BACE1 Inhibition Using 2’-OMePS Steric Blocking Antisense Oligonucleotides
Abstract
:1. Introduction
2. Materials and Methods
2.1. AO Design and Synthesis
2.2. Cell Culture and Transfection
2.3. RNA Extraction and RT-PCR
2.4. Western Blot
3. Results
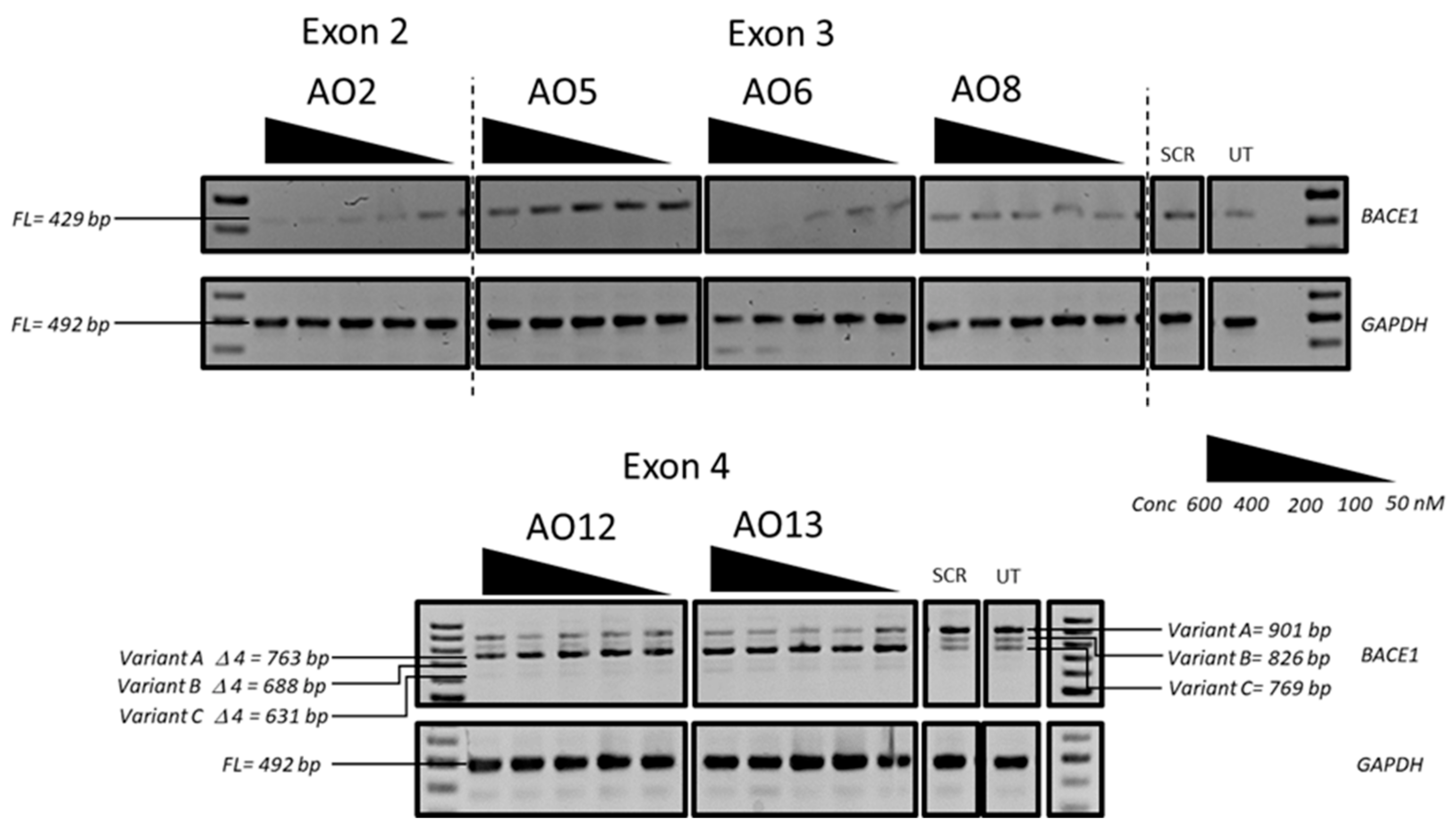
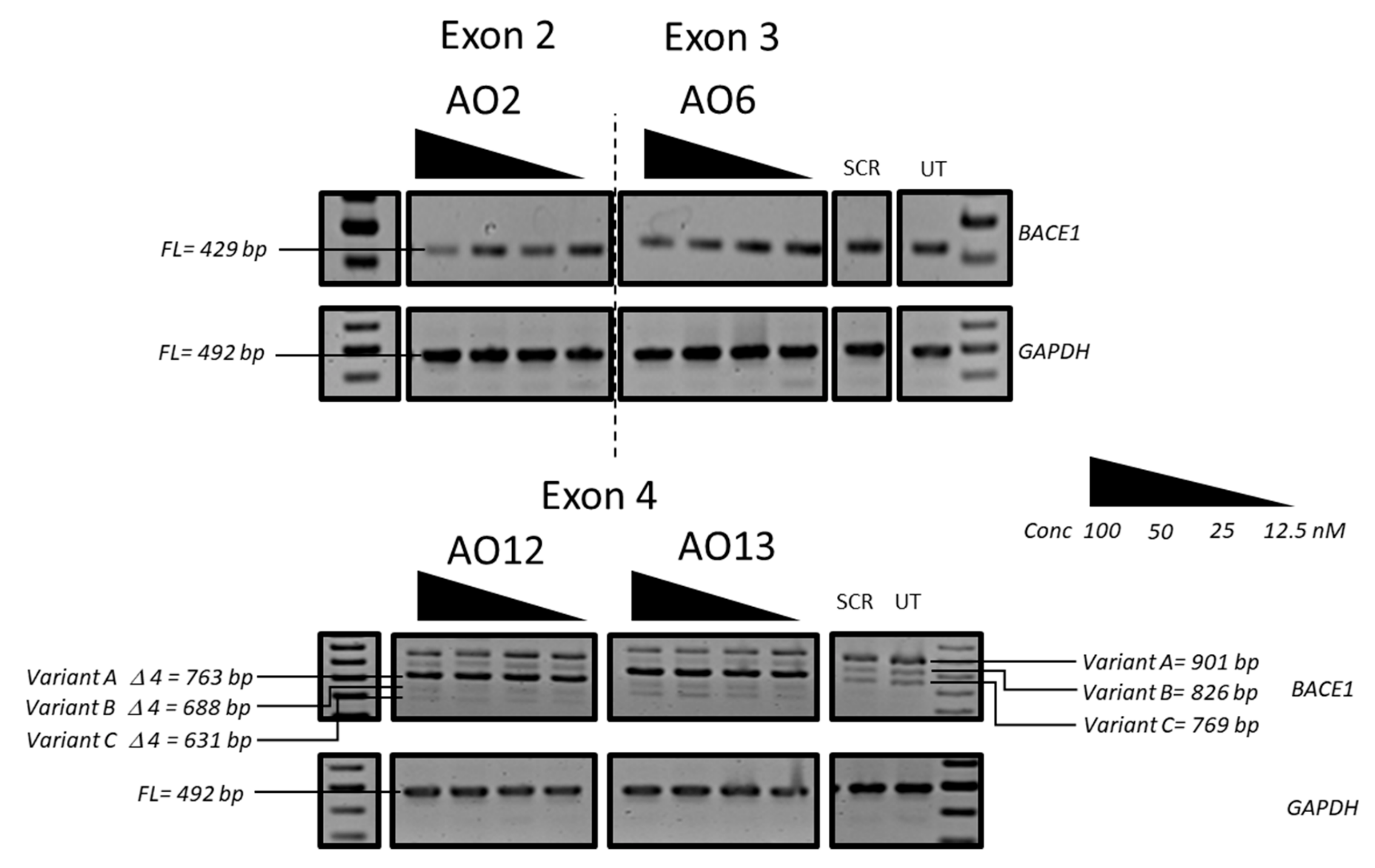
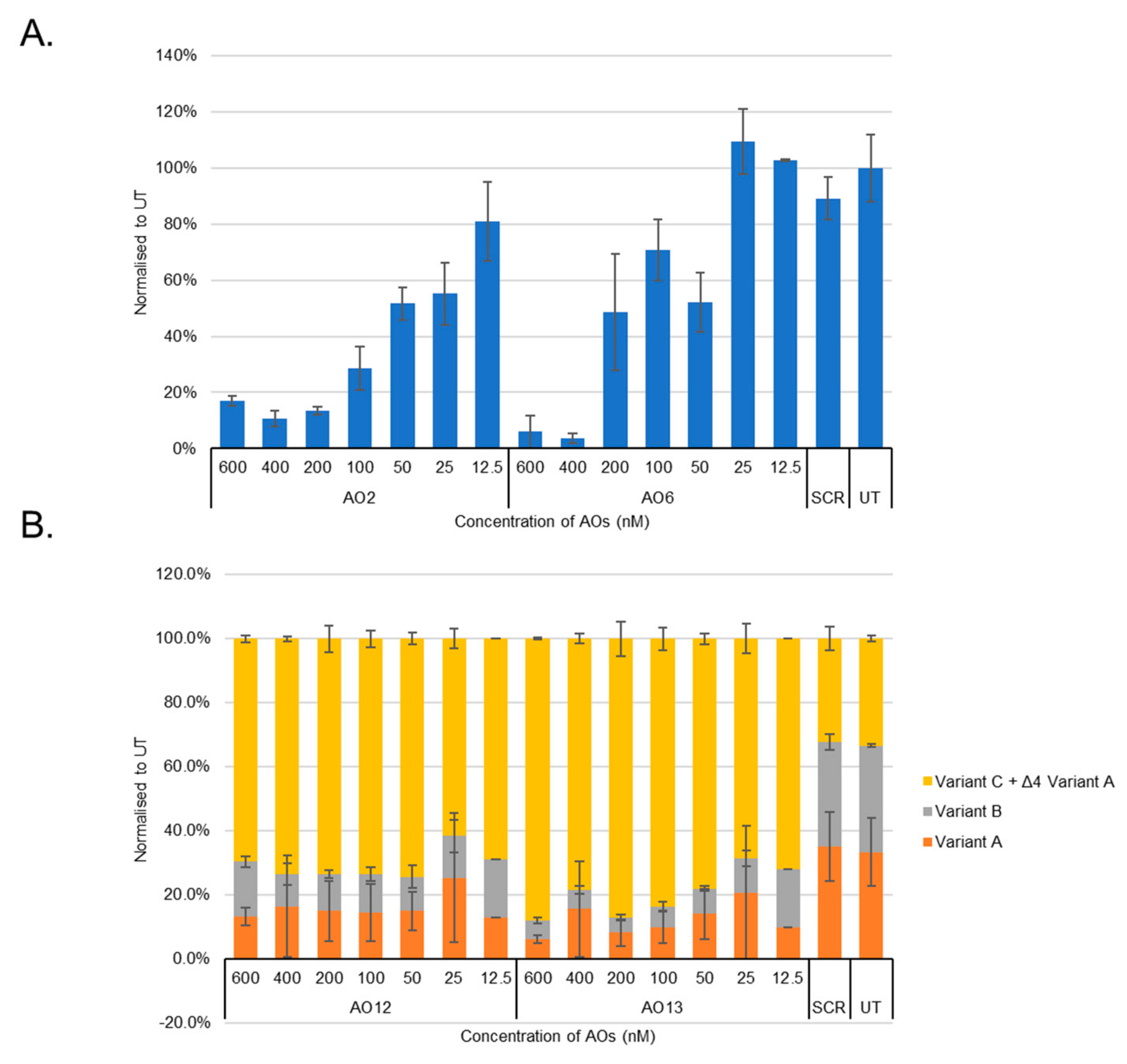
3.1. Evaluation of the Most Efficient 2’-OMePS AOs to Induce Exon-Skipping of the BACE1 Transcript in HEK293 Cells In Vitro
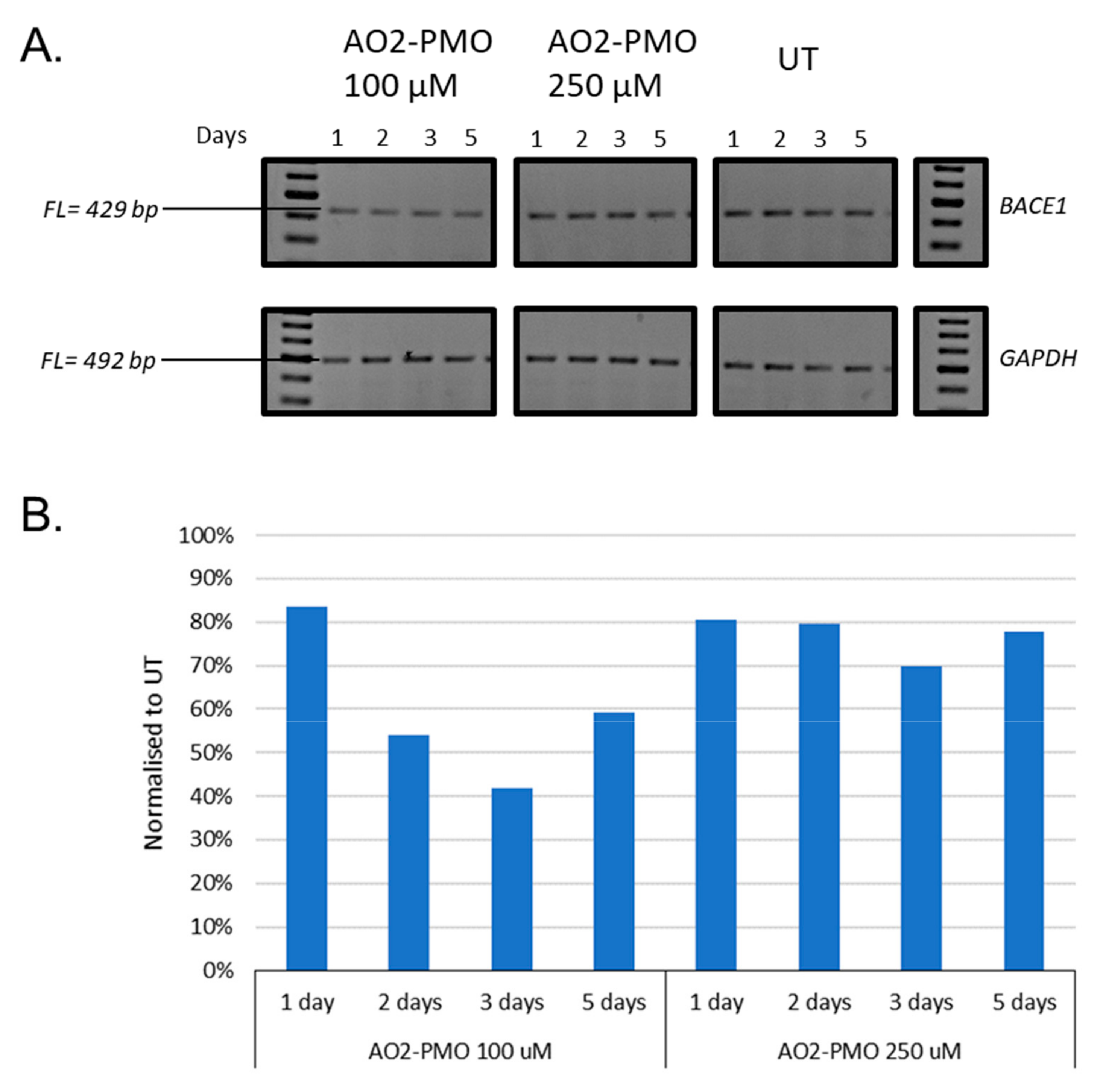
3.2. Evaluation of the Most Efficient 2’-OMePS AOs as a PMO to Induce Exon-Skipping of BACE1 Transcript in HEK293 Cells In Vitro
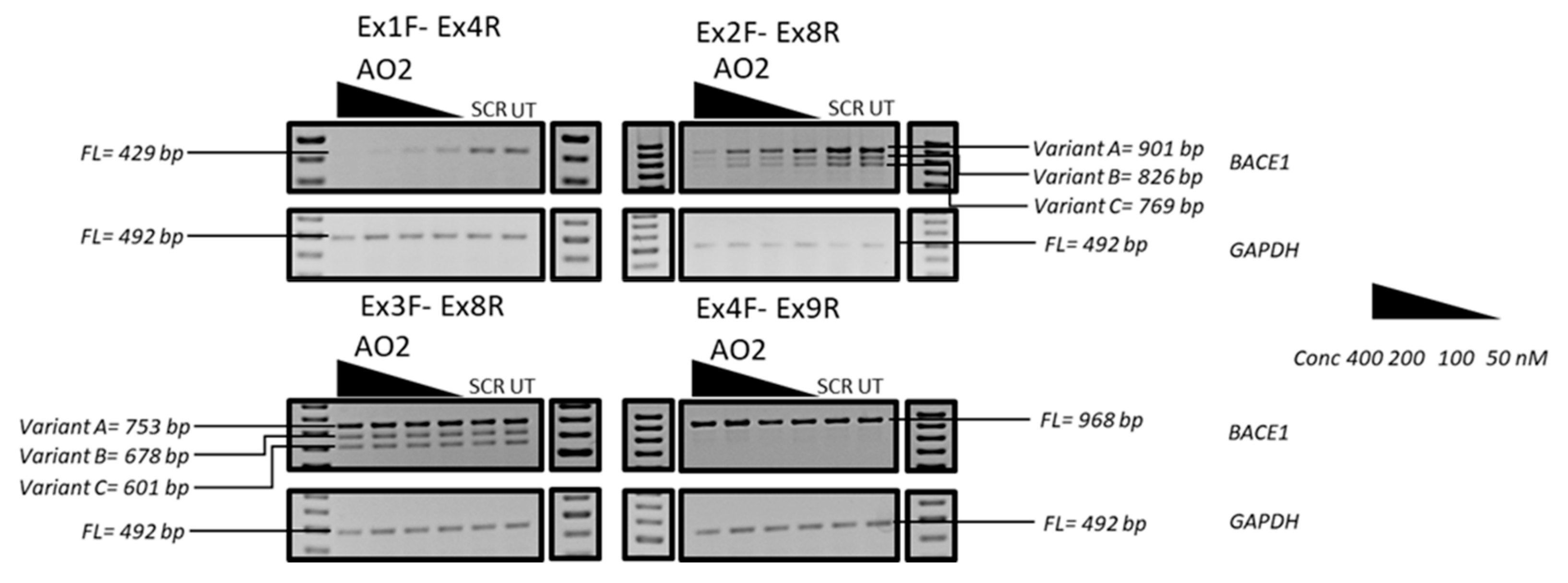
3.3. Evaluation of the Mechanism of Action of AO2
3.4. Evaluation of BACE1 Protein Downregulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Näslund, J. Correlation between elevated levels of amyloid β-peptide in the brain and cognitive decline. JAMA 2000, 283, 1571–1577. [Google Scholar] [CrossRef] [PubMed]
- Gandy, S. The role of cerebral amyloid β accumulation in common forms of alzheimer disease. J. Clin. Investig. 2005, 115, 1121–1129. [Google Scholar] [PubMed]
