Abstract
Wood, the most abundant biomass on Earth, is composed of secondary xylem differentiated from vascular cambium. However, the underlying molecular mechanisms of wood formation remain largely unclear. To gain insight into wood formation, we performed a series of wood-forming tissue-specific transcriptome analyses from a hybrid poplar (Populus alba × P. glandulosa, clone BH) using RNA-seq. Together with shoot apex and leaf tissue, cambium and xylem tissues were isolated from vertical stem segments representing a gradient of secondary growth developmental stages (i.e., immature, intermediate, and mature stem). In a comparative transcriptome analysis of the ‘developing xylem’ and ‘leaf’ tissue, we could identify critical players catalyzing each biosynthetic step of secondary wall components (e.g., cellulose, xylan, and lignin). Several candidate genes involved in the initiation of vascular cambium formation were found via a co-expression network analysis using abundantly expressed genes in the ‘intermediate stem-derived cambium’ tissue. We found that transgenic Arabidopsis plants overexpressing the PtrHAM4-1, a GRAS family transcription factor, resulted in a significant increase of vascular cambium development. This phenotype was successfully reproduced in the transgenic poplars overexpressing the PtrHAM4-1. Taken together, our results may serve as a springboard for further research to unravel the molecular mechanism of wood formation, one of the most important biological processes on this planet.
1. Introduction
Woody biomass represents more than 90% of the total biomass produced within the earth’s terrestrial ecosystems. Roughly 25% of the annual anthropogenic CO2 emissions can be assimilated during woody biomass formation, suggesting that wood formation serves as one of earth’s major long-term terrestrial carbon sinks [1,2]. Woody biomass has the potential to be renewable as well as carbon neutral with regard to its conversion into various forms of energy (e.g., electricity, gas, and liquid energy); thus, it has attracted attention in fields of sustainable energy [3,4,5,6,7,8,9,10].
Wood is primarily produced by woody perennials through a process called secondary growth (i.e., wood formation). Secondary growth is achieved by the vascular cambium, a cylindrical domain of pluripotent stem cells below the organ surface, forming wood (i.e., secondary xylem) inside and bast (i.e., secondary phloem) outside in a strictly bidirectional manner by coordinated cell division and differentiation [11]. However, the underlying molecular mechanisms of vascular cambium initiation/proliferation and vascular patterning remain largely unclear. Since the herbaceous model plant Arabidopsis undergoes secondary growth within the stem, root, and hypocotyl [12,13], most of our current understanding of secondary growth comes from Arabidopsis studies. Recently, the PXY/TDR (PHLOEM INTERCALATED WITH XYLEM/TDIF RECEPTOR) signaling network has been reported [14,15], which is highly conserved among euphyllophytes (i.e., ferns and seed plants) [16]. PXY/TDR binds the CLE (CLV3/EMBRYO SURROUNDING REGION)-related peptide CLE41/CLE44/TDIF and activates the WOX4 and WOX14 transcription factors [17,18]. WOX4, expressed in the cambium, promotes stem cell proliferation by interacting with HAM4 (HAIRY MERISTEM 4), one of four HAMs in Arabidopsis [19]. HAM proteins are members of the GRAS (GAI, RGA, SCR) family of transcription factors and are essential components of a non-cell autonomous signaling pathway for maintenance of shoot meristem identity [20,21]. Additionally, PXY-TDIF signaling reduces Brassinosteroid (BR) signaling associated with xylem differentiation by interacting directly with BIN2 (BRASSINOSTEROID-INSENSITIVE2) [22,23].
Wood comprises of secondary cell walls consisting primarily of three polymers: Cellulose; hemicellulose; and lignin [24]. Cellulose microfibrils provide load-bearing strength to the cell wall by forming scaffolds with other wall polymers, such as xylan and lignin [25]. Xylan is one of the major hemicelluloses found in the secondary cell walls of poplar, and, unlike cellulose, has reducing end oligosaccharides with a variety of side chains [26]. Lignin is a complex phenolic compound providing compression strength and hydrophobicity to secondary walls [4,27]. Biosynthesis of these cell wall components is specifically regulated by multi-layered transcription factor networks [28].
Many large-scale gene expression analyses have been performed to understand these regulatory networks [29,30,31,32,33,34,35,36,37,38]. For example, Yang et al. [33] in their study identified many candidate genes for cell wall biosynthesis using comparative genomics of Arabidopsis and Populus. Cai et al. [34] uncovered cell wall-related genes in Populus using co-expression network analysis. Taylor-Teeples et al. [35] identified 50 genes for xylem cell specification including transcription factors and enzymes involved in cellulose, hemicellulose, and lignin biosynthesis by systematically mapping the regulatory network at cell-type resolution in Arabidopsis. Shi et al. [36] identified multiple transcription factors and wood component genes within Populus trichocarpa using the transcriptomes of five tissues (xylem, phloem, shoot, leaf, and root) and two wood-forming cell types (fibers and vessels). In addition, Sundell et al. [37] reported a high spatial-resolution transcriptome analysis spanning the secondary phloem, vascular cambium, and wood-forming tissues of Populus tremula, which were obtained by longitudinal tangential cryosections, and then provided an interactive Web resource capable of exploring expression profiles and co-expression networks, the AspWood (http://aspwood.popgenie.org). Very recently, Chao et al. [38] reported on the developmental dynamics of the Populus stem transcriptome to identify differentially-expressed transcripts by PacBio full-length sequencing and RNA-seq analysis that are involved in the transition from primary to secondary growth.
Populus is employed as a model system for secondary growth not only due to its inherent massive wood formation but also because it is an important source of biofuel [39,40,41,42]. Within Populus secondary growth, it has been reported that PXY-TDIF signaling is conserved [43]. However, secondary growth of woody perennials seems to differ substantially from that of Arabidopsis root or hypocotyl; thus, it is imperative to identify the genes regulating secondary growth in Populus as a woody model [28,36,44].
In this report, we analyzed a series of wood-forming tissue-specific transcriptomes from a hybrid poplar to understand the molecular mechanism of xylem cell differentiation from cambial cell fate specification. Cambium and xylem cells from a gradient of secondary growth developmental stages (i.e., immature, intermediate, and mature stem) were isolated and RNA-seq libraries were constructed. We found many critical players catalyzing each secondary wall biosynthetic step as well as novel candidate genes possibly involved in the vascular cambium initiation using a proof-of-concept experiment.
