene Expression with qRT-PCR
3. Results
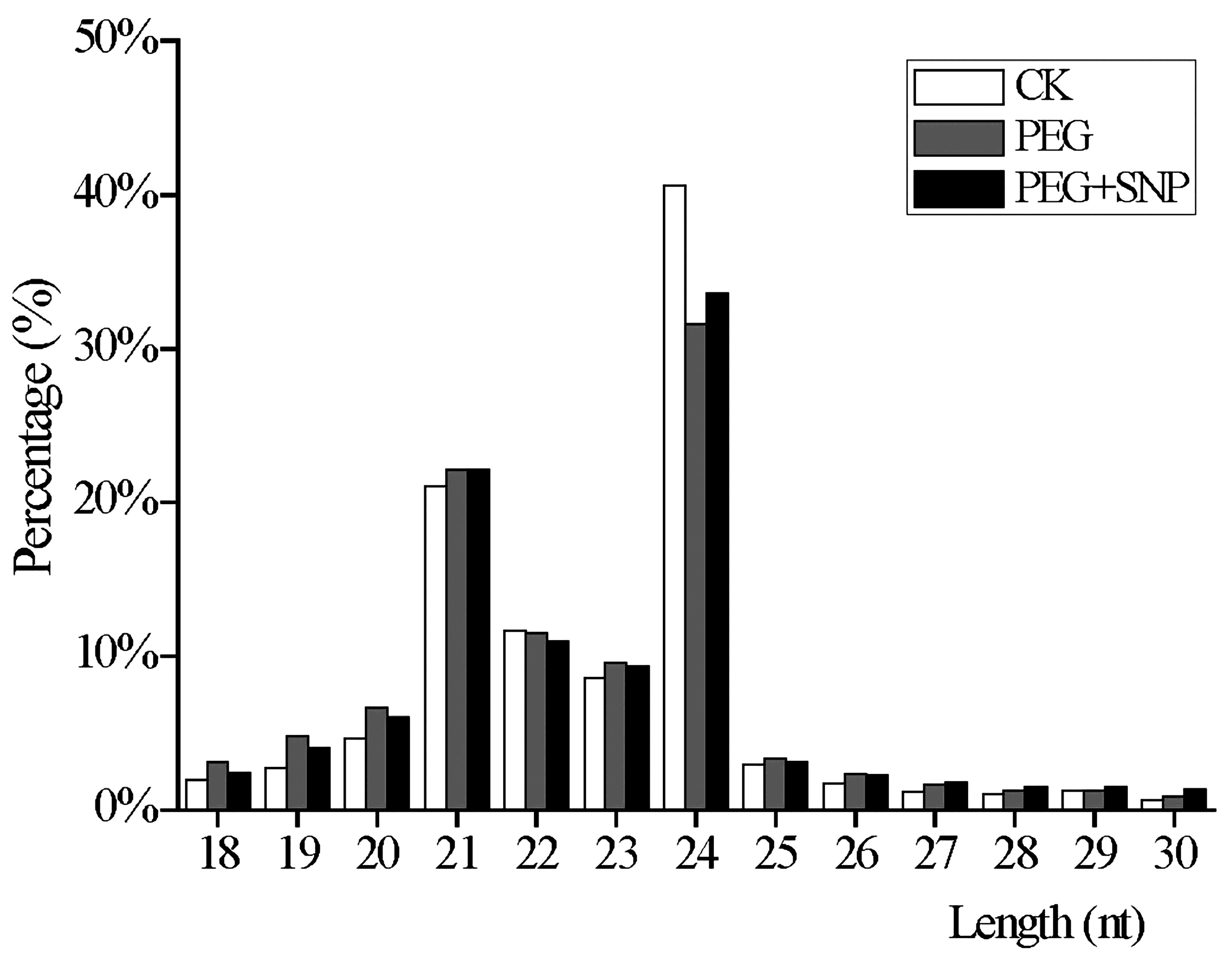
3.1. Deep Sequencing Results of Alfalfa Small RNA (sRNA) Libraries
3.2. Conserved miRNAs from Alfalfa
3.3. Check of Unknown miRNAs in Medicago sativa L. “Sandeli”
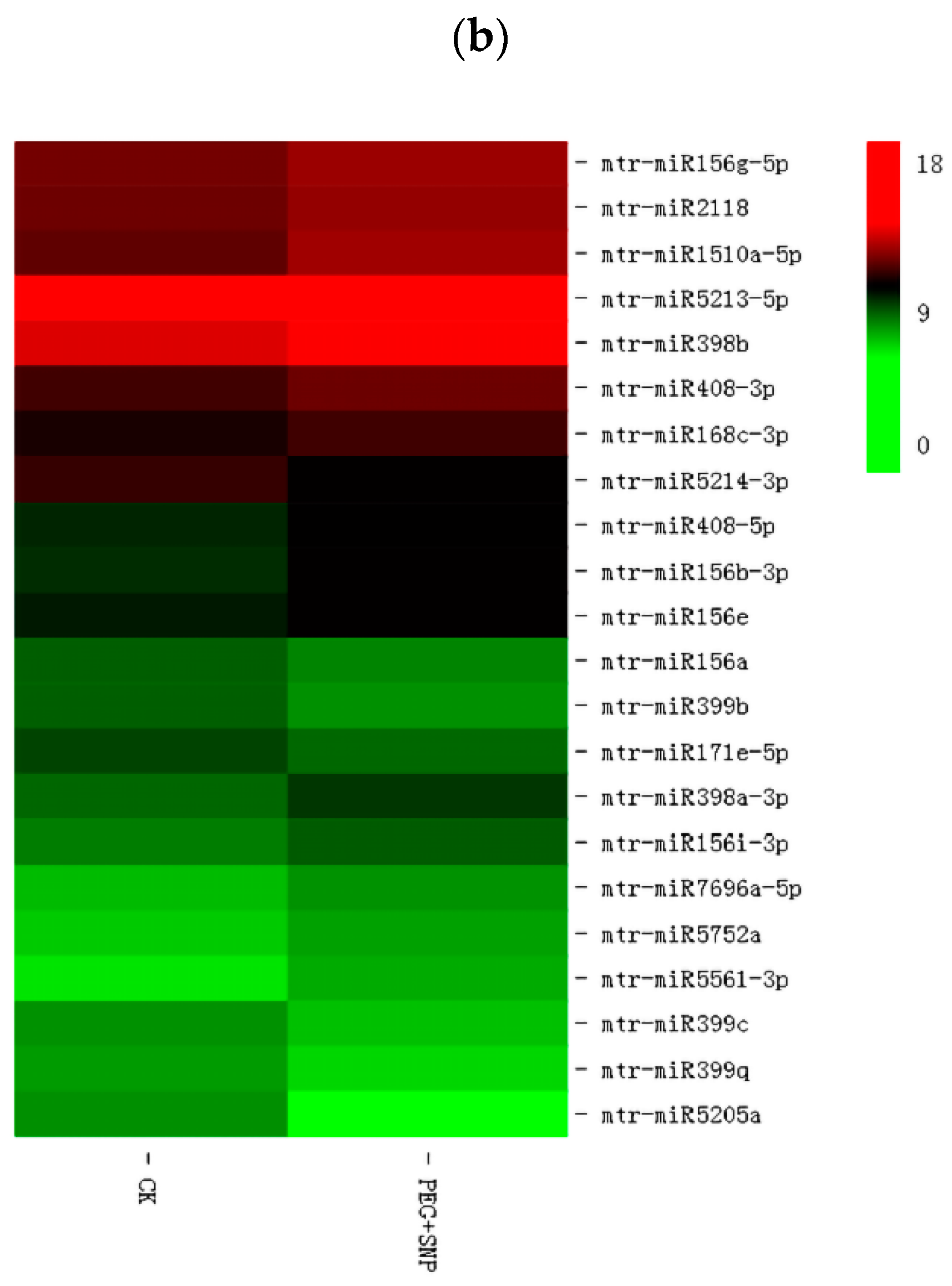
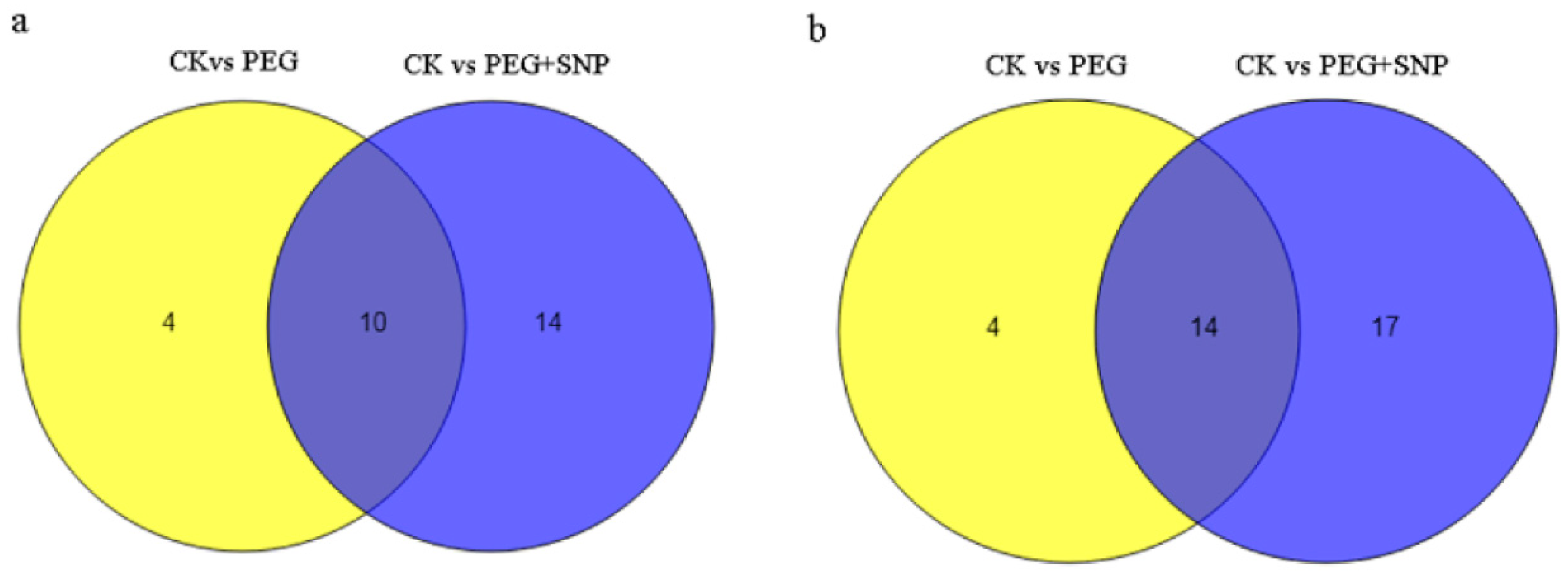
3.4. The Response of miRNA to Drought Stress and Exogenous Nitric Oxide under Drought Stress
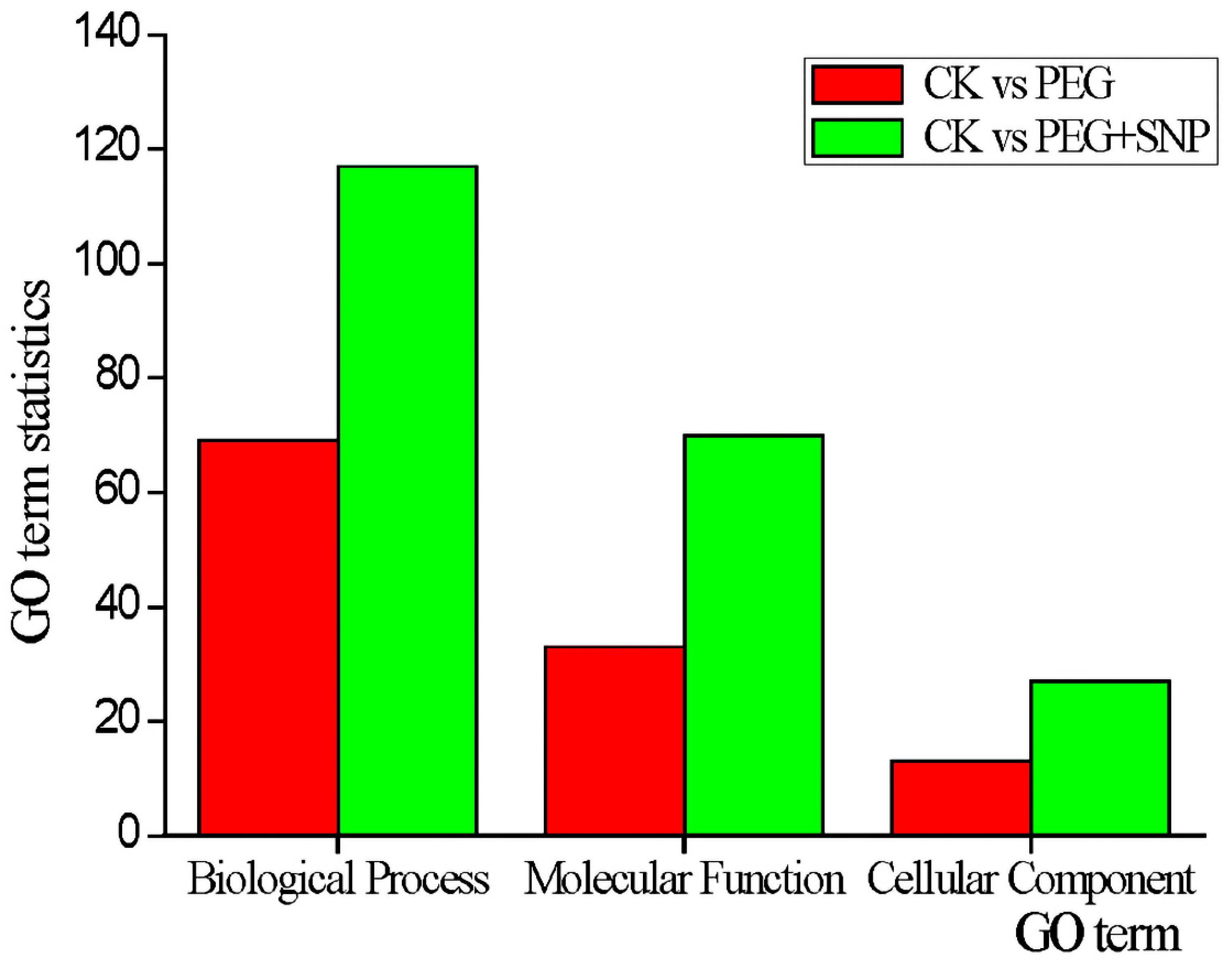
3.5. Prediction and Functional Classification of Target Genes of Drought Stress Response Type and Exogenous Nitric Oxide Reactive miRNA under Drought Stress
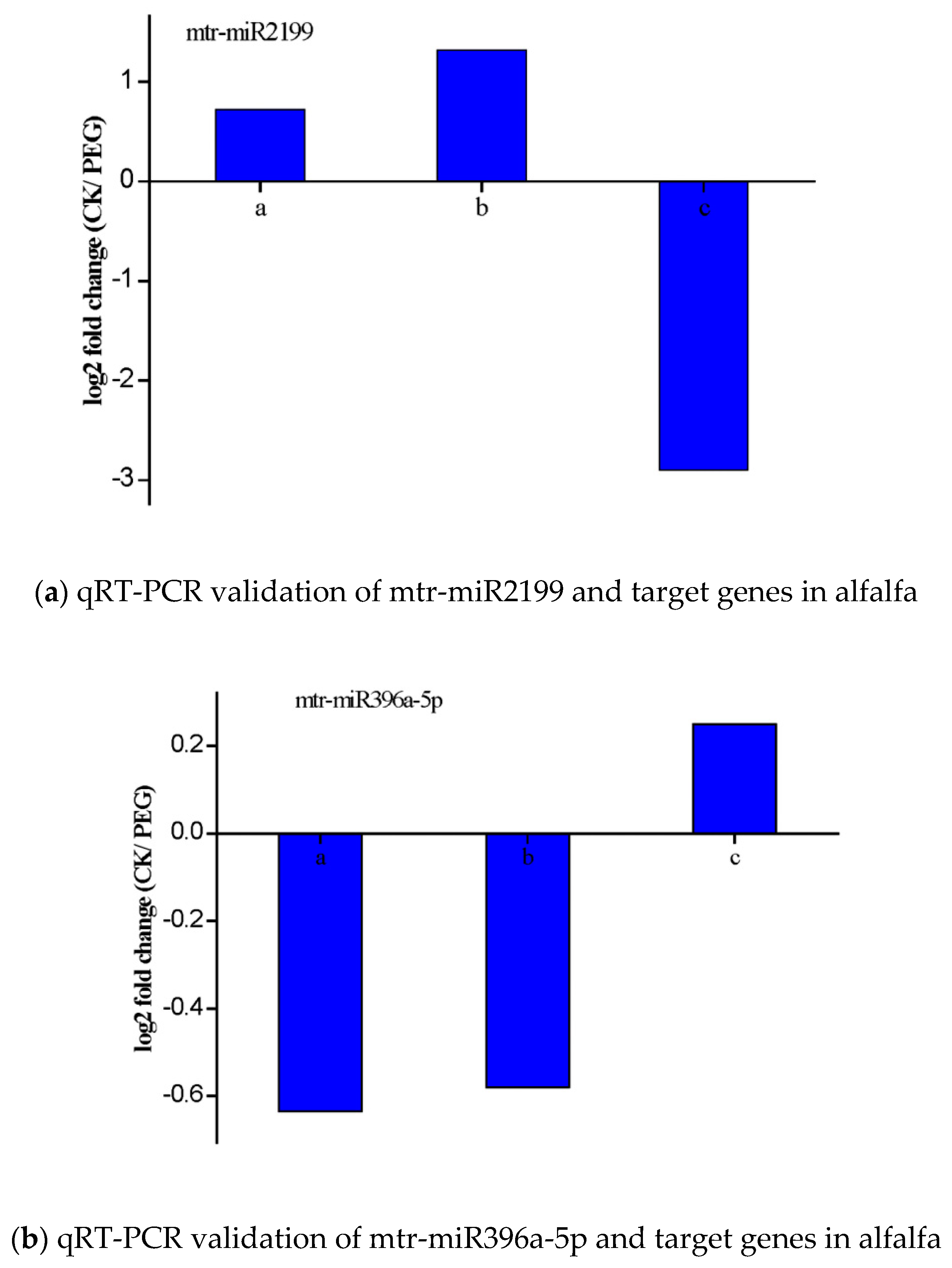
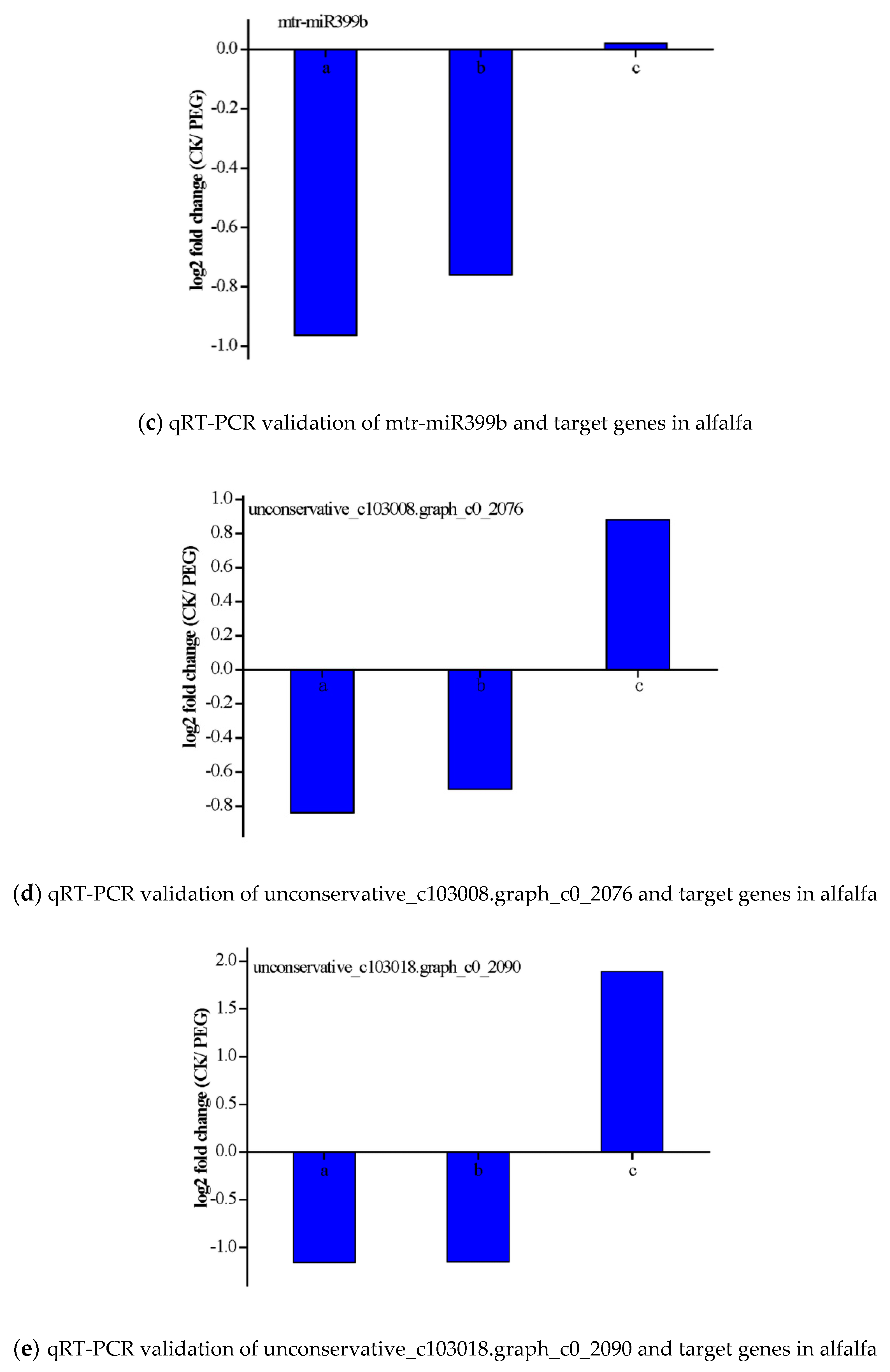
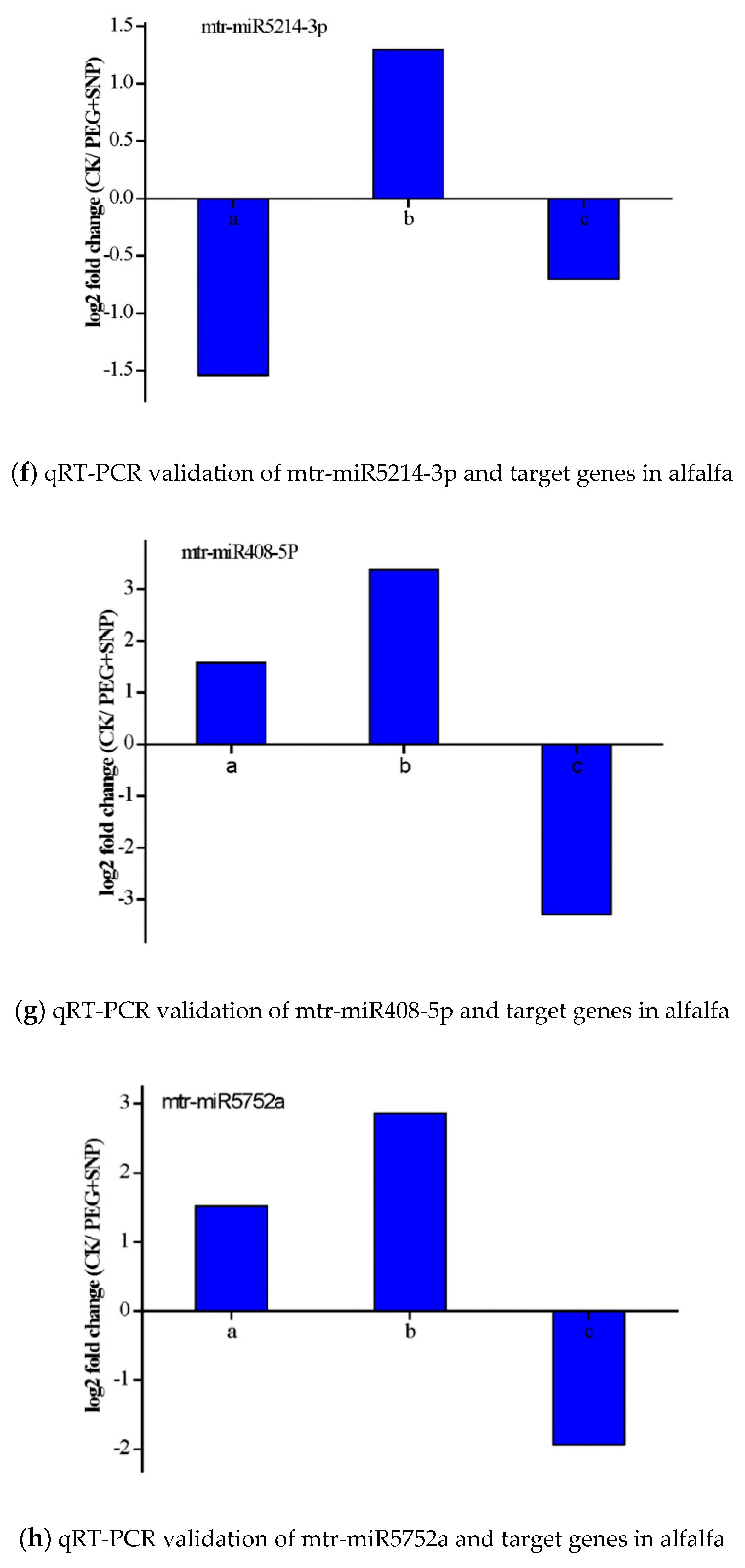
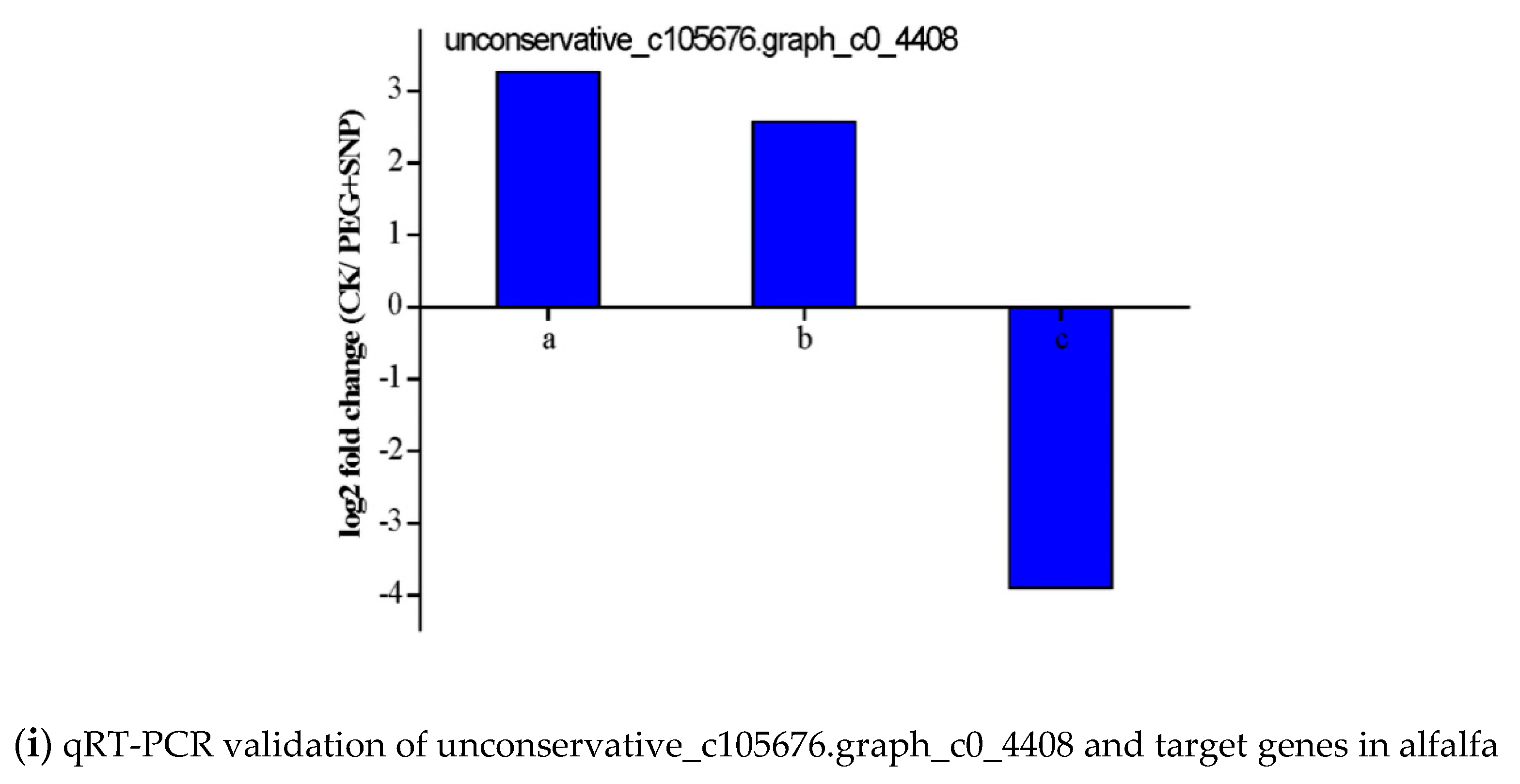
3.6. Validation of the Expression of miRNAs and Their Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shu, Y.; Li, W.; Zhao, J.; Liu, Y.; Guo, C. Transcriptome sequencing and expression profiling of genes involved in the response to abiotic stress in Medicago ruthenica. Genet. Mol. Biol. 2018, 41, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, L.; Lindstrom, O.M.; Markham, J.; Missaoui, A.M. Dissecting key adaptation traits in the polyploid perennial medicago sativa using GBS-SNP mapping. Front. Plant Sci. 2018, 9, 934–953. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liu, M.; Liu, X.L.; Cheng, X.G.; Liang, Z.W. Silicon priming created an enhanced tolerance in alfalfa (Medicago sativa L.) seedlings in response to high alkaline stress. Front. Plant Sci. 2018, 9, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ren, W.; Li, Y.; Xu, Y.; Teng, Y.; Christie, P.; Luo, Y. Nontargeted metabolomic analysis to unravel the impact of di(2-ethylhexyl) phthalate stress on root exudates of alfalfa (Medicago sativa). Sci. Total Environ. 2019, 646, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Cai, H.; Ji, W.; Luo, X.; Wang, Z.; Wu, J.; Wang, X.; Cui, L.; Wang, Y.; Zhu, Y.; et al. Overexpression of GsZFP1 enhances salt and drought tolerance in transgenic alfalfa (Medicago sativa L). Plant Physiol. Biochem. 2013, 71, 22–30. [Google Scholar] [CrossRef]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef]
- Xin, X.Q.; Wei, X.H.; Han, D.; Yue, K.; Zhao, Y. Study on the activities of antioxidant enzymes and isozymes of seeds of alfalfa germination under exogenous NO and reverse regulation of PEG stress. J. Grass Ind. 2018, 27, 105–112. [Google Scholar]
