Wood Transcriptome Profiling Identifies Critical Pathway Genes of Secondary Wall Biosynthesis and Novel Regulators for Vascular Cambium Development in Populus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Growth Conditions, and RNA Sequencing
2.2. Transcript Assembly, Abundance Estimation, and Annotation
2.3. Quantitative Real-Time PCR (qRT-PCR) and Semi-Quantitative RT-PCR
2.4. Vector Construction and Production of Transgenic Plants
2.5. Histological Analysis
3. Results
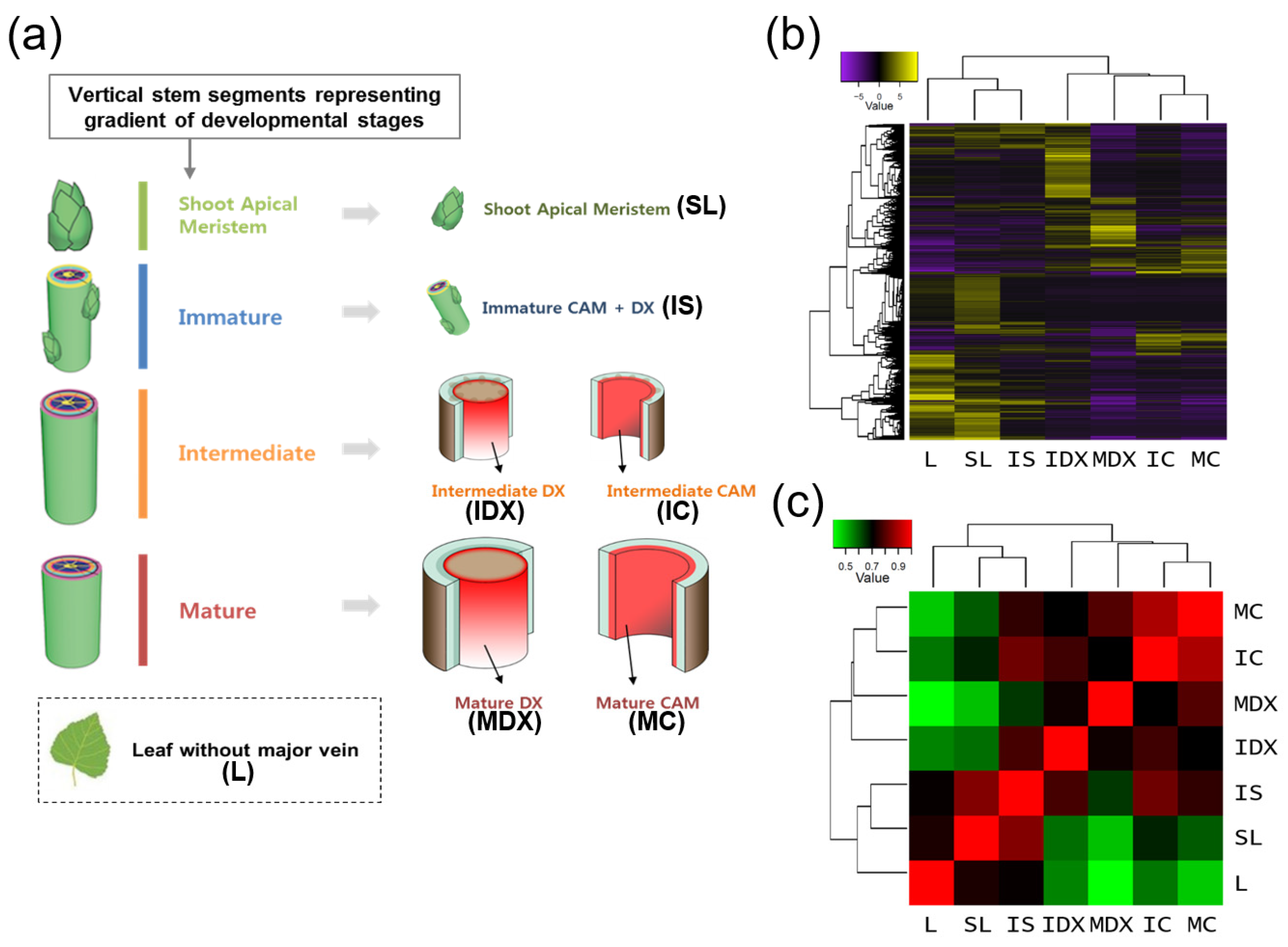
3.1. Generation of a Wood-Forming Tissue-Specific Transcriptome from a Hybrid Poplar
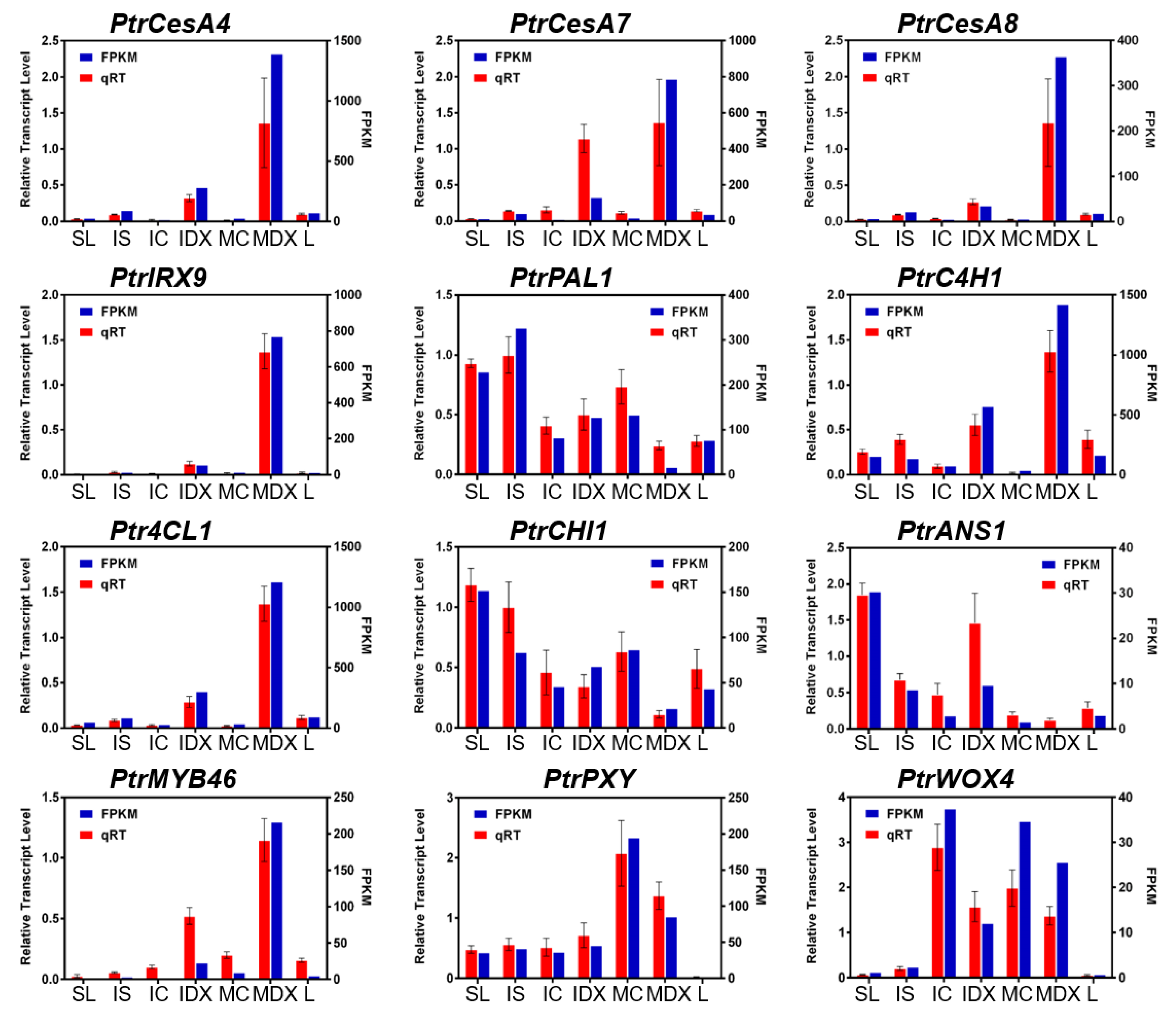
3.2. Reliability of Wood-Forming Tissue-Specific Transcriptome
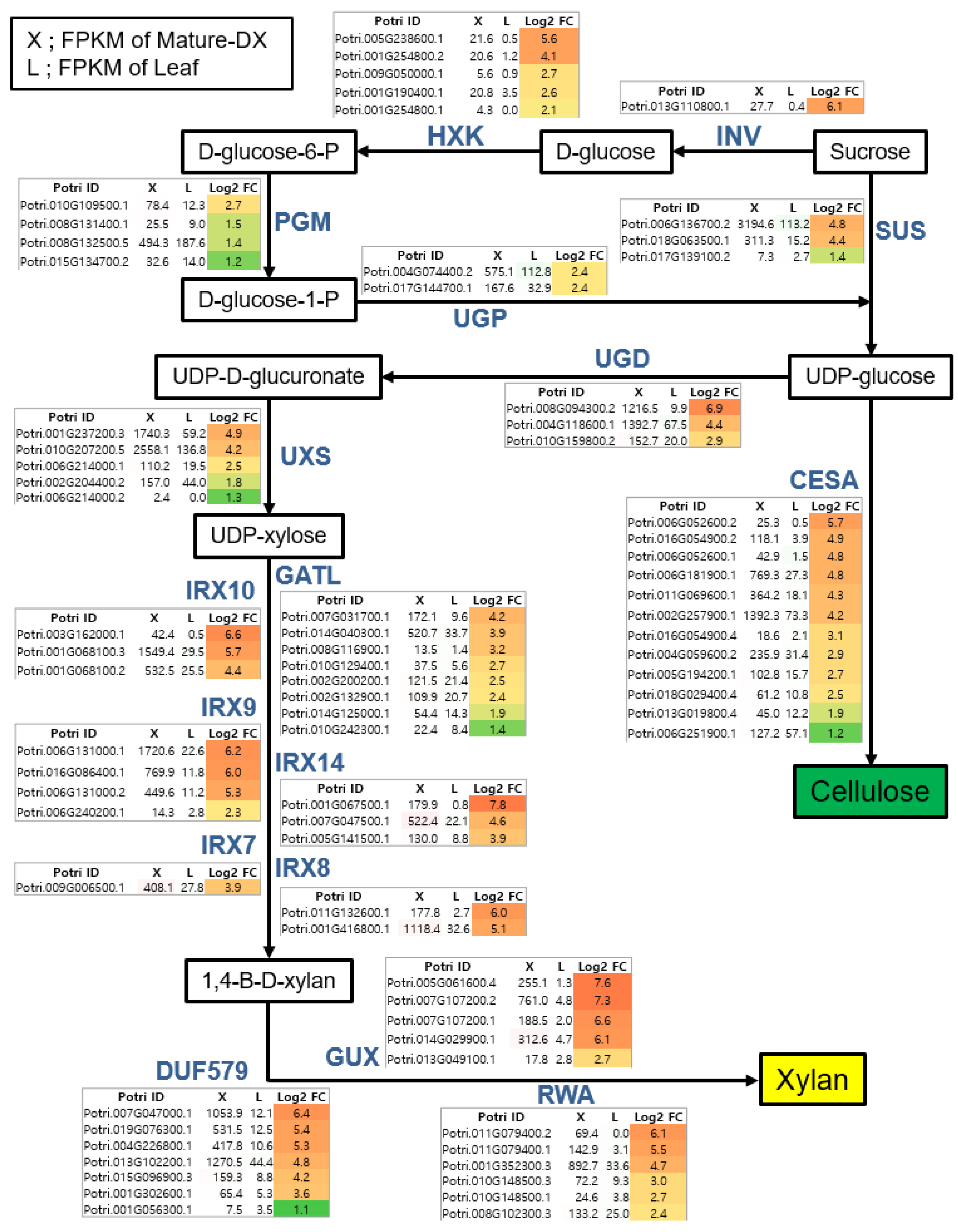
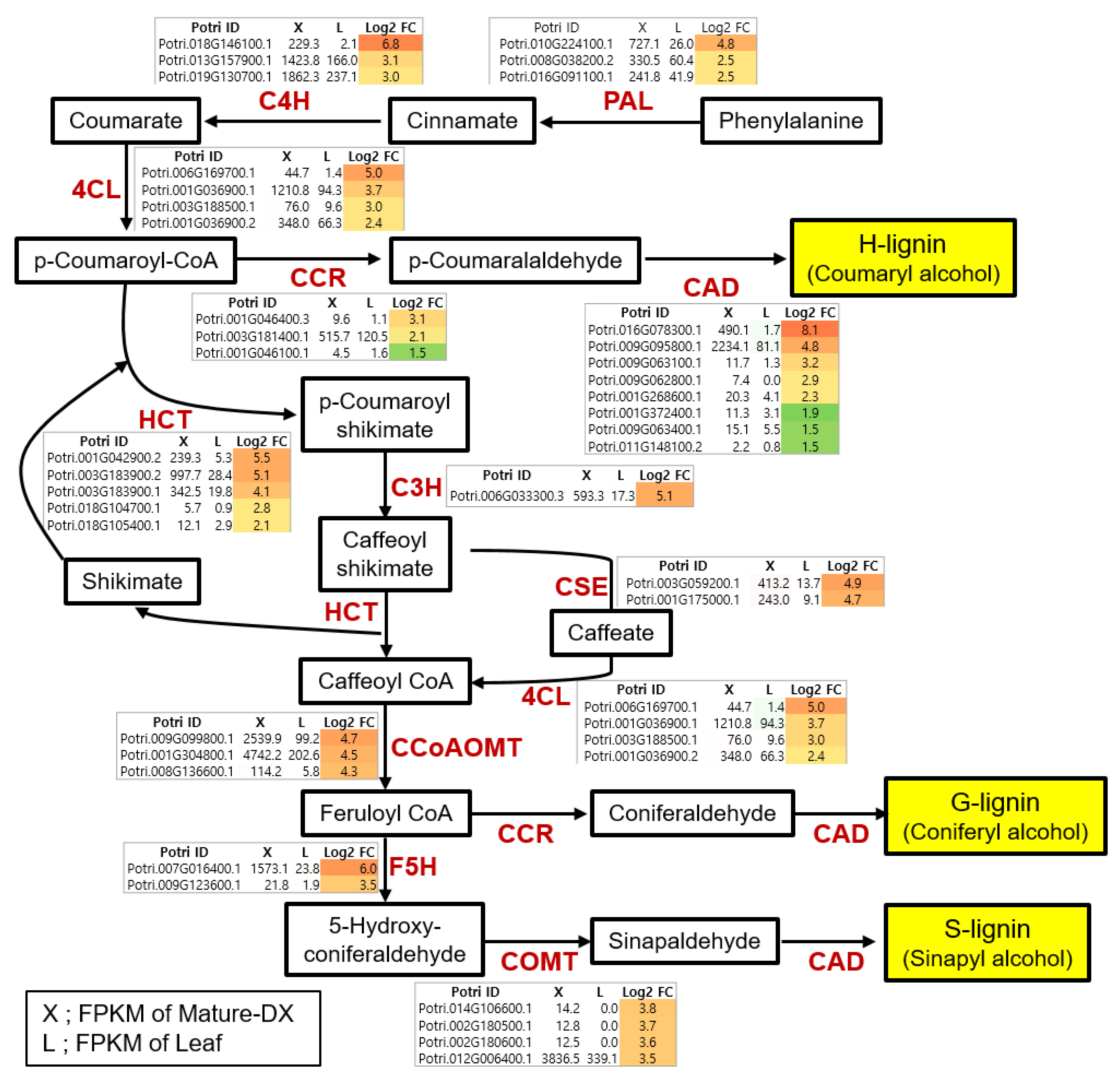
3.3. Identification of Critical Pathway Genes for Secondary Wall Biosynthesis in Populus
3.4. Identification of Genes Involved in Vascular Cambium Initiation and Maintenance
3.5. Overexpression of Ptrham4-1 Enhanced Vascular Cambium Development in Both Transgenic Arabidopsis and Poplar Plants
4. Discussion
4.1. Critical Pathway Genes of Secondary Wall Biosynthesis in Woody Perennials
4.2. Transcriptional Regulators Involved in Vascular Cambium Development
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pan, Y.; Bridsey, R.A.; Fang, J.; Houghton, R.; Kauppi, P.E.; Kurz, W.A.; Phillips, O.L.; Shvidenko, A.; Lewis, S.L.; Canadell, J.G.; et al. A large and persistent carbon sink in the world’s forests. Science 2011, 333, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Lal, R. Managing soils and ecosystems for mitigating anthropogenic carbon emissions and advancing global food security. BioScience 2010, 60, 708–721. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Pan, X.; Zalesny, R.S. Pretreatment of woody biomass for biofuel production: Energy efficiency, technologies, and recalcitrance. Appl. Microbiol. Biotechnol. 2010, 87, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Chundawat, S.P.; Beckham, G.T.; Himmel, M.E.; Dale, B.E. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Serapiglia, M.J.; Humiston, M.C.; Xu, H.; Hogsett, D.A.; Mira de Orduna, R.; Stipanovic, A.J.; Smart, L.B. Enzymatic saccharification of shrub willow genotypes with differing biomass composition for biofuel production. Front. Plant Sci. 2013, 4, 57. [Google Scholar] [CrossRef]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.; Nieves, I.U.; Mullinnix, M.T.; Sagues, W.J.; Hoffman, R.W.; Fernández-Sandoval, M.T.; Tian, Z.; Rockwood, D.L.; Tamang, B.; Ingram, L.O. Optimization of dilutephosphoric-acid steam pretreatment of Eucalyptus benthamii for biofuel production. Appl. Energy 2014, 125, 76–83. [Google Scholar] [CrossRef]
- Albers, S.C.; Berklund, A.M.; Graff, G.D. The rise and fall of innovation in biofuel. Nat. Biotechnol. 2016, 34, 814–821. [Google Scholar] [CrossRef]
- Ko, J.H.; Kim, W.C.; Cho, J.S.; Choi, Y.I.; Park, E.J.; Im, J.H.; Han, O.; Keathley, D.; Han, K.H. EliteTreeTM: An advanced biomass tree crop technology that features greater wood density and accelerated stem growth. Biofuels Bioprod. Biorefin. 2017, 11, 521–533. [Google Scholar] [CrossRef]
- Vo, T.K.; Cho, J.S.; Kim, S.S.; Ko, J.H.; Kim, J. Genetically engineered hybrid poplars for the pyrolytic production of bio-oil: Pyrolysis characteristics and kinetics. Energy Convers. Manag. 2017, 153, 48–59. [Google Scholar] [CrossRef]
- Ragni, L.; Greb, T. Secondary growth as a determinant of plant shape and form. Semin. Cell Dev. Biol. 2018, 79, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Dolan, L.; Roberts, K. Secondary thickening in roots of Arabidopsis thaliana: Anatomy and cell surface changes. New Phytol. 1995, 131, 121–128. [Google Scholar] [CrossRef]
- Ko, J.H.; Han, K.H.; Park, S.; Yang, J. Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol. 2004, 135, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Turner, S.R. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 2010, 137, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. Regulation of vascular development by CLE peptide-receptor systems. J. Integr. Plant Biol. 2010, 52, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Bowman, J.L. A Role of TDIF peptide signaling in vascular cell differentiation is conserved among Euphyllophytes. Front. Plant Sci. 2015, 6, 1048. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213. [Google Scholar] [CrossRef]
- Etchells, J.P.; Provost, C.M.; Mishra, L.; Turner, S.R. WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 2013, 140, 2224–2234. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, X.; Engstrom, E.M.; Nimchuk, Z.L.; Pruneda-Paz, J.L.; Tarr, P.T.; Yan, A.; Kay, S.A.; Meyerowitz, E.M. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 2015, 517, 377–380. [Google Scholar] [CrossRef]