2. Materials and Methods
2.1. Plant Materials, Growth Conditions, and RNA Sequencing
Hybrid poplars (Populus alba × P. glandulosa, clone BH) were grown in a field (latitude 37.2N, longitude 126.9E) after transplanting two-month-old plants grown in a growth room (16 h of light; light intensity, 150 μmol/m2/s; 24 °C). This clone was selected and utilized in this study because it is easy to transform the gene of interest for further study. Tissue samples were collected as described previously [45] and combined from at least 10 one-year-old trees. In brief, shoot apical meristem samples were collected from the top of the shoot, and immature, intermediate, and mature stem samples from the second to third internodes, eighth to fifteenth internodes, and twenty-fifth internodes (diameter 1.0–1.5 cm), respectively. Collected leaf samples were from fully expanded young leaves without major veins. Cambium tissue was collected by scraping the inner part of the detached bark using a double edge razor blade, and developing xylem was collected by scraping the surface of the debarked stems. The total RNA was isolated using the cetyl trimethylammonium bromide (CTAB) method with slight modifications [46]. In brief, fine powder from plant tissues was mixed with a CTAB buffer, followed by a phenol/chloroform/isoamyl alcohol (25:24:1) extraction. Isopropanol was added to the mixture to isolate RNA, the RNA quality was estimated using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). One microgram of total RNA from each sample was used for an RNA sequencing library construction following the Illumina TruSeq RNA library type (Illumina, San Diego, CA, USA). The libraries were quantified using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and paired-end sequencing was performed on an Illumina HiSeq2000 platform (Illumina). The resulting RNA sequencing data were deposited in NCBI with BioProject accession (PRJNA522944; https://dataview.ncbi.nlm.nih.gov/object/PRJNA522944?reviewer=56urghn66puds3q2e274rsiuvl).
2.2. Transcript Assembly, Abundance Estimation, and Annotation
Raw reads generated from RNA sequencing were cleaned by PRINSEQ-Lite 0.20.4 (http://prinseq.sourceforge.net/) using a Phred quality score of 20 or above with minimum lengths of 50 bp or greater. For genome-guided transcriptome assembly, a script in Trinity v2.2.0 was used [47,48]. In brief, the cleaned paired-end reads of each library were mapped to the P. trichocarpa genome v3.0 (http://www.phytozome.org) using Bowtie v. 1.2.2 [49]. The transcript abundance (e.g., read count) was determined by RSEM (RNA-Seq by Expectation Maximization, v. 1.3.0) [50] and represented by a FPKM (Fragments Per Kilobase of transcript per Million mapped reads) value. The edgeR was used for statistical analysis of the differential transcript abundance by input read counts [51] using the following script; $run_DE_analysis.pl --counts.matrix --method edgeR --output edgeR_dir --dispersion 0.1 --samples samples.txt. For transcript annotation, BLASTX (e-value 1e-5) was performed against A. thaliana (Athaliana_167_TAIR10) and P. trichocarpa (Ptrichocarpa_210_v3) protein datasets from Phytozome V12 (https://phytozome.jgi.doe.gov/pz/portal.html), respectively.
2.3. Quantitative Real-Time PCR (qRT-PCR) and Semi-Quantitative RT-PCR
One microgram of poplar total RNA was reverse transcribed to produce first-strand cDNA using a PrimeScript™ RT reagent kit (Takara, Otsu, Japan) following the manufacturer’s instructions. The gene expression patterns were analyzed by quantitative real-time PCR (qRT-PCR) [52]. All primer sequences were designed using the Primer3 program (http://fokker.wi.mit.edu). Poplar Actin2 gene was used as the quantitative control [53]. The qRT-PCR was performed using a CFX96™ Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA) with iQ™ SYBR® Supermix (Bio-Rad). In Arabidopsis samples, the total RNA was extracted using TRIZOL reagent (Ambion, Austin, TX, USA), according to the suggested protocol, with slight modifications. One microgram of the total RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) in a 20 μL reaction. RT-PCR was carried out using 1 μL of the two-fold diluted reaction product as a template. Amplified DNA fragments were separated on 1% agarose gel and visualized with ethidium bromide staining.
2.4. Vector Construction and Production of Transgenic Plants
The full-length cDNA encoding PtrHAM4-1 (Potri.005G125800) was amplified from hybrid poplar (clone BH) by PCR and inserted downstream of the 35S promoter in the pK2GW7 vector [54] using the Gateway cloning system in order to produce 35S::PtrHAM4-1 constructs. Vector constructs were introduced into Agrobacterium tumefaciens strain C58, which was used to transform Arabidopsis thaliana (Columbia; Col-0) and a hybrid poplar by the floral-dip method [55] and leaf disk transformation–regeneration method [56]. All of the constructs used in this study were verified by DNA sequencing.
2.5. Histological Analysis
Rosette level stems of 60-day-old Arabidopsis plants were used to obtain hand-cut cross sections and stained with 0.05% toluidine blue O for 1 min as described previously [56]. Interfascicular cambium-derived tissue (ICD) extensions were measured at all of the interfascicular regions from 60-day-old stem sections immediately above rosette level. In poplar, lengths of cambial layers were measured in a total of eight directions of each stem sections and at least five ramets of each transgenic line (3-month-grown in test tube after subculture) were used for statistical analysis. Images were captured using a microscope (CHB-213; Olympus, Tokyo, Japan) and analyzed by ImageJ software (National Institutes of Health; http://www.nih.gov/).
3. Results
3.1. Generation of a Wood-Forming Tissue-Specific Transcriptome from a Hybrid Poplar
To explore the molecular mechanism of wood formation, we designed a series of tissue type-specific transcriptome analyses using a hybrid poplar (Populus alba × P. glandulosa, clone BH). Each wood-forming tissue was isolated from vertical stem segments representing a gradient of developmental stages with regard to secondary growth (i.e., immature, intermediate, and mature stems; Figure 1a), which was previously confirmed by microscopic observation of the stem cross sections [45]. Thus, a total of seven tissue types from at least 10 actively growing poplars (field grown, one-year-old) was collected and combined, including SL (Shoot apical meristem with Leaf primordia), IS (Immature Stem), IC (Intermediate stem-derived Cambium), IDX (Intermediate stem-derived Developing Xylem), MC (Mature stem-derived Cambium), and MDX (Mature stem-derived Developing Xylem). In addition, L (Leaf without major veins) was included as a negative control for wood formation (Figure 1a). SL includes shoot apex with leaf primordia; IS indicates immature stem tissue of the third to the fifth internodes containing procambium and primary xylem; IC and IDX were collected from intermediate stems (seventh to 10th internodes), while MC and MDX were collected from mature stems (20–25th internodes) with a scraping method after separating bark tissues [45] (see Materials and Methods).