- Wendehenne, D.; Pugin, A.; Klessig, D.F. Nitric oxide: Comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 2001, 6, 177–183. [Google Scholar] [CrossRef]
- Procházková, D.; Haisel, D.; Wilhelmová, N. Effects of exogenous nitric oxide on photosynthesis. Photosynthetica 2013, 51, 483–489. [Google Scholar] [CrossRef]
- Tanou, G.; Job, C.; Rajjou, L.; Arc, E.; Belghazi, M.; Diamantidis, G.; Molassiotis, A.; Job, D. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 2009, 60, 795–804. [Google Scholar] [CrossRef]
- Hao, G.P.; Xing, Y.; Zhang, J.H. Role of nitric oxide dependence on nitric oxide synthase-like activity in the water stress signaling of maize seedling. J. Integr. Plant Biol. 2008, 50, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Indoliya, Y.; Chauha, A.S.; Singh, S.P.; Singh, A.P.; Dwivedi, S.; Tripathi, R.D. Nitric oxide mediated transcriptional modulation enhances plant adaptive responses to arsenic stress. Sci. Rep. 2017, 7, 3592–3605. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, Q.; Wang, Q.; Lu, D.; Zhang, H.; Wang, J.; Fu, R. Transcriptional profiling provides new insights into the role of nitric oxide in enhancing Ganoderma oregonense resistance to heat stress. PLoS ONE 2017, 7, 15694–15708. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.G.; Chen, L.; Zhang, L.L.; Zhang, W.H. Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tzaiolerance in Arabidopsis. Plant Physiol. 2009, 151, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Santisree, P.; Bhatnagar-Mathur, P.; Sharma, K.K. NO to drought-multifunctional role of nitric oxide in plant drought: Do we have all the answers? Plant Sci. 2015, 239, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Yi, Q.; Wei, X.H.; Qiang, X.; Zhao, Y.; Ding, C.F. Investigation into the mechanism of Mo-mediated Ca2+ signaling During seed Germination and antioxidation in Medicago sativa under drought stress. Acta Prataculturae Sin. 2016, 25, 57–65. [Google Scholar]
- Xin, X.Q.; Wei, X.H.; Han, T.; Yue, K.; Zhao, Y. Research on exogenous NO and reverse regulation to the dynamic changes of antioxidant enzymes and isoenzymes in alfalfa seed germination under PEG stress. Acta Prataculturae Sin. 2018, 27, 105–112. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Budak, H.; Akpinar, B.A. Plant miRNAs: Biogenesis, organization and origins. Funct. Integr. Genom. 2015, 15, 523–531. [Google Scholar] [CrossRef]