- Stuurman, J.; Jäggi, F.; Kuhlemeier, C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002, 16, 2213–2218. [Google Scholar] [CrossRef]
- Engstrom, E.M. HAM proteins promote organ indeterminacy: But how? Plant Signal. Behav. 2012, 1, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Caño-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.C.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Ito, T.; Nakagami, H.; Hirakawa, Y.; Saito, M.; Tamaki, T.; Fukuda, H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF–TDR signaling. Nat. Commun. 2014, 5, 3504. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Verbančič, J.; Lunn, J.E.; Stitt, M.; Persson, S. Carbon supply and the regulation of Cell Wall Synthesis. Mol. Plant 2018, 11, 75–94. [Google Scholar] [CrossRef] [PubMed]
- York, W.S.; O’Neill, M.A. Biochemical control of xylan biosynthesis—Which end is up? Curr. Opin. Plant Biol. 2008, 11, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Kim, W.C.; Kim, J.Y.; Ahn, S.J.; Han, K.H. MYB46-mediated transcriptional regulation of secondary wall biosynthesis. Mol. Plant 2012, 5, 961–963. [Google Scholar] [CrossRef] [PubMed]
- D’haeseleer, P.; Liang, S.; Somogyi, R. Genetic network inference: From co-expression clustering to reverse engineering. Bioinformatics 2000, 16, 707–726. [Google Scholar] [CrossRef]
- Hertzberg, M.; Aspeborg, H.; Schrader, J.; Andersson, A.; Erlandsson, R.; Blomqvist, K.; Bhalerao, R.; Uhlén, M.; Teeri, T.T.; Lundeberg, J.; et al. A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. USA 2001, 98, 14732–14737. [Google Scholar] [CrossRef]
- Schrader, J.; Nilsson, J.; Mellerowicz, E.; Berglund, A.; Nilsson, P.; Hertzberg, M.; Sandberg, G. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 2004, 16, 2278–2292. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Beers, E.P.; Han, K.H. Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol. Genet. Genom. 2006, 276, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ye, C.Y.; Bisaria, A.; Tuskan, G.A.; Kalluri, U.C. Identification of candidate genes in Arabidopsis and Populus cell wall biosynthesis using text-mining, co-expression network analysis and comparative genomics. Plant Sci. 2011, 181, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Li, C.H.; Huang, J. Systematic identification of cell-wall related genes in Populus based on analysis of functional modules in co-expression network. PLoS ONE 2014, 9, e95176. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Wang, J.P.; Lin, Y.C.; Li, Q.; Sun, Y.H.; Chen, H.; Sederoff, R.R.; Chiang, V.L. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 2017, 245, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Sundell, D.; Street, N.R.; Kumar, M.; Mellerowicz, E.J.; Kucukoglu, M.; Johnsson, C.; Kumar, V.; Mannapperuma, C.; Delhomme, N.; Nilsson, O.; et al. AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 2017, 29, 1585–1604. [Google Scholar] [CrossRef] [PubMed]
- Chao, Q.; Gao, Z.F.; Zhang, D.; Zhao, B.G.; Dong, F.Q.; Fu, C.X.; Liu, L.J.; Wang, B.C. The developmental dynamics of the Populus stem transcriptome. Plant Biotechnol. J. 2019, 17, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S.; Douglas, C.J. Populus: A model system for plant biology. Annu. Rev. Plant Biol. 2007, 58, 435–458. [Google Scholar] [CrossRef] [PubMed]
- Porth, I.; EI-Kassaby, Y.A. Using Populus as a lignocellulosic feedstock for bioethanol. Biotechnol. J. 2015, 10, 510–524. [Google Scholar] [CrossRef]
- Biswal, A.K.; Atmodjo, M.A.; Pattathil, S.; Amos, R.A.; Yang, X.; Winkeler, K.; Collins, C.; Mohanty, S.S.; Ryno, D.; Tan, L.; et al. Working towards recalcitrance mechanisms: Increased xylan and homogalacturonan production by overexpression of GAlactUronosylTransferase12, GAUT12. causes increased recalcitrance and decreased growth in Populus. Biotechnol. Biofuels 2018, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Movahedi, A.; Sang, M.; Zhang, J.; Mohammadi, K.; Sun, W.; Yaghuti, A.A.Z.; Kadkhodaei, S.; Ruan, H.; Zhuge, Q. Functional analyses of PtROS1-RNAi in Poplars and evaluation of its effect on DNA methylation. J. Plant Biol. 2018, 61, 227–240. [Google Scholar] [CrossRef]
- Etchells, J.P.; Mishra, L.S.; Kumar, M.; Campbell, L.; Turner, S.R. Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 2015, 25, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Groover, A. Transcriptional regulation of secondary growth and wood formation. J. Integr. Plant Biol. 2010, 52, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Kim, H.T.; Hwang, I.; Han, K.H. Tissue-type-specific transcriptome analysis identifies developing xylem-specific promoters in poplar. Plant Biotechnol. J. 2012, 10, 587–596. [Google Scholar] [CrossRef]