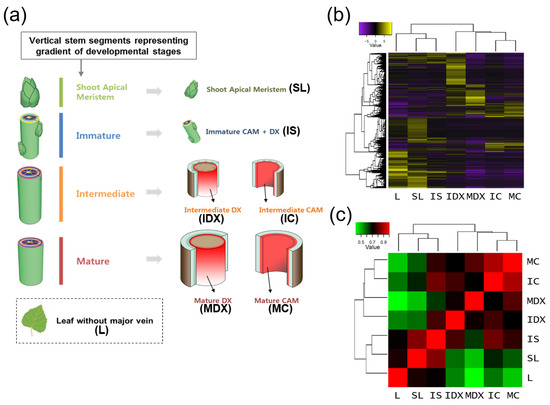
Figure 1.
Tissue-specific transcriptome analysis of a hybrid poplar. RNA-seq analysis of various poplar tissues for tissue-specific study. (a) Strategy for poplar tissue sampling used in this study. Schematic diagram of tissue samples collected. SL (shoot apical meristem with leaf primordia), IS (immature stem), IC (intermediate cambium), IDX (intermediate developing xylem), MC (mature cambium), MDX (mature developing xylem), and ML (mature leaf without major veins). (b,c) Sample correlation by heatmap (b) and correlation matrix (c), which were produced by Trinity (analyze_diff_expr.pl) with log2 fold change value (p-value, 0.005).
The total RNA extracted from the aforementioned tissues was used directly for RNA-seq analysis (Hiseq 2000, Illumina, San Diego, CA, USA). Seven RNA-seq libraries were constructed and sequenced to produce 6.5 to 7.5 million reads per library, corresponding to more than 6 Gb per sample (Supplemental Figure S1). Using genome-guided assembly, about 78.5% of the sequences were mapped to the Populus trichocarpa genome (v3.0) (Supplemental Figure S2), with a total of 38,329 transcripts across all tissues after removal of redundant transcripts (from a total of 84,168 transcripts) (Supplemental Table S1).
3.2. Reliability of Wood-Forming Tissue-Specific Transcriptome
Transcriptional interrelationships of the seven tissue libraries were evaluated by generating a sample correlation heatmap and matrix using the Trinity package [48] (Figure 1b,c). Interestingly, the seven tissues segregated into two groups: One with L, SL and IS; and the other with ID, MD, IC and MC (Figure 1b). In addition, MC and IC as well as SL and IS were closely located within the same clades. As expected, this result suggests that physiologically similar tissues have comparable transcriptomes. For example, in the correlation matrix corresponding to each tissue, MC is highly correlated with IC (Figure 1c), which is convincing, because both MC and IC consist primarily of cambial cells, and next comes MDX, which flanks MC. Interestingly, the next most correlative tissue to IC is IS-instead of IDX, which flanks IC-implying that IC is related to IS in containing immature meristem tissues. These results suggest that the tissue-specific transcriptome data is reliable.
To further verify the RNA-seq data, we performed a real-time quantitative RT-PCR (qRT-PCR) analysis of 12 selected genes, including eight genes involved in secondary cell wall biosynthesis (CesA4, CesA7, CesA8, IRX9, PAL1, C4H1, 4CL3, MYB46), two in anthocyanin biosynthesis (CHI1, ANS1), and two in vascular cambium development (PXY, WOX4) (Figure 2). The tissue-specific transcript levels of all the selected genes are very similar to those of the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values, suggesting that our RNA-seq data with no biological replications is applicable for further in silico analysis as a reference transcriptome.

Figure 2.
Confirmation of RNA-seq results by independent qRT-PCR analysis. Expression of each indicated gene is plotted. In each plot, blue bars (right Y-axis) indicate FPKM (Fragment Per Kilobase of transcript per Million mapped reads) values from RNA-seq data while red bars (left Y-axis) indicate qRT-PCR results. A gene model name for P. trichocarpa (v3.0) is shown in parenthesis as: PtrCesA4 (Potri.002G257900.1); PtrCesA7 (Potri.018G103900.1); PtrCesA8 (Potri.011G069600.1); PtrIRX9 (Potri.016G086400.1); PtrPAL1 (Potri.006G126800.1); PtrC4H1 (Potri.013G157900.1); Ptr4CL1 (Potri.001G036900.1); PtrCHI1 (Potri.010G213000.1); PtrANS1 (Potri.003G119100.1); PtrMYB46 (Potri.009G053900.1); PtrPXY (Potri.003G107600.1); and PtrWOX4 (Potri.014G025300.1). PtrActin2 (Potri.019G010400.1) was used as a reference gene. Error bars indicate S.E. gene-specific primer sequences used in this analysis are shown in Table S2.
3.3. Identification of Critical Pathway Genes for Secondary Wall Biosynthesis in Populus
To identify the key players in the biosynthesis pathways of secondary wall components (i.e., cellulose, xylan, and lignin) from a hybrid poplar, we assembled biosynthetic pathways including genes regulating each step, which are upregulated more than two-fold in “MDX” vs. “L” tissue (Figure 3 and Figure 4).
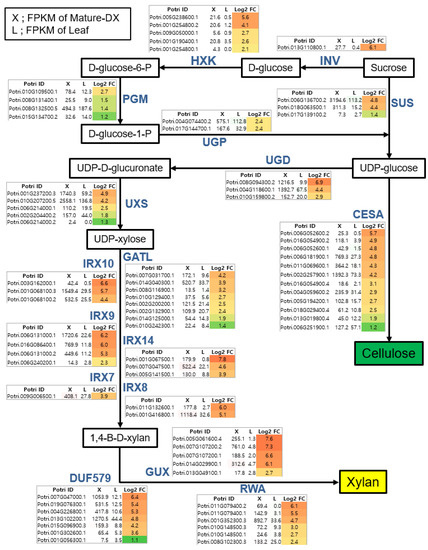
Figure 3.