- Li, R.; Lindholm, K.; Yang, L.B.; Yue, X.; Citron, M.; Yan, R.; Beach, T.; Sue, L.; Sabbagh, M.; Cai, H.; et al. Amyloid peptide load is correlated with increased -secretase activity in sporadic alzheimer’s disease patients. Proc. Natl. Acad. Sci. USA 2004, 101, 3632–3637. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 2001, 81, 741–766. [Google Scholar] [CrossRef]
- Cole, S.L.; Vassar, R. The alzheimer’s disease β-secretase enzyme, bace1. Mol. Neurodegener. 2007, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-B.; Lindholm, K.; Yan, R.; Citron, M.; Xia, W.; Yang, X.-L.; Beach, T.; Sue, L.; Wong, P.; Price, D.; et al. Elevated β-secretase expression and enzymatic activity detected in sporadic alzheimer disease. Nat. Med. 2003, 9, 3–4. [Google Scholar] [CrossRef]
- Zhao, J.; Fu, Y.; Yasvoina, M.; Shao, P.; Hitt, B.; O’Connor, T.; Logan, S.; Maus, E.; Citron, M.; Berry, R.; et al. β-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: Implications for alzheimer’s disease pathogenesis. J. Neurosci. 2007, 27, 3639–3649. [Google Scholar] [CrossRef]