- Logemann, J.; Schell, J.; Willmitzer, L. Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 1987, 163, 16–20. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Zhu, C.; Zhang, C.; Cui, S. Detection of porcine parvovirus using a taqman-based real-time PCR with primers and probe designed for the NS1 gene. Virol. J. 2010, 7, 353. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Kim, M.D.; Choi, Y.I.; Park, S.D.; Yun, D.J.; Noh, E.W.; Lee, H.S.; Kwak, S.S. Transgenic poplar expressing Arabidopsis NDPK2 enhances growth as well as oxidative stress tolerance. Plant Biotechnol. J. 2010, 9, 334–347. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Jeon, H.W.; Cho, J.S.; Park, E.J.; Han, K.H.; Choi, Y.I.; Ko, J.H. Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 2016, 14, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Wilton, O.C. Correlation of cambial activity with flowering and regeneration. Bot. Gaz. 1938, 99, 854–864. [Google Scholar] [CrossRef]
- Melzer, S.; Lens, F.; Gennen, J.; Vanneste, S.; Rohde, A.; Beeckman, T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 2008, 40, 1489–1492. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Geisler, M.; Johansson, H.; Harholt, J.; Scheller, H.V.; Mellerowicz, E.J.; Kleczkowski, L.A. UDP-glucose pyrophosphorylase is not rate limiting, but is essential in Arabidopsis. Plant Cell Physiol. 2009, 50, 998–1011. [Google Scholar] [CrossRef]
- Barratt, D.H.; Derbyshire, P.; Findlay, K.; Pike, M.; Wellner, N.; Lunn, J.; Feil, R.; Simpson, C.; Maule, A.J.; Smith, A.M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 13124–13129. [Google Scholar] [CrossRef]
- Rende, U.; Wang, W.; Gandla, M.L.; Jönsson, L.J.; Niittylä, T. Cytosolic invertase contributes to the supply of substrate for cellulose biosynthesis in developing wood. New Phytol. 2017, 214, 796–807. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, H.E.; Döring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.G.; Howells, R.M.; Huttly, A.K.; Vickers, K.; Turner, S.R. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 2003, 100, 1450–1455. [Google Scholar] [CrossRef] [PubMed]
- Seifert, G.J. Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr. Opin. Plant Biol. 2004, 7, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Zhong, R.; Ye, Z.H. Biochemical characterization of xylan xylosyltransferases involved in wood formation in poplar. Plant Signal. Behav. 2012, 7, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Saleme, M.L.S.; Cesarino, I.; Vargas, L.; Kim, H.; Vanholme, R.; Goeminne, G.; Van Acker, R.; Fonseca, F.C.A.; Pallidis, A.; Voorend, W.; et al. Silencing CAFFEOYL SHIKIMATE EXTERASE affects lignification and improves saccharification in Poplar. Plant Physiol. 2017, 175, 1040–1057. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Matthews, M.L.; Williams, C.M.; Shi, R.; Yang, C.; Tunlaya-Anukit, S.; Chen, H.C.; Li, Q.; Liu, J.; Lin, C.Y.; et al. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 2018, 9, 1579. [Google Scholar] [CrossRef] [PubMed]
- Pineau, C.; Freydier, A.; Ranocha, P.; Jauneau, A.; Turner, S.; Lemonnier, G.; Renou, J.P.; Tarkowski, P.; Sandberg, G.; Jouanin, L.; et al. hca: An Arabidopsis mutant exhibiting unusual cambial activity and altered vascular patterning. Plant J. 2015, 44, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgenes, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 11, 805–815. [Google Scholar] [CrossRef]
- Suer, S.; Agusti, J.; Sanchez, P.; Schwarz, M.; Greb, T. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 2011, 23, 3247–3259. [Google Scholar] [CrossRef] [PubMed]
- Kucukoglu, M.; Nilsson, J.; Zheng, B.; Chaabouni, S.; Nilsson, O. WUSCHEL-RELATED HOMEOBOX4, WOX4.-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 2017, 215, 642–657. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.J.; Li, X.; Huang, J.; Gropp, G.M.; Gjetvaj, B.; Lindsay, D.L.; Wei, S.; Coutu, C.; Chen, Z.; Wan, X.C.; et al. SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme. Nat. Commun. 2015, 6, 7243. [Google Scholar] [CrossRef] [PubMed]
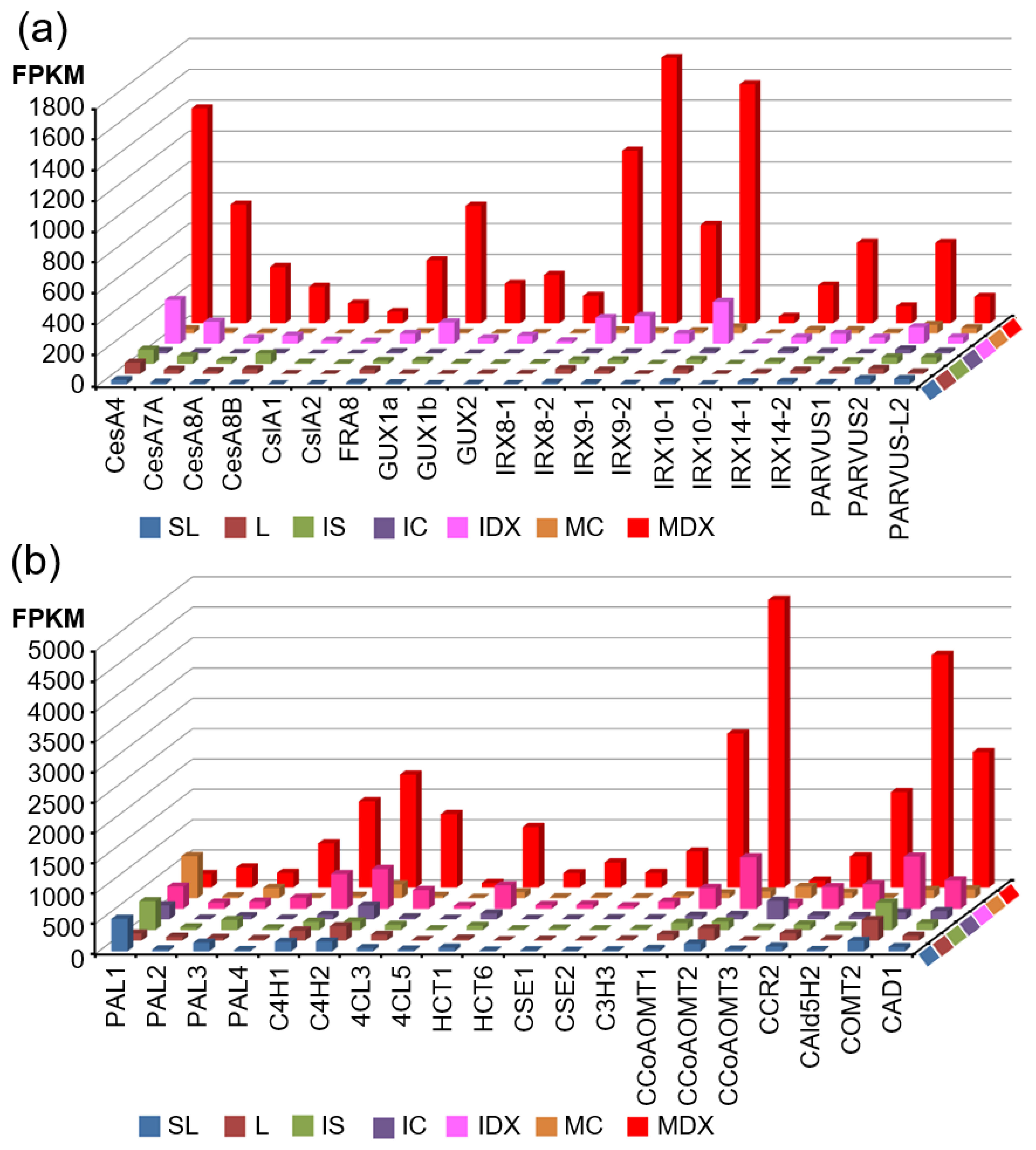
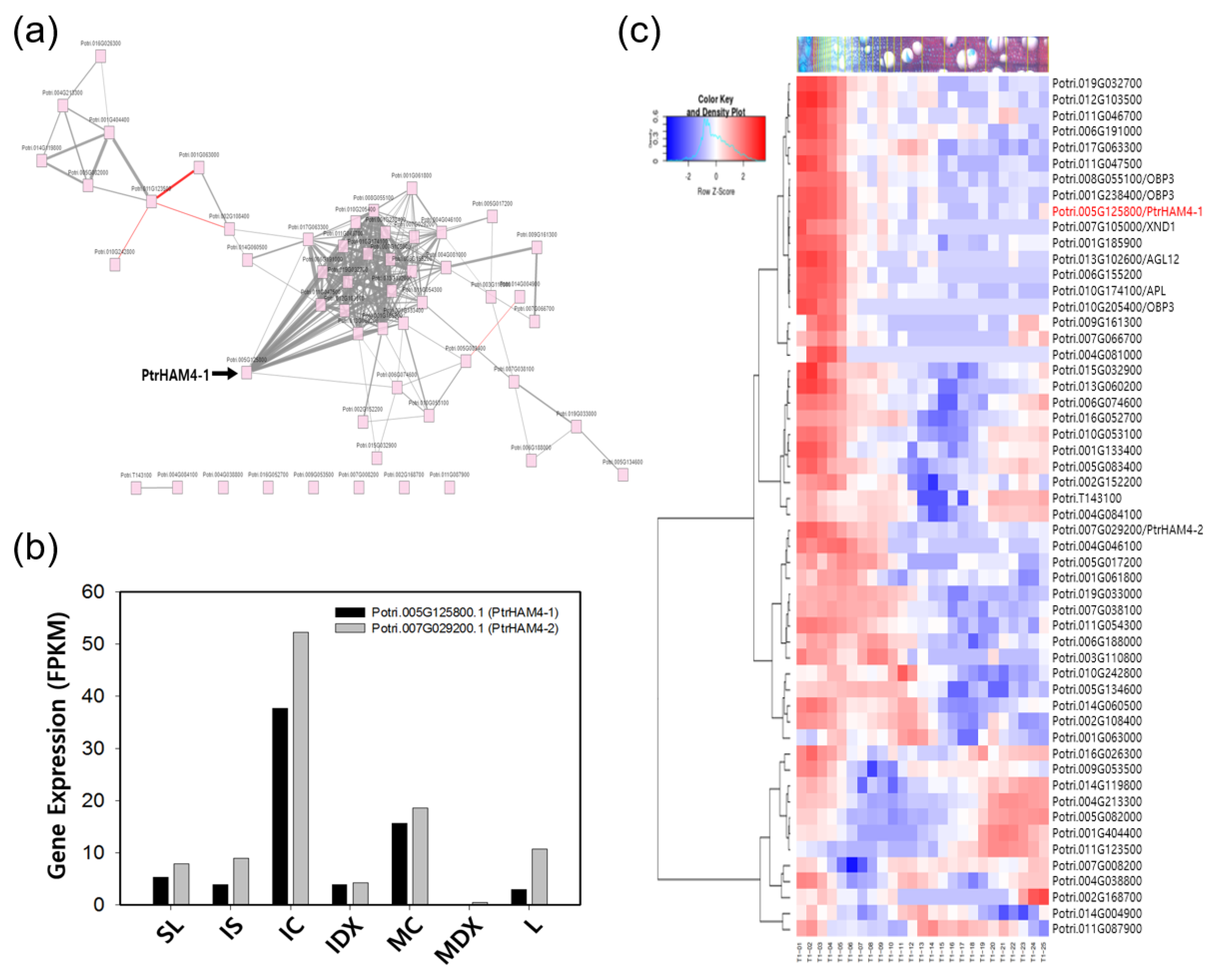
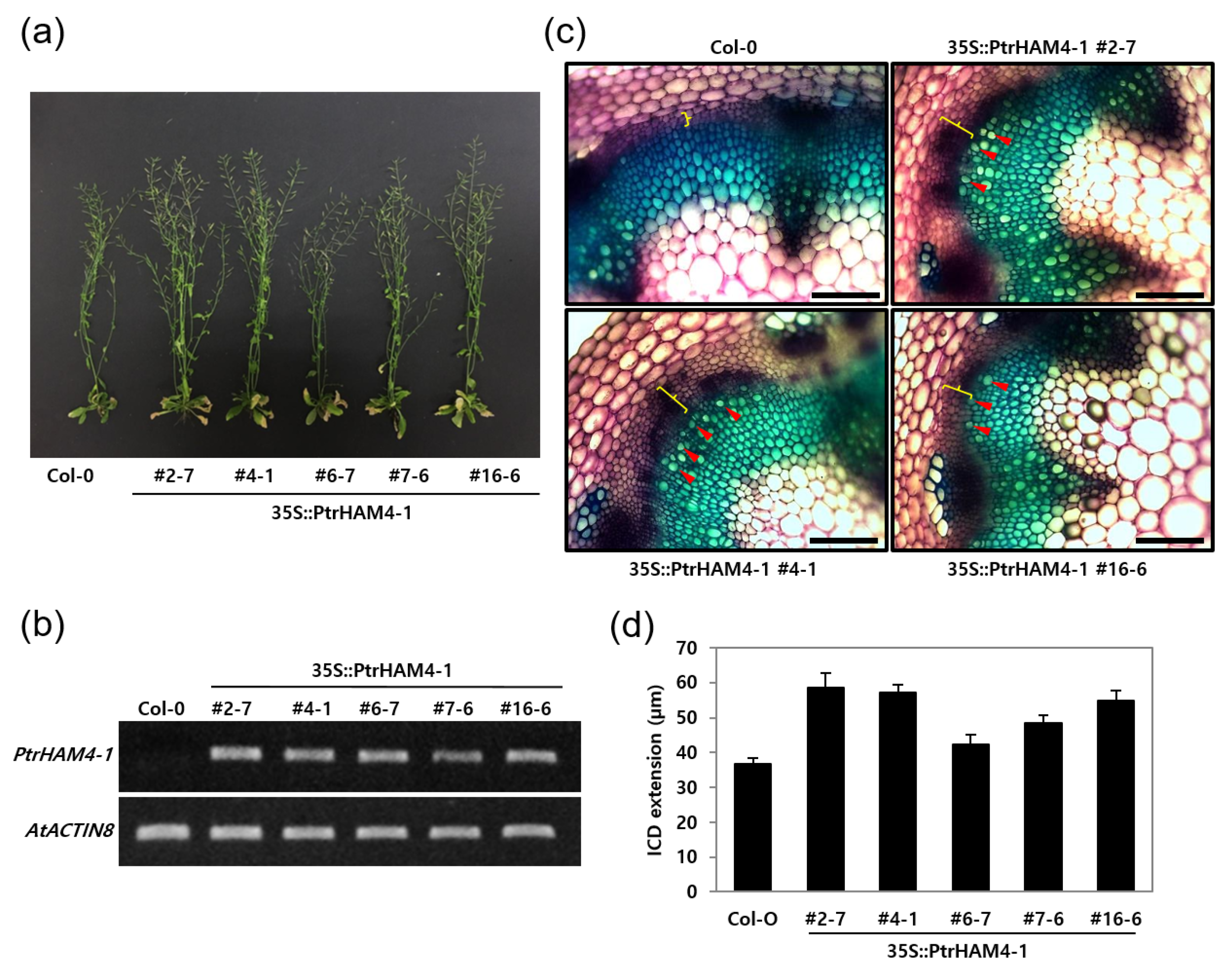
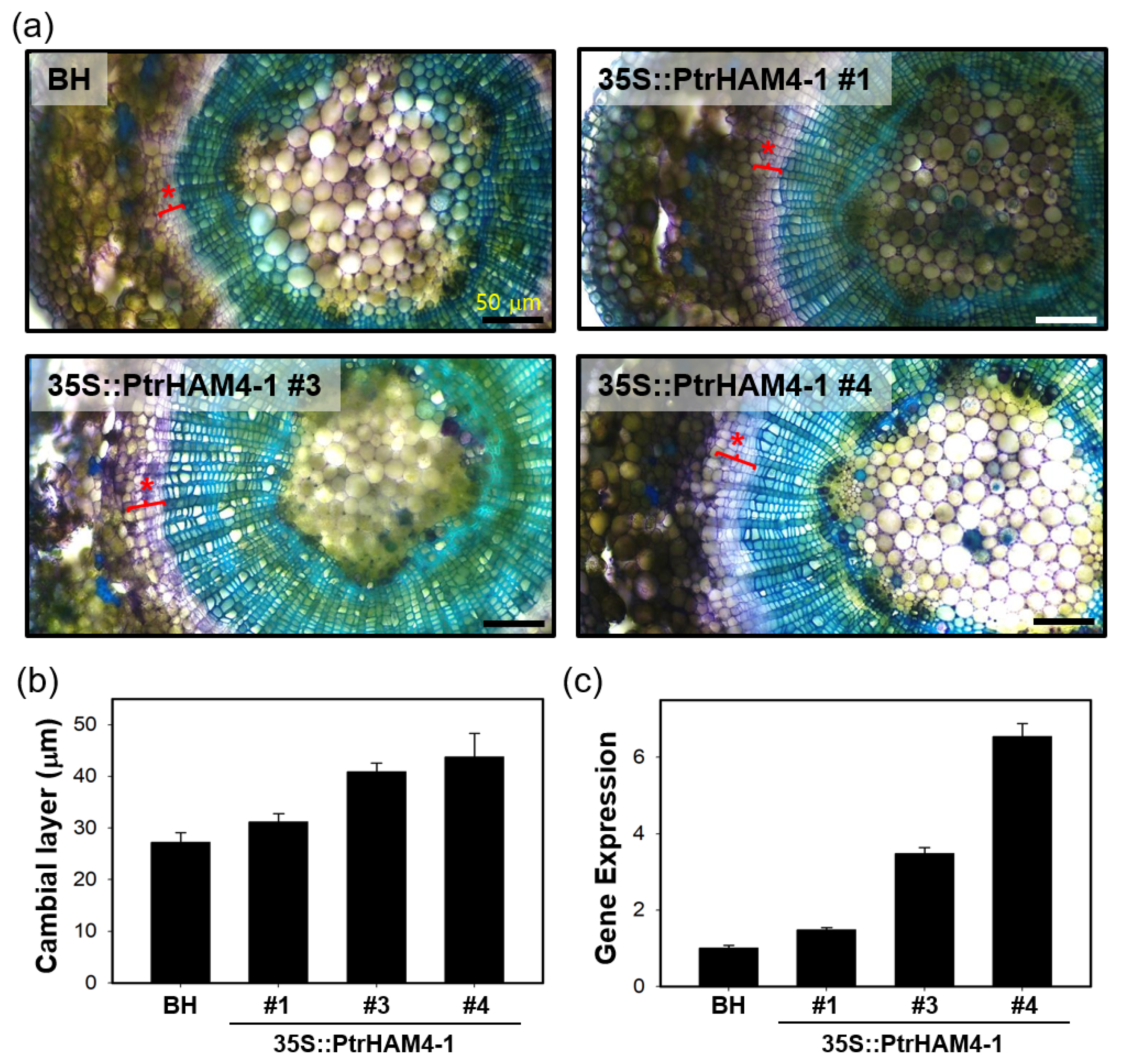
| RSEM_ID a | L | IDX | MDX | SL | IS | IC | MC | Potri ID b | % c | AGI d | % e | Description |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG4654|c0_g1_i1 | 0.4 | 6.4 | 4.9 | 10.1 | 3.7 | 5.9 | 8.2 | Potri.002G024700.1 | 97.4 | AT1G19850.1 | 75.3 | MONOPTEROS/Auxin Response Factor 5 |