Identification of major players within cellulose and xylan biosynthetic pathways for secondary wall formation. Metabolites in each step of the biosynthetic pathway are shown in the box and all related genes are shown to the side. Fold change (X/L) was calculated in Log2. X is a FPKM value of mature developing xylem (MDX) and L (Leaf without major veins) represents the leaf FPKM value. Color gradient according to the fold change values was visualized.

Figure 4.
Identification of major players of the monolignol biosynthetic pathway in secondary wall formation. Metabolites in each step of the biosynthetic pathway are shown in the box and all the related genes are shown to the side. Fold changes (X/L) were calculated in Log2. X is a FPKM value of mature developing xylem (MDX) and L is the leaf FPKM value. Color gradient according to fold change values was visualized.
Figure 3 illustrates the biosynthetic pathways of cellulose and xylan with sucrose as a starting molecule. The monolignol biosynthetic pathway with phenylalanine as a starting molecule is displayed in Figure 4. For each step in the biosynthetic pathway, the responsible enzymes are mostly encoded by multi-genes. However, based on our transcriptome data, we were able to pinpoint the key players within each step. For example, among the eight genes encoding CAD (CINNAMYL ALCOHOL DEHYDROGENASE), Potri.009G095800.1 is likely the most important due to it being the most highly expressed in terms of quantity (2234.1) in MDX, with >27-fold upregulation compared to L. In addition, Potri.016G078300.1 may be considered an important player, as it exhibited the highest specificity (>228-fold upregulation) in MDX compared with specificity of L, although its quantity was rather low (490.1) (Figure 4). Likewise, of the three genes encoding SUS (SUCROSE SYNTHASE), Potri.006G136700.2 is likely the principal gene as indicated by its high quantity and specificity in MDX. Combining all the wood-forming tissue-specific expression analyses, we were able to identify key players in each step of the biosynthesis pathways in terms of secondary wall components (Figure 5).
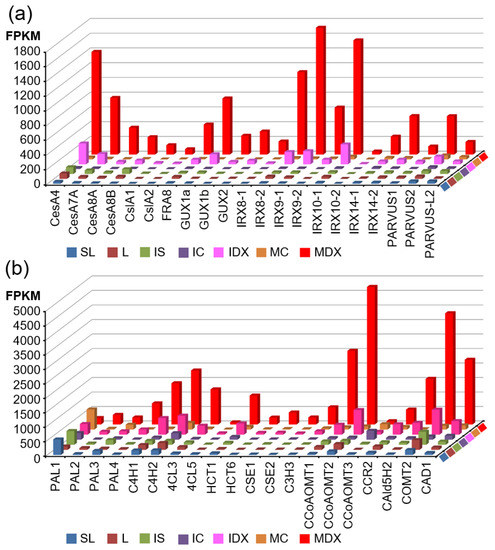
Figure 5.
Wood-forming tissue-specific expression of secondary wall biosynthetic genes in poplar. Transcript abundance of all secondary wall biosynthetic genes in each tissue was plotted. (a) Genes responsible for biosynthesis of cellulose and hemicellulose: CesA4 (Potri.002G257900.1); CesA7-A (Potri.006G181900.1); CesA8-A (Potri.011G069600.1); CesA8-B (Potri.004G059600.2); CslA1 (Potri.008G026400.1); CslA2 (Potri.010G234100.1); FRA8 (Potri.009G006500.1); GUX1a (Potri.007G107200.2); GUX1b (Potri.005G061600.4); GUX2 (Potri.014G029900.1); IRX8-1 (Potri.011G132600.1); IRX8-2 (Potri.001G416800.1); IRX9-1 (Potri.006G131000.1); IRX9-2 (Potri.016G086400.1); IRX10-1 (Potri.001G068100.3); IRX10-2 (Potri.003G162000.1); IRX14-1 (Potri.005G141500.1); IRX14-2 (Potri.007G047500.1); PARVUS-1 (Potri.002G132900.1); PARVUS-2 (Potri.014G040300.1); PARVUS-L-2 (Potri.007G031700.1). (b) Genes responsible for monolignol biosynthesis: PAL1 (Potri.006G126800.1); PAL2 (Potri.008G038200.2); PAL3 (Potri.016G091100.1); PAL4 (Potri.010G224100.1); C4H1 (Potri.013G157900.1); C4H2 (Potri.019G130700.1); 4CL3 (Potri.001G036900.1); 4CL5 (Potri.003G188500.1); HCT1 (Potri.003G183900.2); HCT6 (Potri.001G042900.2); CSE1 (Potri.003G059200.1); CSE2 (Potri.001G175000.1); C3H3 (Potri.006G033300.3); CCoAOMT1 (Potri.009G099800.1); CCoAOMT2 (Potri.001G304800.1); CCoAOMT3 (Potri.008G136600.1); CCR2 (Potri.003G181400.1); CAld5H2 (Potri.007G016400.1); COMT2 (Potri.012G006400.1); CAD1 (Potri.009G095800.1). Transcript abundance on the Y-axis represents FPKM values.
3.4. Identification of Genes Involved in Vascular Cambium Initiation and Maintenance
Next, we analyzed our transcriptome data to identify genes that were involved in vascular cambium formation, of which little information is available to date, especially for woody perennials. Firstly, we checked expression of homologs to Arabidopsis genes known to be involved in cambium formation (Table 1). Interestingly, most of these were highly expressed in either IC or MC, or both IC and MC. For example, poplar genes homologous to PIN4, LOG1, and AtHAM4 were highly expressed in IC; while PIN1, AHK4/CRE1/WOL1, AHP1, CLE41, CLE44, AtAUR1, MOL1, and PXY homologs were highly expressed in MC. In contrast, WOX4, TMO5-Like1, LHW, TMO3/CRF2, AtHB8, SHR, TMO6, and DOF5.6 were abundantly expressed in both IC and MC when compared to other tissues. These results suggest that poplar has a gene regulatory network of cambial development similar to that identified in Arabidopsis.

Table 1.
Expression of genes involved in the cambium initiation and maintenance.