- Sadleir, K.R.; Kandalepas, P.C.; Buggia-Prévot, V.; Nicholson, D.A.; Thinakaran, G.; Vassar, R. Presynaptic dystrophic neurites surrounding amyloid plaques are sites of microtubule disruption, bace1 elevation, and increased aβ generation in alzheimer’s disease. Acta Neuropathol. 2016, 132, 235–256. [Google Scholar] [CrossRef]
- Stein, C.A.; Castanotto, D. Fda-approved oligonucleotide therapies in 2017. Mol. Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef]
- Panza, F.; Lozupone, M.; Solfrizzi, V.; Sardone, R.; Piccininni, C.; Dibello, V.; Stallone, R.; Giannelli, G.; Bellomo, A.; Greco, A.; et al. Bace inhibitors in clinical development for the treatment of alzheimer’s disease. Expert Rev. Neurother. 2018, 18, 847–857. [Google Scholar] [CrossRef]
- Yan, R.; Bienkowski, M.J.; Shuck, M.E.; Miao, H.; Tory, M.C.; Pauley, A.M.; Brashler, J.R.; Stratman, N.C.; Mathews, W.R.; Buhl, A.E. Membrane-anchored aspartyl protease with alzheimer’s disease β-secretase activity. Nature 1999, 402, 533–537. [Google Scholar] [CrossRef]
- Vassar, R. β-secretase cleavage of alzheimer’s amyloid precursor protein by the transmembrane aspartic protease bace. Science 1999, 286, 735–741. [Google Scholar] [CrossRef]
- Mowrer, K.R.; Wolfe, M.S. Promotion of bace1 mrna alternative splicing reduces amyloid β-peptide production. J. Biol. Chem. 2008, 283, 18694–18701. [Google Scholar] [CrossRef]