- Kidner, C.A.; Martienssen, R.A. The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 2005, 8, 38–44. [Google Scholar] [CrossRef]
- Bai, B.; Shi, B.; Hou, N.; Cao, Y.; Meng, Y.; Bian, H.; Zhu, M.; Han, N. microRNAs participate in gene expression regulation and phytohormone cross-talk in barley embryo during seed development and germination. BMC Plant Biol. 2017, 17, 150–166. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2005, 110, 513–520. [Google Scholar] [CrossRef]
- Chen, X. A MicroRNA as a translational repressor of APETALA2 in arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Palatnik, J.F.; Allen, E.; Wu, X.L.; Schommer, C.; Schwab, R.; Carrington, J.C. Control of leaf morphogenesis by micrornas. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Peng, T.; Wang, Q.; Wu, Y.; Chang, J.; Zhang, M.; Tang, G.; Li, C. Development of incompletely fused carpels in maize ovary revealed by miRNA, target gene and phytohormone analysis. Front. Plant Sci. 2017, 8, 463–480. [Google Scholar] [CrossRef]
- Liu, M.M.; Yu, H.Y.; Zhao, G.J.; Huang, Q.F.; Lu, Y.G.; Ouyang, B. Identification of drought-responsive microRNAs in tomato using high-throughput sequencing. Funct. Integr. Genom. 2018, 18, 67–78. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, J.W.; Wang, Z.; Wen, Y.K.; Wang, J.; He, W.H.; Liu, B.L.; Si, H.J.; Wang, D. Identification of novel and conserved microRNAs related to drought stress in potato by deep sequencing. PLoS ONE 2014, 9, e95489. [Google Scholar] [CrossRef]
- Dong, Z.H.; Zhang, J.H.; Zhu, Q.Z.; Zhao, L.F.; Sui, S.X.; Li, Z.S.; Zhang, Y.L.; Wang, H.; Tian, D.L.; Zhao, Y.K. Identification of microRNAs involved in drought stress responses in early-maturing cotton by high-throughput sequencing. Genes Genom. Genes Genom. 2018, 40, 305–314. [Google Scholar] [CrossRef]
- Wang, T.Z.; Chen, L.; Zhao, M.G.; Tian, Q.Y.; Zhang, W.H. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011, 12, 367–378. [Google Scholar] [CrossRef]
- Li, Y.; Wan, L.Q.; Bi, S.Y.; Wan, X.F.; Li, Z.Y.; Cao, J.; Tong, Z.Y.; Xu, H.Y.; He, F.; Li, X.L. Identification of drought-responsive MicroRNAs from roots and leaves of alfalfa by high-throughput sequencing. Genes 2017, 8, 119. [Google Scholar] [CrossRef]
- Arshad, M.; Gruber, M.Y.; Hannoufa, A. Transcriptome analysis of microRNA156 overexpression alfalfa roots under drought stress. Sci. Rep. 2018, 8, 9363–9376. [Google Scholar] [CrossRef] [PubMed]