| GG29733|c1_g1_i1 | 1.1 | 1.0 | 0.0 | 8.6 | 6.4 | 10.5 | 0.4 | Potri.014G146800.5 | 95.3 | AT1G23080.2 | 66.1 | PIN4, Auxin efflux carrier protein |
| GG28573|c0_g1_i1 | 0.7 | 12.1 | 25.6 | 1.2 | 2.4 | 37.5 | 34.7 | Potri.014G025300.1 | 99.5 | AT1G46480.1 | 54.5 | WOX4, WUSCHEL related homeobox 4 |
| GG36578|c7_g1_i3 | 0.5 | 2.7 | 1.1 | 0.8 | 0.3 | 4.7 | 4.4 | Potri.019G089000.1 | 97.0 | AT1G68810.1 | 40.0 | ABS5/T5L1(TMO5-LIKE1), bHLH protein |
| GG25890|c1_g1_i1 | 4.5 | 17.3 | 21.8 | 12.3 | 24.0 | 34.6 | 77.0 | Potri.012G047200.2 | 99.0 | AT1G73590.1 | 73.0 | PIN1, Auxin efflux carrier protein |
| GG19091|c5_g1_i1 | 0.9 | 8.8 | 3.4 | 4.3 | 5.6 | 14.3 | 17.1 | Potri.008G137900.1 | 98.0 | AT2G01830.2 | 72.1 | AHK4/CRE1/WOL, histidine kinase protein |
| GG22514|c1_g1_i1 | 0.3 | 3.2 | 6.1 | 6.5 | 5.6 | 7.6 | 13.7 | Potri.010G102900.1 | 98.8 | AT2G01830.3 | 72.2 | AHK4/CRE1/WOL, histidine kinase protein |
| GG2062|c1_g1_i1 | 14.9 | 18.4 | 12.7 | 57.1 | 24.2 | 33.4 | 46.3 | Potri.001G216900.1 | 98.0 | AT2G27230.2 | 75.3 | LHW, transcription factor-related |
| GG4628|c1_g1_i2 | 8.0 | 15.6 | 7.0 | 6.9 | 5.9 | 56.7 | 34.6 | Potri.002G024000.2 | 98.7 | AT2G28305.1 | 75.3 | LOG1, Lonely Guy1, lysine decarboxylase |
| GG27071|c0_g1_i1 | 0.5 | 8.0 | 13.3 | 8.6 | 6.8 | 12.8 | 37.8 | Potri.013G028300.1 | 97.4 | AT3G21510.1 | 66.4 | AHP1, histidine-containing phosphotransmitter 1 |
| GG708|c1_g1_i1 | 1.2 | 2.8 | 2.6 | 1.8 | 1.5 | 29.2 | 44.6 | Potri.001G075200.1 | 96.7 | AT3G24770.1 | 75.3 | CLE41, CLAVATA3/ESR-RELATED 41 |
| GG24830|c0_g1_i1 | 0.0 | 0.5 | 0.4 | 0.1 | 0.9 | 9.7 | 20.8 | Potri.011G102400.2 | 96.6 | AT4G13195.1 | 75.0 | CLE44, CLAVATA3/ESR-RELATED 44 |
| GG8362|c3_g1_i1 | 4.7 | 3.3 | 3.0 | 10.1 | 10.9 | 10.0 | 14.5 | Potri.003G136300.1 | 97.2 | AT4G23750.2 | 75.3 | TMO3/CRF2, Target of MONOPTEROS 3 |
| GG16016|c0_g1_i1 | 1.1 | 7.1 | 4.6 | 7.9 | 8.6 | 9.7 | 18.8 | Potri.006G235000.1 | 98.0 | AT4G32830.1 | 90.4 | AtAUR1, ataurora1 |
| GG34739|c4_g1_i1 | 0.7 | 43.3 | 54.9 | 6.3 | 17.5 | 29.6 | 31.7 | Potri.018G045100.2 | 98.9 | AT4G32880.1 | 82.2 | AtHB8, homeobox protein 8 |
| GG12514|c1_g1_i1 | 2.5 | 1.6 | 0.0 | 2.9 | 4.3 | 20.1 | 8.3 | Potri.005G125800.1 | 98.0 | AT4G36710.1 | 75.3 | ATHAM4, GRAS transcription factor |
| GG16746|c2_g1_i1 | 10.8 | 4.2 | 0.5 | 8.0 | 9.0 | 52.3 | 18.7 | Potri.007G029200.1 | 98.0 | AT4G36710.1 | 52.7 | ATHAM4, GRAS transcription factor |
| GG17654|c1_g1_i1 | 0.1 | 3.5 | 0.6 | 4.2 | 6.5 | 14.5 | 22.1 | Potri.007G132000.1 | 98.0 | AT4G37650.1 | 54.5 | SHORTROOT, GRAS transcription factor |
| GG12811|c0_g1_i1 | 0.4 | 7.0 | 0.4 | 14.1 | 5.5 | 18.5 | 33.2 | Potri.005G161500.3 | 98.0 | AT4G40060.1 | 38.0 | AtHB16, homeobox protein 16 |
| GG35388|c0_g1_i1 | 3.5 | 7.5 | 24.9 | 7.2 | 9.7 | 35.1 | 104.3 | Potri.018G113000.1 | 98.4 | AT5G51350.1 | 57.0 | MOL1, Leucine-rich repeat receptor kinase |
| GG12545|c1_g1_i1 | 0.3 | 1.1 | 2.8 | 1.3 | 1.9 | 21.4 | 22.4 | Potri.005G134200.1 | 98.0 | AT5G60200.1 | 75.3 | TMO6, Target of MONOPTEROS 6 |
| GG8087|c4_g1_i1 | 1.8 | 45.5 | 85.2 | 35.8 | 41.3 | 36.3 | 194.8 | Potri.003G107600.1 | 98.7 | AT5G61480.1 | 75.3 | PXY, Leucine-rich repeat receptor kinase |
| GG30939|c0_g1_i1 | 3.2 | 1.5 | 0.0 | 5.9 | 1.8 | 9.1 | 17.7 | Potri.015G077100.1 | 96.8 | AT5G62940.1 | 56.4 | DOF5.6/HCA2, Dof-type zinc finger protein |
| GG26232|c1_g1_i1 | 2.1 | 1.4 | 0.5 | 3.3 | 3.0 | 14.7 | 20.4 | Potri.012G081300.1 | 96.2 | AT5G62940.1 | 42.7 | DOF5.6/HCA2, Dof-type zinc finger protein |
| RSEM_ID a | L | IDX | MDX | SL | IS | IC | MC | Potri ID b | % c | AGI d | % e | Description |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG30488|c4_g1_i2 | 3.7 | 4.9 | 3.3 | 4.0 | 4.7 | 15.7 | 8.2 | Potri.015G032900.1 | 97.0 | AT1G14685.1 | 49.5 | basic pentacysteine 2 |
| GG11484|c2_g2_i1 | 0.0 | 1.3 | 1.0 | 0.8 | 2.6 | 9.7 | 5.5 | Potri.005G017200.1 | 96.8 | AT1G23380.1 | 75.3 | KNAT6. KNOTTED1-like homeobox gene 6 |
| GG585|c4_g1_i1 | 2.3 | 7.4 | 6.9 | 5.0 | 5.2 | 37.5 | 19.2 | Potri.001G061800.1 | 97.9 | AT1G28050.1 | 75.3 | B-box type zinc finger protein with CCT domain |
| GG17054|c0_g1_i2 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | 6.8 | 2.3 | Potri.007G066700.1 | 98.0 | AT1G31320.1 | 80.8 | LOB domain-containing protein 4 |
| GG25587|c0_g1_i1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 16.8 | 2.7 | Potri.T054700.1 | 92.0 | AT1G43950.1 | 35.7 | auxin response factor 23 |
| GG38344|c0_g1_i1 | 0.0 | 0.3 | 0.7 | 0.2 | 0.0 | 6.5 | 1.9 | Potri.T054900.1 | 95.8 | AT1G34390.1 | 50.0 | auxin response factor 22 |