- Peters, F.; Salihoglu, H.; Rodrigues, E.; Herzog, E.; Blume, T.; Filser, S.; Dorostkar, M.; Shimshek, D.R.; Brose, N.; Neumann, U. Bace1 inhibition more effectively suppresses initiation than progression of β-amyloid pathology. Acta Neuropathol. 2018, 135, 695–710. [Google Scholar] [CrossRef]
- Vassar, R. Implications for bace1 inhibitor clinical trials: Adult conditional bace1 knockout mice exhibit axonal organization defects in the hippocampus. J. Prev. Alzheimer’s Dis. 2019, 6, 78–84. [Google Scholar]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The imagej ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef]
- Chen, S.; Le, B.; Rahimizadeh, K.; Shaikh, K.; Mohal, N.; Veedu, R. Synthesis of a morpholino nucleic acid (mna)-uridine phosphoramidite, and exon skipping using mna/2′-o-methyl mixmer antisense oligonucleotide. Molecules 2016, 21, 1582. [Google Scholar] [CrossRef]
- Chen, S.; Le, B.T.; Chakravarthy, M.; Kosbar, T.R.; Veedu, R.N. Systematic evaluation of 2′-fluoro modified chimeric antisense oligonucleotide-mediated exon skipping in vitro. Sci. Rep. 2019, 9, 6078. [Google Scholar] [CrossRef] [PubMed]