- Long, R.C.; Li, M.N.; Kang, J.M.; Zhang, T.J.; Sun, Y.; Yang, Q.C. Small RNA deep sequencing identifies novel and salt-stress-regulated microRNAs from roots of Medicago sativa and Medicago truncatula. Physiol. Plant 2015, 154, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.N.; Zhang, S.H.; Du, H.Q.; Sun, X.G.; Shi, Y.H.; Wang, C.Z. Genome-wide identification of different dormant Medicago sativa L. MicroRNAs in response to fall dormancy. PLoS ONE 2014, 9, e114612. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.J.; Liu, Y.; Li, W.; Song, L.L.; Zhang, J.; Guo, C.H. Genome-wide investigation of MicroRNAs and their targets in response to freezing stress in Medicago sativa L. Based on High-Throughput Sequencing. G3 2016, 6, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Xu, H.Y.; Li, Y.; Wan, X.F.; Ma, Z.; Cao, J.; Li, Z.S.; He, F.; Wang, Y.F.; Wan, L.Q.; et al. Analysis of physiological and miRNA responses to Pi deficiency in alfalfa (Medicago sativa L.). Plant Mol. Biol. 2018, 96, 473–492. [Google Scholar] [CrossRef]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Saminathan, T.; Bodunrin, A.; Singh, N.V.; Devarajan, R.; Nimmakayala, P.; Jeff, M.; Aradhya, M.; Reddy, U.K. Genome-wide identification of microRNAs in pomegranate (Punica granatum L.) by high-throughput sequencing. BMC Plant Biol. 2016, 16, 122. [Google Scholar] [CrossRef]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef]
- Lopez-Arredondo, D.L.; Leyva-Gonzalez, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition:improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef]
- Cao, C.Y.; Long, R.C.; Zhang, T.J.; Junmei Kang, J.M.; Wang, Z.; Wang, P.Q.; Sun, H.; Yu, J.; Yang, Q.C. Genome-wide identification of microRNAs in response to salt/alkali stress in Medicago truncatula through high-throughput sequencing. Int. J. Mol. Sci. 2018, 19, 4076. [Google Scholar] [CrossRef]
- Devers, E.A.; Branscheid, A.; May, P.; Krajinski, F. Stars and symbiosis: microRNA- and microRNA-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 2011, 156, 1990–2010. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Jeong, D.H.; De, P.E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [PubMed]