To identify novel genes involved in vascular cambium initiation and maintenance within poplar, we attempted to isolate a group of genes preferentially expressed in either IC or MC. To do so, we selected genes preferentially upregulated (>3-fold) in either IC or MC compared to other tissues, which resulted in identification of 885 and 798 transcripts, respectively (Supplemental Tables S3 and S4). Among these, we initially focused on transcriptional regulators (64 and 58 transcripts) (Table 2 and Table 3). Table 2 shows abundantly expressed transcriptional regulators in IC, including homologs to Arabidopsis genes involved in cytokinin signaling (e.g., ARR11 and ARR12), negative regulation of xylem vessel formation (e.g., ANAC083/VNI2 and ANAC104/XND1), and meristem differentiation (e.g., PNF, WOX1, AtHAM3, and AtHAM4). Table 3 shows abundantly expressed transcriptional regulators in MC, including genes related to meristem maintenance (e.g., KNAT6), asymmetric stem cell division (e.g., SCHIZORIZA), and cell proliferation (e.g., AINTEGUMENTA, AINTEGUMENTA-like 5, GRF5). Additionally, abaxial and adaxial cell patterning-related genes (e.g., KANADI1, Dof5.1) are listed. Thus, the transcripts listed in Table 2 and Table 3 include many genes known to be involved in vascular cambium initiation and maintenance. These results suggest that this listing is reliable and can be used to identify novel regulators of cambium initiation and maintenance in poplar. For example, seven poplar genes homologous to either Arabidopsis OBP3 or OBP4 are listed in Table 2, but their functional significance in cambial development remains unknown. Previously, it was found that flowering and development of cambium are inversely correlated [57,58]. Interestingly, transcription factors involved in the negative regulation of flowering were also found (e.g., EFM, NTL8, MYR2) (Table 2).

Table 2.
Transcriptional regulators preferentially expressed in the intermediate cambium (IC) tissue.

Table 3.
Transcriptional regulators preferentially expressed in the mature cambium (MC) tissue.
To discover potential novel regulators involved in the initiation of vascular cambium, we performed a co-expression network analysis using the AspWood website [37] by querying the abundantly expressed transcriptional regulators of IC in Table 2. We found that most of the genes were interconnected with high correlation (Figure 6a). Among them, several genes showed a high degree of prominent centrality based on number of neighbors [37] including Potri.005G125800.1. Both Potri.005G125800.1 and its closest homolog, Potri.007G029200.1, are preferentially expressed in IC (Figure 6b) and show a higher similarity to AtHAM4 compared to other AtHAMs, which were found from both the amino acid sequence alignment (Figure S3a) and phylogenic analysis (Figure S3b). AtHAM4 was recently reported as a candidate for regulating cambium initiation in Arabidopsis by interacting with WOX4 [19]. Thus, we designated Potri.005G125800.1 and Potri.007G029200.1 as PtrHAM4-1 and PtrHAM4-2, respectively. The high spatial-resolution wood formation data [37] further support their preferential expression in the phloem and cambial tissues (Figure 6c).

Figure 6.
Identification of PtrHAM4-1, preferentially expressed in cambium tissue based on in Silico analysis. (a) Co-expression network of the intermediate cambium (IC) tissue preferentially expressed transcriptional regulators. The co-expression network was obtained from the AspWood website (http://aspwood.popgenie.org/aspwood-v3.0/) by querying a total of 64 poplar genes (Potri. ID) in the Table 2. PtrHAM4-1 (Potri.005G125800) was relocated to emphasize. (b) Tissue-specific expression of both Potri.005G125800.1/PtrHAM4-1 and its closest homolog, Potri.007G029200.1/PtrHAM4-2. This diagram was reconstructed from our RNAseq data. (c) PtrHAM4-1 is highly expressed in the phloem and cambial tissues. To obtain a gene expression profile by exploiting the high spatial-resolution wood formation data [37] the list of genes from Table 2 was queried to the AspWood website. The resulting heatmap showed that PtrHAM4-1 (indicated by red letters) is highly expressed in the phloem and cambial tissues.
3.5. Overexpression of Ptrham4-1 Enhanced Vascular Cambium Development in Both Transgenic Arabidopsis and Poplar Plants
To test whether PtrHAM4-1 is functionally involved in cambium formation, we generated transgenic Arabidopsis plants overexpressing PtrHAM4-1 using the CaMV 35S promoter (i.e., 35S::PtrHAM4-1). Overall, the 35S::PtrHAM4-1 Arabidopsis plants grew normally, but with a dramatic increase of cambial cell proliferation in the inflorescent stems of all five independent T3 homozygous lines (Figure 7). We observed the secondary xylem vessels in the interfascicular region of the 35S::PtrHAM4-1 Arabidopsis plants, which are not found in wild type (WT) plants (Figure 7c). This fact indicates that the vascular cambium was developed in the 35S::PtrHAM4-1 Arabidopsis plants. Accordingly, the 35S::PtrHAM4-1 Arabidopsis exhibited significantly increased interfascicular cambium-derived tissues (ICD) extension of >60% compared to WT plants (Figure 7c,d).

Figure 7.
Overexpression of PtrHAM4-1 in transgenic Arabidopsis increased cambium development. (a) Overall growth phenotypes of transgenic Arabidopsis plants. Five independent T3 homozygote transgenic lines (e.g., 2–7, 4–1, 6–7, 7–6, and 16–6) are shown. (b) Expression of the PtrHAM4-1 gene in the independent transgenic Arabidopsis lines compared with Col-0. First-strand cDNA was synthesized from the total RNA extracted from stem tissues and used as a template in semi-quantitative RT-PCR experiments. (c) Observation of cambium development in stem cross sections from 60-day-old transgenic Arabidopsis and Col-0. Yellow braces indicate the ICD (interfascicular cambium-derived tissue) extension and red arrowheads point to secondary xylem vessels within the interfascicular region. Scale bars represent 100 µm. (d) Quantification of ICD extension in 60-day-old transgenic Arabidopsis plants compared to Col-0. ICD extensions were measured in all interfascicular regions of rosette level stem sections from five independent transgenic lines. Error bars indicate S.E. (n = 5).