| GG24361|c1_g1_i1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.5 | 0.0 | Potri.011G046700.1 | 95.6 | AT1G61110.1 | 55.4 | ANAC025, NAC domain protein 25 |
| GG9929|c0_g1_i1 | 0.0 | 0.0 | 0.0 | 1.4 | 0.8 | 8.2 | 4.1 | Potri.004G081000.1 | 98.4 | AT1G65910.1 | 75.3 | ANAC028, NAC domain protein 28 |
| GG9939|c4_g1_i1 | 7.3 | 6.7 | 3.8 | 4.9 | 3.5 | 34.1 | 18.1 | Potri.004G084100.2 | 99.2 | AT1G66140.1 | 75.3 | zinc finger protein 4 |
| GG38162|c0_g1_i1 | 2.9 | 1.4 | 0.8 | 2.1 | 1.0 | 15.9 | 7.2 | Potri.T143100.1 | 99.0 | AT1G66140.1 | 39.1 | zinc finger protein 4 |
| GG22021|c2_g1_i1 | 0.8 | 1.5 | 2.7 | 2.2 | 0.7 | 28.0 | 11.3 | Potri.010G053100.1 | 98.0 | AT1G67710.1 | 46.5 | ARR11, response regulator 11 |
| GG27777|c1_g1_i1 | 0.7 | 2.2 | 1.1 | 0.1 | 1.2 | 31.5 | 8.5 | Potri.013G102600.1 | 96.6 | AT1G71692.1 | 64.2 | AGL12, AGAMOUS-like 12 |
| GG23195|c1_g1_i1 | 7.5 | 2.8 | 0.5 | 4.3 | 7.2 | 52.8 | 17.5 | Potri.010G174100.1 | 99.2 | AT1G79430.2 | 60.0 | APL, ALTERED PHLOEM DEVELOPMENT |
| GG20358|c1_g1_i1 | 9.4 | 9.7 | 12.7 | 4.6 | 7.6 | 49.6 | 19.5 | Potri.004G010000.1 | 98.0 | AT2G02060.1 | 75.3 | Homeodomain-like superfamily protein |
| GG15222|c0_g1_i1 | 0.6 | 1.0 | 0.3 | 1.0 | 2.0 | 15.9 | 8.2 | Potri.006G155200.1 | 98.0 | AT2G03500.1 | 51.6 | EFM, EARLY FLOWERING MYB PROTEIN |
| GG15988|c0_g1_i1 | 0.0 | 1.1 | 0.7 | 0.3 | 0.0 | 5.5 | 1.2 | Potri.006G188000.1 | 98.0 | AT2G25180.1 | 53.8 | ARR12, response regulator 12 |
| GG21457|c0_g1_i1 | 2.6 | 1.0 | 0.8 | 5.2 | 3.8 | 19.2 | 12.4 | Potri.009G161300.1 | 98.0 | AT2G27300.1 | 56.0 | ANAC040/NTL8, NTM1-like 8 |
| GG11094|c2_g1_i1 | 0.0 | 19.4 | 14.2 | 14.6 | 12.6 | 73.1 | 29.3 | Potri.004G213300.1 | 95.6 | AT2G27990.1 | 75.3 | PNF, BEL1-like homeodomain 8 |
| GG29464|c1_g1_i1 | 4.1 | 14.8 | 9.7 | 18.4 | 9.5 | 85.0 | 33.9 | Potri.014G119800.1 | 96.4 | AT2G34830.1 | 60.4 | WRKY35/MEE24, WRKY transcription factor |
| GG10107|c0_g1_i1 | 0.5 | 2.5 | 0.0 | 1.1 | 1.3 | 7.9 | 1.1 | Potri.004G102600.1 | 94.7 | AT2G37630.1 | 75.3 | AS1/MYB91/PHANTASTICA-like1 |
| GG5942|c1_g1_i1 | 3.8 | 8.0 | 2.2 | 3.2 | 2.7 | 29.6 | 12.5 | Potri.002G152200.1 | 96.0 | AT2G45680.1 | 75.3 | TCP9, TCP transcription factor9 |
| GG27378|c2_g1_i1 | 4.1 | 0.8 | 0.0 | 0.9 | 4.1 | 21.9 | 6.6 | Potri.013G060200.4 | 98.6 | AT3G04030.3 | 59.4 | MYR2, Homeodomain transcription factor |
| GG36059|c1_g1_i1 | 5.8 | 2.0 | 0.4 | 1.0 | 2.5 | 77.0 | 10.6 | Potri.019G032700.2 | 96.3 | AT3G04030.3 | 58.5 | MYR2, Homeodomain transcription factor |
| GG36093|c1_g1_i1 | 0.9 | 0.4 | 0.0 | 0.0 | 0.4 | 5.3 | 4.4 | Potri.019G036300.1 | 97.3 | AT3G13540.1 | 37.9 | myb domain protein 5 |
| GG3783|c2_g1_i1 | 1.1 | 2.3 | 0.7 | 0.3 | 2.1 | 12.2 | 1.1 | Potri.001G404400.1 | 98.9 | AT3G15510.1 | 75.3 | ANAC056, AtNAC2 |
| GG25019|c1_g1_i1 | 2.4 | 2.0 | 0.1 | 2.1 | 1.4 | 10.4 | 0.4 | Potri.011G123500.1 | 98.1 | AT3G15510.1 | 53.5 | ANAC056, AtNAC2 |
| GG32655|c7_g1_i2 | 3.6 | 4.5 | 3.2 | 1.7 | 2.4 | 16.6 | 7.4 | Potri.015G039100.1 | 48.5 | AT3G18010.1 | 39.2 | WOX1, WUSCHEL related homeobox 1 |
| GG35905|c0_g2_i1 | 0.0 | 2.0 | 1.9 | 0.8 | 3.3 | 11.8 | 11.5 | Potri.T050600.1 | 100.0 | AT3G23240.1 | 51.3 | ethylene response factor 1 |
| GG24276|c7_g1_i1 | 0.6 | 2.8 | 0.0 | 0.5 | 1.7 | 8.6 | 4.4 | Potri.016G026300.1 | 53.9 | AT3G42170.1 | 24.6 | DAYSLEEPER, BED zinc finger, HAT family |
| GG2779|c0_g1_i2 | 0.0 | 1.2 | 1.4 | 1.2 | 2.4 | 9.7 | 3.9 | Potri.001G295700.1 | 96.6 | AT3G49940.1 | 75.3 | LOB domain-containing protein 38 |
| GG2261|c1_g1_i1 | 0.9 | 0.4 | 0.7 | 3.9 | 3.2 | 18.3 | 8.9 | Potri.001G238400.2 | 97.6 | AT3G55370.1 | 75.3 | OBP3/OBF-binding protein 3, Dof family |
| GG18276|c1_g1_i1 | 1.4 | 0.9 | 1.0 | 4.9 | 4.6 | 16.5 | 10.8 | Potri.008G055100.2 | 98.0 | AT3G55370.2 | 42.1 | OBP3/OBF-binding protein 3, Dof family |
| GG23518|c0_g1_i1 | 0.4 | 1.5 | 0.5 | 0.8 | 1.4 | 6.4 | 1.8 | Potri.010G205400.2 | 98.2 | AT3G55370.2 | 54.5 | OBP3/OBF-binding protein 3, Dof family |
| GG32069|c2_g1_i1 | 3.1 | 5.9 | 3.8 | 1.8 | 1.4 | 31.6 | 25.1 | Potri.016G052700.1 | 96.8 | AT3G57670.1 | 60.6 | NTT, C2H2 zinc finger protein |
| GG8104|c1_g1_i1 | 2.6 | 0.5 | 0.2 | 4.1 | 3.7 | 14.7 | 1.8 | Potri.003G110800.1 | 97.6 | AT4G00150.1 | 75.3 | ATHAM3, GRAS transcription factor |
| GG28884|c1_g2_i1 | 14.8 | 3.5 | 2.9 | 6.8 | 7.5 | 78.0 | 21.7 | Potri.014G060500.2 | 97.3 | AT4G00150.1 | 62.7 | ATHAM3, GRAS transcription factor |
| GG12113|c0_g1_i1 | 0.4 | 2.8 | 0.9 | 0.9 | 5.2 | 100.2 | 19.3 | Potri.005G083400.1 | 98.0 | AT4G14540.1 | 75.3 | NF-YB3, nuclear factor Y, subunit B3 |