- Eyles, R.P.; Williams, P.H.; Ohms, S.J.; Weiller, G.F.; Ogilvie, H.A.; Djordjevic, M.A.; Imin, N. microRNA profiling of root tissues and root forming explant cultures in Medicago truncatula. Planta 2013, 238, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Cohu, C.M.; Abdel-Ghany, S.E.; Gogolin-Reynolds, K.A.; Onofrio, A.M.; Bodecker, J.R.; Kimbrel, J.A.; Niyogi, K.K.; Pilon, M. Copper Delivery by the Copper Chaperone for Chloroplast and Cytosolic Copper/Zinc-Superoxide Dismutases: Regulation and Unexpected Phenotypes in an Arabidopsis Mutant. Mol. Plant 2009, 2, 1336–1350. [Google Scholar] [CrossRef] [PubMed]
- Gao, J. Genome-wide identification and function analysis of SBP gene family in maize. Acta Agron. Sin. 2016, 42, 201–211. [Google Scholar]
- Liu, J.; Cheng, X.L.; Liu, P.; Sun, J.Q. miR156-Targeted SBP-Box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 2017, 174, 1931–1948. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016, 11, e1117723. [Google Scholar] [CrossRef]
- Liu, J.Y.; Osbourn, A.; Pengda, M. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Castroverde, C.D.M. The AREB1-ADA2b-GCN5 complex regulates gene expression during drought stress. Plant Cell 2018, 31, 559–560. [Google Scholar] [CrossRef]
- Batlang, U.; Baisakh, N.; Ambavaram, M.M.; Pereira, A. Phenotypic and physiological evaluation for drought and salinity stress responses in rice. Methods Mol. Biol. 2013, 956, 209–225. [Google Scholar] [PubMed]
- Mohammadi, P.P.; Moieni, A.; Komatsu, S. Comparative proteome analysis of drought-sensitive and drought-tolerant rapeseed roots and their hybrid F1 line under drought stress. Amino Acids 2012, 43, 2137–2152. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Li, L.Z. Multiple regression analysis reveals microRNA regulatory networks in oryza sativa under drought stress. Int. J. Genom. 2018, 2018, 9395261. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.J.; Zhu, S.W.; Liu, F.; Wang, W.; Wang, X.W.; Han, G.M.; Cheng, B.J. Identification of arbuscular mycorrhiza fungi responsive microRNAs and their regulatory network in maize. Int. J. Mol. Sci. 2018, 19, 3201. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.S.; Zeng, H.Q.; Liu, Z.P.; Yang, Z.M. Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ. 2012, 35, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Candar-Cakir, B.; Arican, E.; Zhang, B. Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol. J. 2016, 14, 1727–1746. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.E.; Kim, W.; Lee, A.R.; Jin, S.; Jun, A.R.; Kim, N.K.; Lee, J.H.; Ahn, J.H. Base-pair opening dynamics of the microRNA precursor pri-miR156a affect temperature-responsive flowering in Arabidopsis. Biochem. Biophys. Res. Commun. 2017, 484, 839–844. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.X.; Cui, W.X.; Guan, C.Y.; Mao, W.J.; Zhang, Z.H. Improvement of fruit quality by overexpressing miR399 in woodland strawberry. J. Agric. Food Chem. 2017, 65, 7361–7370. [Google Scholar] [CrossRef]
- Gaxiola, R.A.; Li, J.; Undurraga, S.; Dang, L.M.; Allen, G.J.; Alper, S.L.; Fink, G.R. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. USA 2001, 98, 11444–11449. [Google Scholar] [CrossRef]
- Zhao, F.Y.; Zhang, X.J.; Li, P.H.; Zhao, Y.X.; Zhang, H. Co-expression of thesuaeda salsa ssnhx1andarabidopsis avp1confer greater salt tolerance to transgenic rice than the singleSsnhx1. Mol. Breed. 2006, 17, 341–353. [Google Scholar] [CrossRef]
- Ferreira, T.H.; Gentile, A.; Vilela, R.D.; Costa, G.G.; Dias, L.I.; Endres, L.; Menossi, M. microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS ONE 2012, 7, e46703. [Google Scholar] [CrossRef] [PubMed]
- Ghorecha, V.; Zheng, Y.; Liu, L.; Sunkar, R.; Krishnayya, N.S.R. MicroRNA dynamics in a wild and cultivated species of Convolvulaceae exposed to drought stress. Physiol. Mol. Biol. Plants 2017, 23, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Trindade, I.; Capitao, C.; Dalmay, T.; Fevereiro, M.; Santos, D. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Ghorecha, V.; Patel, K.; Ingle, S.; Sunkar, R.; Krishnayya, N.S. Analysis of biochemical variations and microRNA expression in wild (Ipomoea campanulata) and cultivated (Jacquemontia pentantha) species exposed to in vivo water stress. Physiol. Mol. Biol. Plants 2014, 20, 57–67. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Bian, B.T.; Gong, T.Y.; Liao, W.B. Comparative proteomic analysis of key proteins during abscisic acid-hydrogen peroxide-induced adventitious rooting in cucumber (Cucumis sativus L.) under drought stress. J. Plant Physiol. 2018, 229, 185–194. [Google Scholar] [CrossRef]
- Huang, K.; Peng, L.; Liu, Y.Y.; Yao, R.D.; Liu, Z.B.; Li, X.F.; Yang, Y.; Wang, J.M. Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 2018, 498, 92–98. [Google Scholar] [CrossRef]
- Geigenberger, P.; Stitt, M. Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J. 2000, 23, 795–806. [Google Scholar] [CrossRef]