- Le, B.T.; Adams, A.M.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Rational design of short locked nucleic acid-modified 2′-o-methyl antisense oligonucleotides for efficient exon-skipping in vitro. Mol. Ther. Nucleic Acids 2017, 9, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Le, B.T.; Chen, S.; Abramov, M.; Herdewijn, P.; Veedu, R.N. Evaluation of anhydrohexitol nucleic acid, cyclohexenyl nucleic acid and d-altritol nucleic acid-modified 2′-o-methyl RNA mixmer antisense oligonucleotides for exon skipping in vitro. Chem. Commun. 2016, 52, 13467–13470. [Google Scholar] [CrossRef] [PubMed]
- Le, B.T.; Filichev, V.V.; Veedu, R.N. Investigation of twisted intercalating nucleic acid (tina)-modified antisense oligonucleotides for splice modulation by induced exon-skipping in vitro. RSC Adv. 2016, 6, 95169–95172. [Google Scholar] [CrossRef]
- Le, B.T.; Hornum, M.; Sharma, P.K.; Nielsen, P.; Veedu, R.N. Nucleobase-modified antisense oligonucleotides containing 5-(phenyltriazol)-2′-deoxyuridine nucleotides induce exon-skipping in vitro. RSC Adv. 2017, 7, 54542–54545. [Google Scholar] [CrossRef]