| GG594|c0_g1_i2 | 0.2 | 3.6 | 2.8 | 1.0 | 1.8 | 10.8 | 5.9 | Potri.001G063000.1 | 96.6 | AT4G20970.1 | 75.3 | basic helix-loop-helix (bHLH) protein |
| GG28018|c0_g2_i1 | 29.8 | 34.3 | 13.4 | 22.1 | 44.2 | 167.8 | 32.9 | Potri.013G129800.1 | 95.0 | AT4G20970.1 | 37.6 | basic helix-loop-helix (bHLH) protein |
| GG24134|c0_g2_i1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 10.4 | 1.7 | Potri.002G168700.1 | 34.2 | AT4G23810.1 | 75.3 | WRKY53, WRKY transcription factor |
| GG24745|c5_g1_i4 | 4.5 | 2.7 | 0.4 | 1.9 | 4.2 | 14.0 | 4.6 | Potri.011G087900.3 | 96.3 | AT4G26640.2 | 57.6 | WRKY20, WRKY transcription factor |
| GG1779|c1_g1_i4 | 3.9 | 1.4 | 0.3 | 1.2 | 2.9 | 32.4 | 3.7 | Potri.001G185900.3 | 98.0 | AT4G29100.1 | 75.3 | basic helix-loop-helix (bHLH) protein |
| GG36060|c1_g1_i1 | 1.1 | 9.8 | 4.6 | 10.4 | 4.8 | 52.2 | 45.6 | Potri.019G033000.1 | 94.9 | AT4G32890.1 | 39.6 | GATA transcription factor 9 |
| GG12514|c1_g1_i1 | 2.5 | 1.6 | 0.0 | 2.9 | 4.3 | 20.1 | 8.3 | Potri.005G125800.1 | 98.0 | AT4G36710.1 | 53.0 | ATHAM4, GRAS transcription factor |
| GG16746|c2_g1_i1 | 10.8 | 4.2 | 0.5 | 8.0 | 9.0 | 52.3 | 18.7 | Potri.007G029200.1 | 98.0 | AT4G36710.1 | 52.7 | ATHAM4, GRAS transcription factor |
| GG12582|c1_g1_i4 | 0.0 | 0.6 | 0.3 | 1.1 | 1.8 | 6.4 | 0.5 | Potri.005G134600.1 | 98.0 | AT4G37180.1 | 75.3 | Homeodomain-like superfamily protein |
| GG12688|c0_g1_i1 | 0.0 | 0.7 | 1.1 | 2.1 | 2.4 | 22.4 | 6.1 | Potri.005G147100.2 | 98.0 | AT4G37790.1 | 75.3 | homeobox protein HAT22 |
| GG16541|c0_g1_i2 | 1.5 | 1.6 | 1.4 | 0.4 | 0.6 | 7.3 | 0.5 | Potri.007G008200.1 | 98.0 | AT4G37790.1 | 63.6 | homeobox protein HAT22 |
| GG24113|c2_g3_i1 | 9.1 | 9.8 | 5.7 | 9.1 | 23.3 | 159.4 | 37.1 | Potri.010G242800.5 | 53.6 | AT5G04840.1 | 50.0 | bZIP protein |
| GG15591|c1_g1_i3 | 2.5 | 1.1 | 0.1 | 0.2 | 1.1 | 23.9 | 1.9 | Potri.006G191000.2 | 98.0 | AT5G06800.1 | 52.6 | myb-like HTH transcriptional regulator |
| GG20492|c0_g1_i1 | 4.3 | 5.1 | 3.5 | 0.8 | 2.6 | 26.1 | 11.3 | Potri.009G053500.1 | 98.0 | AT5G12850.1 | 47.4 | CCCH-type zinc finger protein |
| GG26437|c1_g1_i1 | 0.0 | 0.2 | 0.5 | 3.2 | 3.6 | 16.2 | 2.4 | Potri.012G103500.2 | 97.5 | AT5G13180.1 | 48.6 | ANAC083/VNI2, NAC domain protein 83 |
| GG33423|c0_g1_i1 | 22.5 | 23.1 | 3.6 | 19.6 | 29.7 | 91.9 | 21.4 | Potri.017G063300.1 | 96.4 | AT5G13180.1 | 41.6 | ANAC083/VNI2, NAC domain protein 83 |
| GG1261|c0_g1_i1 | 2.0 | 0.7 | 1.5 | 1.1 | 0.0 | 13.9 | 3.5 | Potri.001G133400.1 | 98.0 | AT5G45580.1 | 75.3 | Homeodomain-like superfamily protein |
| GG14451|c0_g2_i1 | 1.3 | 0.0 | 0.0 | 1.2 | 0.0 | 5.6 | 0.5 | Potri.006G074600.1 | 98.0 | AT5G57150.1 | 48.7 | basic helix-loop-helix (bHLH) protein |
| GG9514|c0_g1_i2 | 1.3 | 2.4 | 4.4 | 2.6 | 2.4 | 20.2 | 7.8 | Potri.004G038800.1 | 97.1 | AT5G60850.1 | 75.3 | OBP4/OBF-binding protein 4, Dof family |
| GG9580|c1_g1_i1 | 0.3 | 1.6 | 1.2 | 3.7 | 1.5 | 15.8 | 6.8 | Potri.004G046100.1 | 97.8 | AT5G60850.1 | 75.3 | OBP4/OBF-binding protein 4, Dof family |
| GG24368|c0_g1_i1 | 10.4 | 3.1 | 0.3 | 10.5 | 10.8 | 115.5 | 10.3 | Potri.011G047500.1 | 97.1 | AT5G60850.1 | 64.7 | OBP4/OBF-binding protein 4, Dof family |
| GG24425|c2_g1_i1 | 1.7 | 1.6 | 2.9 | 3.1 | 2.9 | 34.4 | 20.3 | Potri.011G054300.1 | 97.2 | AT5G60850.1 | 67.9 | OBP4/OBF-binding protein 4, Dof family |
| GG17394|c0_g1_i1 | 0.1 | 3.3 | 1.5 | 1.5 | 1.9 | 24.6 | 8.0 | Potri.007G105000.1 | 98.0 | AT5G64530.1 | 54.6 | ANAC104/XND1, NAC domain protein 104 |
| GG12096|c0_g1_i1 | 1.2 | 12.3 | 0.7 | 6.7 | 11.0 | 51.3 | 15.5 | Potri.005G082000.5 | 98.0 | AT5G65210.1 | 75.3 | TGA1, bZIP transcription factor |
| GG16813|c0_g1_i1 | 3.0 | 10.2 | 15.5 | 6.6 | 17.3 | 79.8 | 77.9 | Potri.007G038100.1 | 98.0 | AT5G65590.1 | 58.7 | SCAP1/STOMATAL CARPENTER1, Dof family |
| GG5524|c2_g1_i1 | 0.8 | 12.1 | 6.4 | 11.9 | 8.1 | 63.1 | 41.3 | Potri.002G108400.1 | 98.8 | AT5G65640.1 | 75.3 | bHLH93, basic helix-loop-helix protein 93 |
| GG4560|c1_g1_i1 | 0.0 | 1.4 | 1.7 | 0.0 | 0.0 | 12.9 | 1.5 | Potri.014G004900.4 | 37.5 | AT5G67580.1 | 56.7 | telomeric DNA binding protein |
| RSEM_ID a | L | IDX | MDX | SL | IS | IC | MC | Potri ID b | % c | AGI d | % e | Description |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG5056|c1_g1_i1 | 0.0 | 0.6 | 0.7 | 0.0 | 0.0 | 0.6 | 6.5 | Potri.002G009700.2 | 35.7 | AT1G20640.1 | 75.3 | NLP4, NIN-like protein 4 |
| GG11457|c0_g2_i1 | 0.0 | 0.0 | 0.0 | 1.3 | 0.0 | 0.0 | 5.5 | Potri.005G014200.1 | 96.3 | AT1G23380.1 | 75.3 | KNAT6, KNOTTED1-like homeobox gene 6 |
| GG1392|c4_g1_i1 | 0.5 | 1.4 | 11.4 | 1.5 | 1.0 | 5.9 | 35.5 | Potri.012G087100.1 | 98.0 | AT1G23380.1 | 53.4 | KNAT6, KNOTTED1-like homeobox gene 6 |