- Tiessen, A.; Prescha, K.; Branscheid, A.; Palacios, N.; McKibbin, R.; Halford, N.G.; Geigenberger, P. Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signaling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 2003, 35, 490–500. [Google Scholar] [CrossRef]
- Koster, K.L. Glass formation and desiccation tolerance in seeds. Plant Physiol. 1996, 96, 302–304. [Google Scholar] [CrossRef]


| Category | CK | PEG | PEG + SNP | |||
|---|---|---|---|---|---|---|
| Count | Percentage | Count | Percentage | Count | Percentage | |
| Raw reads | 26,849,326 | 100.00% | 24,252,135 | 100.00% | 20,514,941 | 100.00% |
| Clean reads | 23,528,741 | 87.63% | 18,046,004 | 74.41% | 15,076,692 | 73.49% |
| Mapped to genome | 10,366,134 | 38.61% | 7,651,367 | 31.55% | 6,487,853 | 31.63% |
| miRNAs | 1,267,908 | 4.72% | 1,180,088 | 4.87% | 871,935 | 4.25% |
| rRNA | 2,931,163 | 10.92% | 3,527,200 | 14.54% | 1,974,535 | 9.62% |
| snoRNA | 3746 | 0.01% | 7001 | 0.03% | 10,497 | 0.05% |
| tRNA | 234,634 | 0.87% | 333,859 | 1.38% | 417,378 | 2.03% |
| Without annotation | 20,273,991 | 75.51% | 14,113,193 | 58.19% | 12,602,624 | 61.43% |
| Name | Sequence | Score_Total | Expressional Reads | G + C Contents | Hairpin Energy | Length |
|---|---|---|---|---|---|---|
| c104339.graph_c0_3174 | UUUCAUUCCAUAUGUCAUCUAG | 17,723.5 | 14,392 | 31.82% | −53.6 | 22 |
| c111080.graph_c1_10087 | GGCGGAUGUAGCCAAGUGGA | 1,721,964.4 | 730,326 | 60.00% | −53.2 | 20 |
| c114875.graph_c0_14536 | UUCAUUUCUAAAAUAGGCAUUG | 9543.7 | 7780 | 27.27% | −59.2 | 22 |
| c116131.graph_c0_16318 | CGGGAUCGGAGAUUAGAGAAU | 146,625.1 | 106,408 | 47.62% | −73.0 | 21 |
| c86020.graph_c0_42677 | UACGUCUCUGUCUUUCGGGUUG | 16,890.1 | 13,750 | 50.00% | −74.6 | 22 |
| c99918.graph_c0_48282 | UUAAUCAAGGAAAUCACAGUC | 13,503.9 | 13,046 | 33.33% | −55.0 | 21 |
| c102504.graph_c0_1594 | AUGGUUCUUGUUCAGUAGAGU | 30,812.8 | 6815 | 38.10% | −78.4 | 21 |
| c91116.graph_c0_44142 | UUGUGGAACAUAGAAGCACGUG | 9771.1 | 2317 | 45.45% | −63.6 | 22 |
| c121624.graph_c0_25405 | UUGAUUCUCAUCACAACUUGG | 14,278.7 | 7926 | 38.10% | −66.2 | 21 |
| c111105.graph_c0_10123 | UUUGGCAUUCUGUCCACCUCC | 8328.7 | 7532 | 52.38% | −55.6 | 21 |
| miRNAs | Sequences | Target Gene or Protein | Description of Function | Reference |
|---|---|---|---|---|
| mtr-miR156a | tgacagaagagagagagcaca | SQUAMOSA promoter-binding-like protein (SPL) genes. | Anthocyanin biosynthesis; vegetative phase transition. | [30,36,37] |