- Le, B.T.; Murayama, K.; Shabanpoor, F.; Asanuma, H.; Veedu, R.N. Antisense oligonucleotide modified with serinol nucleic acid (sna) induces exon skipping in mdx myotubes. RSC Adv. 2017, 7, 34049–34052. [Google Scholar] [CrossRef]
- Disterer, P.; Kryczka, A.; Liu, Y.; Badi, Y.E.; Wong, J.J.; Owen, J.S.; Khoo, B. Development of therapeutic splice-switching oligonucleotides. Hum. Gene Ther. 2014, 25, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Järver, P.; O’Donovan, L.; Gait, M.J. A chemical view of oligonucleotides for exon skipping and related drug applications. Nucleic Acid Ther. 2014, 24, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-F.; Imam, J.S.; Wilkinson, M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007, 76, 51–74. [Google Scholar] [CrossRef] [PubMed]
- Roßner, S.; Sastre, M.; Bourne, K.; Lichtenthaler, S.F. Transcriptional and translational regulation of bace1 expression—Implications for alzheimer’s disease. Prog. Neurobiol. 2006, 79, 95–111. [Google Scholar] [CrossRef]
- Singer, O.; Marr, R.A.; Rockenstein, E.; Crews, L.; Coufal, N.G.; Gage, F.H.; Verma, I.M.; Masliah, E. Targeting bace1 with sirnas ameliorates alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005, 8, 1343–1349. [Google Scholar] [CrossRef]