| GG5866|c1_g1_i2 | 0.8 | 1.0 | 1.0 | 1.3 | 0.5 | 1.8 | 5.4 | Potri.002G142400.1 | 97.7 | AT1G27360.1 | 75.3 | SPL11, squamosa promoter-binding protein like |
| GG12249|c1_g1_i1 | 0.6 | 0.4 | 0.4 | 0.8 | 1.2 | 6.6 | 16.5 | Potri.005G097800.1 | 98.0 | AT1G31320.1 | 75.3 | LOB domain-containing protein 4 |
| GG1322|c3_g1_i2 | 0.5 | 0.3 | 0.2 | 0.9 | 0.7 | 2.9 | 6.8 | Potri.001G137600.1 | 98.0 | AT1G32240.1 | 75.3 | KANADI 2, Homeodomain-like protein |
| GG7970|c2_g1_i2 | 0.0 | 2.7 | 1.1 | 1.2 | 0.3 | 5.8 | 14.4 | Potri.003G096300.1 | 97.4 | AT1G32240.1 | 75.3 | KANADI 2, Homeodomain-like protein |
| GG2584|c1_g1_i1 | 0.1 | 1.1 | 2.9 | 0.9 | 1.5 | 3.2 | 17.7 | Potri.001G273700.1 | 98.3 | AT1G46264.1 | 75.3 | Heat Shock TF B4, SCHIZORIZA |
| GG5686|c0_g1_i1 | 0.2 | 1.9 | 3.5 | 8.2 | 2.7 | 3.0 | 30.4 | Potri.002G124800.1 | 98.9 | AT1G46264.1 | 75.3 | Heat Shock TF B4, SCHIZORIZA |
| GG28582|c0_g1_i1 | 0.4 | 1.1 | 3.5 | 5.5 | 2.0 | 3.2 | 28.6 | Potri.014G027100.1 | 99.5 | AT1G46264.1 | 55.3 | Heat Shock TF B4, SCHIZORIZA |
| GG34793|c2_g1_i2 | 1.1 | 2.1 | 3.0 | 3.1 | 0.7 | 8.3 | 11.8 | Potri.018G052200.1 | 97.9 | AT1G55110.1 | 55.3 | AtIDD7, indeterminate (ID)-domain 7 |
| GG10199|c7_g3_i1 | 1.6 | 1.4 | 2.2 | 1.9 | 0.8 | 3.0 | 7.1 | Potri.004G081000.1 | 34.2 | AT1G65910.1 | 75.3 | ANAC028, NAC domain protein 28 |
| GG22519|c1_g1_i1 | 3.7 | 5.7 | 6.2 | 11.2 | 5.0 | 19.6 | 36.0 | Potri.010G102700.1 | 95.8 | AT1G66140.1 | 49.7 | zinc finger protein 4 |
| GG11684|c1_g1_i1 | 0.5 | 5.3 | 21.4 | 9.2 | 3.6 | 22.9 | 77.7 | Potri.005G039800.1 | 99.6 | AT1G72210.1 | 75.3 | bHLH96, basic helix-loop-helix (bHLH) protein |
| GG27052|c1_g1_i1 | 0.0 | 0.9 | 8.7 | 1.7 | 1.0 | 7.3 | 29.9 | Potri.013G025900.1 | 98.6 | AT1G72210.1 | 47.0 | bHLH96, basic helix-loop-helix (bHLH) protein |
| GG15139|c6_g2_i2 | 0.0 | 0.0 | 1.8 | 0.0 | 0.0 | 1.6 | 9.0 | Potri.006G145100.4 | 98.0 | AT1G72830.1 | 40.8 | NF-YA3, nuclear factor Y, subunit A3 |
| GG4774|c0_g1_i1 | 0.9 | 2.7 | 0.0 | 0.0 | 0.0 | 25.6 | 35.1 | Potri.002G035000.1 | 97.9 | AT1G75250.2 | 75.3 | RAD-like 6 |
| GG4733|c8_g1_i3 | 0.0 | 0.4 | 0.6 | 2.8 | 2.6 | 3.8 | 16.4 | Potri.002G030900.1 | 98.7 | AT1G75430.1 | 75.3 | BLH11, BEL1-like homeodomain 11 |
| GG6382|c2_g1_i1 | 1.5 | 0.0 | 1.6 | 2.1 | 1.2 | 2.6 | 7.3 | Potri.006G126300.7 | 41.7 | AT2G37025.1 | 49.3 | TRF-like 8 |
| GG797|c0_g1_i1 | 0.0 | 7.1 | 12.8 | 2.1 | 3.5 | 86.3 | 102.5 | Potri.001G083700.1 | 94.4 | AT2G46680.1 | 75.3 | homeobox 7 |
| GG3069|c5_g1_i1 | 2.1 | 7.3 | 8.6 | 4.6 | 1.1 | 11.9 | 27.6 | Potri.017G107500.2 | 50.0 | AT3G02380.1 | 60.4 | COL2, CONSTANS-like 2 |
| GG36688|c0_g2_i1 | 1.2 | 1.2 | 0.0 | 0.0 | 1.0 | 7.1 | 7.3 | Potri.010G143500.1 | 27.9 | AT3G03450.1 | 29.8 | RGL2, RGA-like 2, GRAS family |
| GG2395|c0_g1_i1 | 0.8 | 0.0 | 0.9 | 0.0 | 0.7 | 6.1 | 13.9 | Potri.016G122500.1 | 38.5 | AT3G03450.1 | 24.8 | RGL2, RGA-like 2, GRAS family |
| GG11822|c1_g1_i1 | 1.7 | 7.3 | 6.8 | 4.2 | 3.3 | 16.7 | 26.4 | Potri.005G055300.1 | 99.1 | AT3G04670.1 | 75.3 | WRKY39, WRKY DNA-binding protein 39 |
| GG27332|c0_g1_i1 | 0.0 | 0.8 | 1.4 | 3.2 | 1.4 | 3.4 | 11.4 | Potri.013G056400.1 | 91.4 | AT3G13540.1 | 56.5 | myb domain protein 5 |
| GG34915|c2_g1_i1 | 0.0 | 1.2 | 2.7 | 2.3 | 0.4 | 1.4 | 8.9 | Potri.018G065400.1 | 95.4 | AT3G13960.1 | 25.9 | GRF5, growth-regulating factor 5 |
| GG25802|c0_g1_i1 | 0.5 | 0.2 | 0.3 | 1.4 | 1.7 | 7.1 | 9.9 | Potri.012G038100.1 | 99.2 | AT3G17730.1 | 78.1 | ANAC057, NAC domain protein 57 |
| GG3512|c3_g1_i1 | 1.1 | 1.5 | 1.6 | 0.7 | 0.7 | 2.5 | 5.6 | Potri.001G373300.2 | 98.7 | AT3G18380.1 | 75.3 | DTF2, DNA-binding transcription factor2 |
| GG23364|c0_g1_i1 | 0.0 | 2.7 | 3.2 | 0.9 | 1.2 | 5.6 | 19.0 | Potri.010G191500.1 | 95.5 | AT3G18990.1 | 34.5 | VRN1, REDUCED VERNALIZATION RESPONSE1 |
| GG13987|c1_g1_i1 | 3.5 | 5.2 | 3.8 | 4.2 | 1.1 | 13.5 | 23.6 | Potri.001G299300.2 | 98.0 | AT3G19500.1 | 75.3 | basic helix-loop-helix (bHLH) protein |
| GG16812|c2_g1_i1 | 0.1 | 18.2 | 30.2 | 11.1 | 14.1 | 42.1 | 135.6 | Potri.007G036400.1 | 98.0 | AT3G50410.1 | 54.1 | OBP1, OBF binding protein 1 |
| GG12474|c0_g1_i1 | 0.7 | 0.5 | 0.5 | 1.9 | 0.8 | 5.1 | 9.0 | Potri.005G122700.1 | 98.0 | AT3G50870.1 | 75.3 | GATA18/MNP/HAN, GATA family |
| GG17903|c0_g1_i1 | 0.7 | 7.6 | 6.6 | 1.3 | 2.0 | 7.5 | 27.6 | Potri.008G011900.2 | 98.0 | AT3G54320.1 | 60.2 | WRI1, WRINKLED 1 |
| GG27921|c0_g1_i1 | 0.7 | 1.3 | 3.7 | 3.0 | 0.7 | 8.4 | 12.9 | Potri.013G117600.1 | 96.3 | AT3G56770.1 | 40.1 | basic helix-loop-helix (bHLH) protein |
| GG9055|c0_g1_i1 | 0.2 | 0.8 | 0.5 | 1.1 | 0.6 | 9.8 | 17.6 | Potri.014G114300.2 | 35.9 | AT3G60030.1 | 57.5 | SPL12, squamosa promoter-binding protein like |