The same construct (35S::PtrHAM4-1) was introduced to a hybrid poplar to further confirm the phenotypic significance of 35S::PtrHAM4-1 Arabidopsis plants. We generated a total of 25 independent transgenic poplar lines and found the increased cambial development (e.g., ICD extension) in many lines compared to the wild-type BH clone (i.e., BH) (Figure 8a). Subsequent quantification and gene expression analysis further showed that the phenotypic significance is nicely correlated with the expression level of the PtrHAM4-1 gene (Figure 8). Taken together, our results suggest that PtrHAM4-1 may function as an important player in the initiation of vascular cambium formation in poplar.

Figure 8.
Transgenic poplar overexpressing PtrHAM4-1 resulted in an increased cambium development. (a) Observation of cambium development in stem cross sections of 3-month-old transgenic poplars and wild type BH clone (BH) grown in test tube. Red braces under the red star indicate the cambial layers. Scale bars represent 50 µm. (b) Quantification of cambial layers of transgenic poplars compared to BH. The length of cambial layers was measured in stem sections from five ramets of each transgenic lines described in (a). Error bars indicate S.E. (n = 5). (c) Expression of the PtrHAM4-1 gene in the independent transgenic poplar lines compared to BH. First-strand cDNA was synthesized from the total RNA extracted from stem tissues and used as a template in the qRT-PCR experiments.
4. Discussion
Secondary growth is one of the most important biological processes on earth. However, our knowledge concerning the underlying molecular mechanisms of vascular cambium initiation/proliferation and vascular patterning is still fragmented, especially for woody perennials. We hypothesized that genes abundantly and specifically expressed in wood-forming tissues may be important players in wood formation. Recently, many extensive wood-forming tissue-type transcriptome analyses of Populus have been reported. Shi et al. [36] in their study used four tissues (shoot tip, leaf, xylem, and phloem) and two wood-forming cell types (fibers and vessels) of P. trichocarpa. Sundell et al. [37] presented high spatial-resolution transcriptome data spanning the phloem, vascular cambium, early/developing xylem, and mature xylem with successive longitudinal tangential cryosections of P. tremula stem. Chao et al. [38] used three tissues from a hybrid poplar (Populus deltoids x P. euramericana cv. ‘Nanlin895’): Shoot apex; internodes 1–3 (IN1–3); and internodes 4–5 (IN4–5).
Here, we emphasized the transcriptome analysis of cambium differentiation and xylem cell fate specification by designing a gradient of secondary growth developmental stages, including cambium and xylem cell types, within a total of seven tissues: SL; IS; IC; IDX; MC; MDX; and L (Figure 1a). To overcome the limitations of our single run transcriptome data (i.e., no biological replication), we tried; (1) to minimize the biological variation of the samples by combining tissue samples from at least 10 poplars; (2) to check the reliability of our wood-forming tissue-specific transcriptome data by employing the sample correlation heatmap and matrix data (Figure 1c); (3) to validate our transcriptome data using well-known tissue-specific marker genes (Figure 2). We performed not only an extensive transcriptome analysis but also a proof of concept experiment to investigate the molecular mechanism(s) underlying wood formation in poplar.
4.1. Critical Pathway Genes of Secondary Wall Biosynthesis in Woody Perennials
Utilizing our series of poplar wood-forming tissue transcriptomes, we sought to uncover the key players for each step in the biosynthesis pathways of secondary wall components (i.e., cellulose, xylan, and lignin) (Figure 3, Figure 4 and Figure 5). Synthesis of secondary wall components depends on the import of sucrose and its subsequent metabolism. However, our understanding of cell wall precursor biosynthesis in developing wood is still limited [24].
Cellulose is a major constituent of plant cell walls and provides load-bearing strength by forming scaffolds with other cell wall polymers, such as xylan and lignin. Arabidopsis has six SUS (SUCROSE SYNTHASE) genes; SUS is believed to be a main route of carbon entry from sucrose to cellulose via production of UDP-Glc, a substrate for the cellulose synthase [25]. Of the seven SUS genes within our transcriptome data, three are highly expressed in MDX compared to expression in L tissue (Figure 3), indicating that these genes are important for cellulose biosynthesis in xylem. Among them, Potri.006G136700, the closest homolog to Arabidopsis SUS4 (AT3G43190), can be regarded as a key player as it exhibits the highest expression and specificity in MDX. An alternative pathway to production of UDP-Glc from sucrose is through invertases and UGP (UDP-GLC PYROPHOSPHORYLASE). However, UGP is not known to be rate-limiting for cell wall synthesis [59]. We found the gene Potri.013G110800 within the 13 invertases in our data, which is a homolog of Arabidopsis CINV2 (CYTOSOLIC INVERTASE) (AT4G09510) with a strong specificity to xylem. It has been reported that loss of cytosolic invertase affects cell wall synthesis in Arabidopsis [60]. Accordingly, Rende et al. [61] suggested that Potri.013G110800 (reported as CIN12) is responsible for supplying UDP-Glc for cellulose biosynthesis in the development of wood of a hybrid aspen (Populus tremula × tremuloides). Cellulose is produced at the cell surface by CESAs (CELLULOSE SYNTHASE A) [62]. Out of the 13 CESAs in our data, 10 exhibited higher expression in MDX, including: Potri.002G257900.1; Potri.011G069600.1; and Potri.006G181900.1, which are homologous to the well-known secondary wall CESAs; CESA4 (AT5G44030), CESA8 (AT4G18780), and CESA7 (AT5G17420), respectively [63].
Xylans contain xylose subunits as a backbone and are one of the major hemicelluloses found in the secondary cell walls of poplar. Unlike cellulose, xylan has reducing end oligosaccharides, which may act as primers or terminators as well as a variety of side chains [24,26]. UGD (UDP-GLUCOSE DEHYDROGENASE) is responsible for directing xylan biosynthesis from UDP-Glc and for catalyzing the production of UDP-glucuronate [64] (Figure 3). Among the four UGDs within our transcriptome data, three were more highly expressed in MDX—including Potri.008G094300.2—which is highly specific to MDX. UXS (UDP-XYLOSE SYNTHASE) utilizes UDP-glucuronate as a substrate to produce UDP-xylose, a subunit of the xylan backbone. Both Potri.010G207200.5 and Potri.001G237200.3 have higher expression and specificity within MDX, thus, these genes are likely the major UXS genes catalyzing this step. The xylosyl backbone is synthesized by the IRX9, IRX10, and IRX14 genes [65]. Our data suggest that Potri.006G131000.1, Potri.001G068100.3, and Potri.001G067500.1 are key players and are homologous to IRX9, IRX10, and IRX14, respectively. In addition, the poplar genes homologous to those in Arabidopsis that synthesize the reducing end oligosaccharide of xylan, such as IRX7/FRA8, IRX8, and GATL/PARVUS (GALACTURONOSYL TRANSFERASE-like), and those responsible for xylan modification, such as GUX, DUF579/GXM, and RWA, are listed in Figure 3. Based on MDX expression and specificity compared to L tissue, we can predict the essential players of each step.