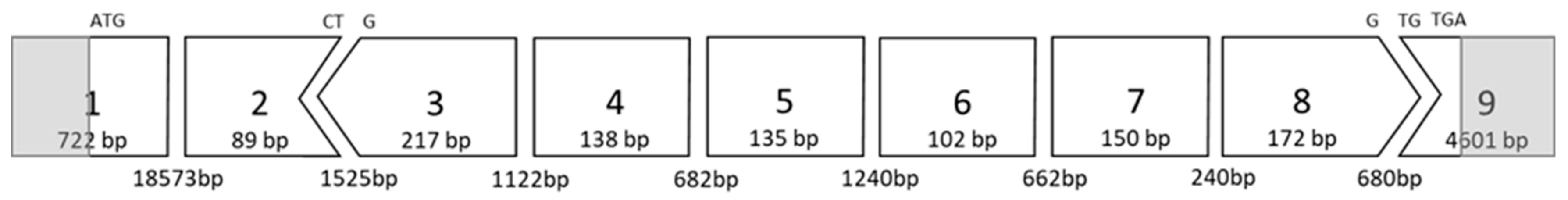

| AO Coordinates | Target | Sequence(3′--> 5′) | AO Number |
|---|---|---|---|
| BACE1 1E2A(+ 10 + 34) | Exon 2 | AGTTACTGCTGCCTGTATCCACCAG | AO1 |
| BACE1 1E2A(+ 38 + 62) | Exon 2 | AAGGGGTGGGGGGCAGCACCCACTG | AO2 |
| BACE1 1E2A(+ 65 + 89) | Exon 2 | AGCTGCCTCTGGTAGTAGCGATGCA | AO3 |
| BACE1 1E3A(+ 16 + 40) | Exon 3 | CACATACACACCCTTCCGGAGGTCC | AO4 |
| BACE1 1E3A(+ 41 + 65) | Exon 3 | CTTCCCACTTGCCCTGGGTGTAGGG | AO5 |
| BACE1 1E3A(+ 89 + 113) | Exon 3 | TGACGTTGGGGCCATGGGGGATGCT | AO6 |
| BACE1 1E3A(+ 141 + 165) | Exon 3 | TTGATGAAGAACTTGTCTGATTCAG | AO7 |
| BACE1 1E3A(+ 193 + 217) | Exon 3 | CCTGGCAATCTCAGCATAGGCCAGC | AO8 |
| BACE1 1E4A(+ 1 + 25) | Exon 4 | AGAAAGGCTCCAGGGAGTCGTCAGG | AO9 |
| BACE1 1E4A(+ 31 + 55) | Exon 4 | GAACGTGGGTCTGCTTTACCAGAGA | AO10 |
| BACE1 1E4A(+ 61 + 85) | Exon 4 | CACCACAAAGCTGCAGGGAGAAGAG | AO11 |
| BACE1 1E4A(+ 88 + 112) | Exon 4 | CTTCAGACTGGTTGAGGGGGAAGCC | AO12 |
| BACE1 1E4A(+ 114 + 138) | Exon 4 | CATGCTCCCTCCGACAGAGGCCAGC | AO13 |
| BACE1 1E6A(+ 4 + 28) | Exon 6 | TGTCCACAATGCTCTTGTCATAGTT | AO14 |
| BACE1 1E6A(+ 36 + 60) | Exon 6 | TTTCTTGGGCAAACGAAGGTTGGTG | AO15 |