| GG6170|c1_g2_i1 | 0.4 | 0.5 | 2.4 | 1.0 | 1.2 | 5.2 | 9.9 | Potri.002G174300.1 | 99.6 | AT3G61850.4 | 75.3 | DAG1, Dof affecting germination1 |
| GG30346|c1_g1_i5 | 0.6 | 0.7 | 1.4 | 0.2 | 0.2 | 2.3 | 5.4 | Potri.014G075200.1 | 23.4 | AT4G00730.1 | 68.4 | ANL2, ANTHOCYANINLESS 2 |
| GG594|c0_g1_i1 | 0.6 | 2.9 | 8.2 | 3.7 | 5.7 | 26.9 | 28.9 | Potri.001G063000.1 | 97.3 | AT4G20970.1 | 75.3 | basic helix-loop-helix (bHLH) protein |
| GG24745|c5_g1_i6 | 1.4 | 2.6 | 2.1 | 2.5 | 2.1 | 6.5 | 7.8 | Potri.011G087900.1 | 90.9 | AT4G26640.2 | 57.6 | WRKY20, WRKY family protein |
| GG28635|c2_g2_i1 | 0.9 | 3.4 | 1.8 | 1.7 | 1.1 | 3.7 | 10.2 | Potri.014G036900.1 | 81.2 | AT4G33280.1 | 41.1 | AP2/B3-like family protein |
| GG28579|c0_g2_i3 | 0.0 | 0.9 | 1.0 | 0.0 | 0.0 | 2.8 | 6.8 | Potri.014G025800.1 | 93.1 | AT4G36930.1 | 50.7 | SPATULA, basic helix-loop-helix (bHLH) protein |
| GG5586|c3_g1_i2 | 0.7 | 1.5 | 0.0 | 0.0 | 0.0 | 2.6 | 8.7 | Potri.002G114800.1 | 96.5 | AT4G37750.1 | 75.3 | AINTEGUMENTA, AP2/ERF protein |
| GG16531|c0_g2_i1 | 0.1 | 1.9 | 1.4 | 2.8 | 2.3 | 5.3 | 9.4 | Potri.007G007400.1 | 98.0 | AT4G37750.1 | 45.0 | AINTEGUMENTA, AP2/ERF protein |
| GG28425|c2_g1_i1 | 0.3 | 0.8 | 0.6 | 2.2 | 1.5 | 5.3 | 7.5 | Potri.014G008100.1 | 98.6 | AT4G37750.1 | 53.2 | AINTEGUMENTA, AP2/ERF protein |
| GG32690|c0_g1_i1 | 0.0 | 0.2 | 0.8 | 0.1 | 0.0 | 2.1 | 6.4 | Potri.016G120800.1 | 98.2 | AT5G01310.1 | 67.5 | APRATAXIN-like |
| GG14540|c1_g1_i1 | 1.1 | 0.7 | 0.3 | 3.1 | 3.9 | 6.2 | 11.9 | Potri.006G084200.1 | 98.0 | AT5G02460.1 | 43.4 | Dof5.1, Dof zinc finger protein |
| GG7630|c0_g1_i1 | 0.0 | 0.6 | 2.4 | 1.3 | 0.3 | 3.3 | 9.7 | Potri.003G064600.1 | 97.5 | AT5G14750.1 | 75.3 | MYB66/WER |
| GG436|c0_g1_i1 | 0.6 | 0.7 | 2.0 | 1.3 | 2.3 | 7.2 | 9.2 | Potri.001G048200.1 | 98.6 | AT5G25190.1 | 75.3 | ESE3, ethylene and salt inducible3 |
| GG34721|c0_g1_i1 | 0.0 | 0.6 | 0.5 | 0.0 | 0.5 | 6.2 | 6.5 | Potri.018G044900.2 | 98.5 | AT5G25830.1 | 49.5 | GATA transcription factor 12 |
| GG15888|c0_g1_i1 | 0.0 | 3.3 | 1.8 | 2.9 | 4.1 | 3.9 | 17.0 | Potri.006G221500.1 | 98.0 | AT5G35550.1 | 52.1 | MYB123/TT2, TRANSPARENT TESTA2 |
| GG4851|c0_g1_i1 | 0.0 | 2.8 | 8.2 | 3.5 | 0.5 | 4.4 | 26.1 | Potri.002G043300.1 | 98.1 | AT5G44210.1 | 75.3 | AP2/ERF protein, ERF9 |
| GG7893|c3_g1_i2 | 0.0 | 3.4 | 6.2 | 1.1 | 0.8 | 6.6 | 27.6 | Potri.003G089800.1 | 97.7 | AT5G46590.1 | 75.3 | ANAC096, NAC domain protein 96 |
| GG5152|c2_g1_i1 | 0.0 | 0.0 | 0.9 | 2.3 | 4.2 | 6.3 | 21.2 | Potri.002G198100.1 | 29.6 | AT5G49330.1 | 75.3 | MYB111, myb domain protein 111 |
| GG25652|c4_g1_i1 | 1.3 | 2.4 | 2.1 | 1.3 | 0.5 | 5.4 | 10.2 | Potri.012G100700.3 | 36.8 | AT5G50670.1 | 45.1 | SPL13B, squamosa promoter-binding protein like |
| GG14378|c1_g1_i2 | 0.0 | 0.9 | 0.7 | 0.5 | 0.6 | 1.5 | 16.4 | Potri.006G066400.1 | 98.0 | AT5G52600.1 | 49.3 | MYB82, myb domain protein 82 |
| GG15367|c3_g1_i3 | 0.0 | 3.3 | 5.7 | 7.3 | 1.9 | 13.7 | 34.2 | Potri.006G167700.1 | 98.0 | AT5G57390.1 | 54.5 | AINTEGUMENTA-like 5 |
| GG35176|c0_g2_i1 | 0.0 | 1.4 | 8.8 | 5.0 | 6.0 | 20.0 | 64.1 | Potri.018G091600.1 | 97.6 | AT5G57390.1 | 53.6 | AINTEGUMENTA-like 5 |
| GG26499|c3_g2_i1 | 0.0 | 0.0 | 4.6 | 0.2 | 0.0 | 0.6 | 14.0 | Potri.012G108500.1 | 96.1 | AT5G61890.1 | 70.0 | EBE, AP2/ERF protein |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.-H.; Cho, J.-S.; Jeon, H.-W.; Sangsawang, K.; Shim, D.; Choi, Y.-I.; Park, E.-J.; Lee, H.; Ko, J.-H. Wood Transcriptome Profiling Identifies Critical Pathway Genes of Secondary Wall Biosynthesis and Novel Regulators for Vascular Cambium Development in Populus. Genes 2019, 10, 690. https://doi.org/10.3390/genes10090690
Kim M-H, Cho J-S, Jeon H-W, Sangsawang K, Shim D, Choi Y-I, Park E-J, Lee H, Ko J-H. Wood Transcriptome Profiling Identifies Critical Pathway Genes of Secondary Wall Biosynthesis and Novel Regulators for Vascular Cambium Development in Populus. Genes. 2019; 10(9):690. https://doi.org/10.3390/genes10090690
Chicago/Turabian StyleKim, Min-Ha, Jin-Seong Cho, Hyung-Woo Jeon, Kanidta Sangsawang, Donghwan Shim, Young-Im Choi, Eung-Jun Park, Hyoshin Lee, and Jae-Heung Ko. 2019. "Wood Transcriptome Profiling Identifies Critical Pathway Genes of Secondary Wall Biosynthesis and Novel Regulators for Vascular Cambium Development in Populus" Genes 10, no. 9: 690. https://doi.org/10.3390/genes10090690
APA StyleKim, M.-H., Cho, J.-S., Jeon, H.-W., Sangsawang, K., Shim, D., Choi, Y.-I., Park, E.-J., Lee, H., & Ko, J.-H. (2019). Wood Transcriptome Profiling Identifies Critical Pathway Genes of Secondary Wall Biosynthesis and Novel Regulators for Vascular Cambium Development in Populus. Genes, 10(9), 690. https://doi.org/10.3390/genes10090690