Lignin is a phenolic compound providing compression strength and hydrophobicity to cell walls, and is a highly heterogeneous and complex polymer composed of p-hydroxylphenyl (H), guaiacyl (G), and syringyl (S) units. The lignin monomer biosynthetic pathway has been extensively investigated and well described [27]. In this pathway, 4CL (4-COUMARATE:CoA LIGASE) catalyzes to produce p-coumaroyl-CoA, a precursor for the biosynthesis of all three of the aforementioned monomers. Potri.001G036900.1 (similar to Arabidopsis 4CL2, AT3G21240.1) seems to be a key gene for this step (Figure 4). S units are synthesized by the action of F5H (FERULATE 5-HYDROXYLASE) and COMT (CAFFEIC ACID O-METHYLTRANSFERASE). Potri.007G016400.1 and Potri.012G006400.1 (similar to Arabidopsis F5H, AT4G36220.1 and COMT, AT5G54160.1, respectively) are prominent within these steps in poplar (Figure 4). Recently, the CSE (CAFFEOYL SHIKIMATE ESTERASE, AT1G52760.1) enzyme has been added to the lignin pathway of Arabidopsis. CSE provides an alternative route to caffeoyl-CoA by catalyzing caffeoyl shikimate [6]. The poplar genome has two CSEs (Potri.001G175000.1 and Potri.003G059200.1) and both exhibit high expression in MDX (Figure 4). Since Arabidopsis cse mutants deposit less lignin but display a four-fold increase in saccharification yield without pretreatment [6], CSE is a promising target for the development of improved lignocellulosic biomass. Recently, Saleme et al. [66] reported that silencing CSE in a hybrid poplar that is morphologically indistinguishable from WT poplar resulted in up to 25% and 62% reduced lignin deposition and increased glucose release, respectively, without pretreatment.
This analysis was able to identify all the essential genes within each step of the biosynthesis pathways for secondary wall components (Figure 3, Figure 4 and Figure 5), with these genes being potential targets for biotechnological improvement for the purpose of ascertaining quality of woody biomass. Very recently, Wang et al. [67] reported a multi-omics quantitative integrative analysis of lignin biosynthesis by perturbing 21 pathway genes to advance the strategic engineering of wood utilization, and these 21 target genes are exactly matched to key players identified from our analysis, with the exception of CSE (Figure 5b). A similar approach could be applicable to other pathways (i.e., those for cellulose and xylan) in combination in order to produce desirable woody biomass.
4.2. Transcriptional Regulators Involved in Vascular Cambium Development
Previously, Pineau et al. [68] reported that the hca (high cambium activity) mutant showed premature and numerous cambial cell divisions in both the fascicular and interfascicular regions. To determine the influence of the hca mutation on global gene expression they performed transcriptome profiling and found AtHAM4/SCL15 (At4g36710), the most upregulated transcription factor belonging to a member of the HAM family, which plays an essential role in shoot meristem maintenance in a non-cell-autonomous manner [20,21]. Zhou et al. [19] demonstrated that HAM family members act as conserved interacting cofactors with WUS/WOX (WUSCHEL/WUSCHEL-RELATED HOMEOBOX) proteins. WUS is a homeodomain transcription factor expressed in the rib meristem of the SAM (shoot apical meristem), and is a key regulatory factor controlling stem cell populations in Arabidopsis [69]. WOX4, expressed in Arabidopsis procambial cells, defines the vascular stem cell niche and regulates cambial cell proliferation [15,18,70]. In particular, Zhou et al. [19] showed that HAM4 and WOX4 physically interact in vivo and were tightly co-regulated in both a spatial and temporal manner. For example, HAM4 and WOX4 are co-expressed in provascular or procambial cell types of various tissues and, in the stem transverse section, HAM4 is expressed specifically in the procambium and overlaps with WOX4 expression. Recently, Kucukoglu et al. [71] suggested that WOX4-like genes regulate cambial cell division activity and secondary growth by using PttWOX4a/b RNAi in Populus trees.
In our co-expression network analysis (Figure 6a), we found several transcription factors highly expressed in IC tissue. Among them, we focused on PtrHAM4-1, a homolog of AtHAM4, which is specifically expressed in the cambial tissues (Figure 6). Indeed, overexpression of PtrHAM4-1 in transgenic Arabidopsis plants resulted in a dramatic increase of the vascular cambium with clear differentiation of secondary xylem vessels, which were known to be produced only from the vascular cambium (Figure 7). This phenotype was further confirmed by overexpressing the PtrHAM4-1 in hybrid poplars (Figure 8). To our knowledge, this is the first evidence that PtrHAM4-1 acts positively in vascular cambium development in plants. Taken together with a report from Kucukoglu et al. [71], our results suggest that a HAM4-WOX4 regulatory module may be conserved in Populus to achieve cambial meristem initiation and proliferation during secondary growth. However, the underlying molecular regulatory mechanism remains unknown. Chromatin modifications, including histone acetylation, have been implicated in meristem activity. Thus, interaction of HAM4/SCL15 with HDA19 (HISTONE DEACETYLASE19) [72] may provide a clue for the regulation of downstream gene expression, which will be a priority for our future studies.