| BACE1 1E6A(+ 75 + 99) | Exon 6 | GGAGGCTGCCTTGATGGATTTGACT | AO16 |
| BACE1 1E8A(+ 6 + 30) | Exon 8 | CTGCGGAAGGATGGTGATGCGGAAG | AO17 |
| BACE1 1E8A(+ 33 + 57) | Exon 8 | AAACTTGTAACAGTCGTCTTGGGAC | AO18 |
| BACE1 1E8A(+ 63 + 87) | Exon 8 | AACAGTGCCCGTGGATGACTGTGAG | AO19 |
| BACE1 1E8A(+ 109 + 133) | Exon 8 | CCCGATCAAAGACAACGTAGAAGCC | AO20 |
| BACE1 1E8A(+ 148 + 172) | Exon 8 | CATGGCAAGCGCTGACAGCAAAGCC | AO21 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakravarthy, M.; Veedu, R.N. BACE1 Inhibition Using 2’-OMePS Steric Blocking Antisense Oligonucleotides. Genes 2019, 10, 705. https://doi.org/10.3390/genes10090705
Chakravarthy M, Veedu RN. BACE1 Inhibition Using 2’-OMePS Steric Blocking Antisense Oligonucleotides. Genes. 2019; 10(9):705. https://doi.org/10.3390/genes10090705
Chicago/Turabian StyleChakravarthy, Madhuri, and Rakesh N Veedu. 2019. "BACE1 Inhibition Using 2’-OMePS Steric Blocking Antisense Oligonucleotides" Genes 10, no. 9: 705. https://doi.org/10.3390/genes10090705
APA StyleChakravarthy, M., & Veedu, R. N. (2019). BACE1 Inhibition Using 2’-OMePS Steric Blocking Antisense Oligonucleotides. Genes, 10(9), 705. https://doi.org/10.3390/genes10090705