| mtr-miR399a | tgccaaaggagatttgcccag | Phosphate transporter; high affinity inorganic phosphate transporter. | Pi uptake. | [38,39] |
| mtr-miR399c | tgccaaaggagatttg | |||
| mtr-miR399q | ccctgtgccaaaggagagctgctctt | |||
| mtr-miR5213-5p | tacgtgtgtcttcacctctgaa | Disease resistance protein. | Response to salt/alkali stress. | [40,41] |
| mtr-miR5752a | cattgtttggtttagtacaaa | Starch synthase; amino acid binding; metal ion binding. | Starch, galactolipid, amylopectin biosynthetic; response to hypoxia; regulation of ethylene-activated; metabolic process; metal ion transport. | [42] |
| mtr-miR7696a-5p | tcaagttctcataattcaaaa | Chitin binding; protein kinase. | Innate immune response; protein phosphorylation; cell wall macromolecule catabolic. | [43] |
| mtr-miR398a-5p | ggagtgacactgagaacacaag | Cu/Zn-superoxide dismutase copper chaperone. | Defense against reactive oxygen toxicity. | [44] |
| mtr-miR5232 | tacatgtcgctctcacctgaa | Glycoside hydrolase; type IIB calcium ATPase protein kinase. | Participate in metabolism. | [41] |
| mtr-miR5559-5p | tacttggtgaattgttggatc | inorganic diphosphatase magnesium ion binding. | Response to cadmium ion; phosphate-containing compound metabolic. | [29] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Ma, W.; Wei, X.; Long, Y.; Zhao, Y.; Su, M.; Luo, Q. Identification of Exogenous Nitric Oxide-Responsive miRNAs from Alfalfa (Medicago sativa L.) under Drought Stress by High-Throughput Sequencing. Genes 2020, 11, 30. https://doi.org/10.3390/genes11010030
Zhao Y, Ma W, Wei X, Long Y, Zhao Y, Su M, Luo Q. Identification of Exogenous Nitric Oxide-Responsive miRNAs from Alfalfa (Medicago sativa L.) under Drought Stress by High-Throughput Sequencing. Genes. 2020; 11(1):30. https://doi.org/10.3390/genes11010030
Chicago/Turabian StyleZhao, Yaodong, Wenjing Ma, Xiaohong Wei, Yu Long, Ying Zhao, Meifei Su, and Qiaojuan Luo. 2020. "Identification of Exogenous Nitric Oxide-Responsive miRNAs from Alfalfa (Medicago sativa L.) under Drought Stress by High-Throughput Sequencing" Genes 11, no. 1: 30. https://doi.org/10.3390/genes11010030
APA StyleZhao, Y., Ma, W., Wei, X., Long, Y., Zhao, Y., Su, M., & Luo, Q. (2020). Identification of Exogenous Nitric Oxide-Responsive miRNAs from Alfalfa (Medicago sativa L.) under Drought Stress by High-Throughput Sequencing. Genes, 11(1), 30. https://doi.org/10.3390/genes11010030