In summary, a comprehensive wood-forming tissue-specific transcriptome analysis from a hybrid poplar successfully pinpointed many essential genes involved in the biosynthetic pathways of secondary wall components. These genes could be focal points for the biotechnological improvement of wood properties within the production of biomaterials and/or biofuels. Furthermore, the transcriptional regulators involved in vascular cambium development were isolated and demonstrated their validity via functional characterization of PtrHAM4-1 using a heterologous expression. Thus, our results may offer insights for disentangling the complex mechanisms of wood formation, one of the most important biological processes on this planet.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/9/690/s1, Figure S1: RNA-seq raw data processing, Figure: S2. Mapping of the sequence reads to the genome of Populus trichocarpa, Figure S3: PtrHAM4-1 is homologous to AtHAM4, Table S1: A total of 38,330 transcripts across all tissues, after removing redundant transcripts from a total of 85,209 transcripts, Table S2: List of primers used in this study, Table S3: List of transcripts preferentially expressed in IC (885 transcripts), Table S4: List of transcripts preferentially expressed in MC (798 transcripts).
Author Contributions
M.-H.K., J.-S.C., H.-W.J. and J.-H.K.: Conceived the project; M.-H.K., J.-S.C., H.-W.J., K.S., D.S., Y.-I.C., E.-J.P., H.L. and J.-H.K.: Performed the experiments and analyzed the data; M.-H.K. and J.-H.K.: Wrote the paper.
Funding
This research was funded by the Forest Resources Genome Project (2014071G10-1722-AA04) and the National Research Foundation of Korea (NRF-2018R1A2B2005362 and NRF-2018R1A6A3A01013476).
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Pan, Y.; Bridsey, R.A.; Fang, J.; Houghton, R.; Kauppi, P.E.; Kurz, W.A.; Phillips, O.L.; Shvidenko, A.; Lewis, S.L.; Canadell, J.G.; et al. A large and persistent carbon sink in the world’s forests. Science 2011, 333, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Lal, R. Managing soils and ecosystems for mitigating anthropogenic carbon emissions and advancing global food security. BioScience 2010, 60, 708–721. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Pan, X.; Zalesny, R.S. Pretreatment of woody biomass for biofuel production: Energy efficiency, technologies, and recalcitrance. Appl. Microbiol. Biotechnol. 2010, 87, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Chundawat, S.P.; Beckham, G.T.; Himmel, M.E.; Dale, B.E. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Serapiglia, M.J.; Humiston, M.C.; Xu, H.; Hogsett, D.A.; Mira de Orduna, R.; Stipanovic, A.J.; Smart, L.B. Enzymatic saccharification of shrub willow genotypes with differing biomass composition for biofuel production. Front. Plant Sci. 2013, 4, 57. [Google Scholar] [CrossRef]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.; Nieves, I.U.; Mullinnix, M.T.; Sagues, W.J.; Hoffman, R.W.; Fernández-Sandoval, M.T.; Tian, Z.; Rockwood, D.L.; Tamang, B.; Ingram, L.O. Optimization of dilutephosphoric-acid steam pretreatment of Eucalyptus benthamii for biofuel production. Appl. Energy 2014, 125, 76–83. [Google Scholar] [CrossRef]
- Albers, S.C.; Berklund, A.M.; Graff, G.D. The rise and fall of innovation in biofuel. Nat. Biotechnol. 2016, 34, 814–821. [Google Scholar] [CrossRef]
- Ko, J.H.; Kim, W.C.; Cho, J.S.; Choi, Y.I.; Park, E.J.; Im, J.H.; Han, O.; Keathley, D.; Han, K.H. EliteTreeTM: An advanced biomass tree crop technology that features greater wood density and accelerated stem growth. Biofuels Bioprod. Biorefin. 2017, 11, 521–533. [Google Scholar] [CrossRef]
- Vo, T.K.; Cho, J.S.; Kim, S.S.; Ko, J.H.; Kim, J. Genetically engineered hybrid poplars for the pyrolytic production of bio-oil: Pyrolysis characteristics and kinetics. Energy Convers. Manag. 2017, 153, 48–59. [Google Scholar] [CrossRef]
- Ragni, L.; Greb, T. Secondary growth as a determinant of plant shape and form. Semin. Cell Dev. Biol. 2018, 79, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Dolan, L.; Roberts, K. Secondary thickening in roots of Arabidopsis thaliana: Anatomy and cell surface changes. New Phytol. 1995, 131, 121–128. [Google Scholar] [CrossRef]
- Ko, J.H.; Han, K.H.; Park, S.; Yang, J. Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol. 2004, 135, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Turner, S.R. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 2010, 137, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. Regulation of vascular development by CLE peptide-receptor systems. J. Integr. Plant Biol. 2010, 52, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Bowman, J.L. A Role of TDIF peptide signaling in vascular cell differentiation is conserved among Euphyllophytes. Front. Plant Sci. 2015, 6, 1048. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213. [Google Scholar] [CrossRef]
- Etchells, J.P.; Provost, C.M.; Mishra, L.; Turner, S.R. WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 2013, 140, 2224–2234. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, X.; Engstrom, E.M.; Nimchuk, Z.L.; Pruneda-Paz, J.L.; Tarr, P.T.; Yan, A.; Kay, S.A.; Meyerowitz, E.M. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 2015, 517, 377–380. [Google Scholar] [CrossRef]
- Stuurman, J.; Jäggi, F.; Kuhlemeier, C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002, 16, 2213–2218. [Google Scholar] [CrossRef]
- Engstrom, E.M. HAM proteins promote organ indeterminacy: But how? Plant Signal. Behav. 2012, 1, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Caño-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.C.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Ito, T.; Nakagami, H.; Hirakawa, Y.; Saito, M.; Tamaki, T.; Fukuda, H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF–TDR signaling. Nat. Commun. 2014, 5, 3504. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Verbančič, J.; Lunn, J.E.; Stitt, M.; Persson, S. Carbon supply and the regulation of Cell Wall Synthesis. Mol. Plant 2018, 11, 75–94. [Google Scholar] [CrossRef] [PubMed]
- York, W.S.; O’Neill, M.A. Biochemical control of xylan biosynthesis—Which end is up? Curr. Opin. Plant Biol. 2008, 11, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Kim, W.C.; Kim, J.Y.; Ahn, S.J.; Han, K.H. MYB46-mediated transcriptional regulation of secondary wall biosynthesis. Mol. Plant 2012, 5, 961–963. [Google Scholar] [CrossRef] [PubMed]
- D’haeseleer, P.; Liang, S.; Somogyi, R. Genetic network inference: From co-expression clustering to reverse engineering. Bioinformatics 2000, 16, 707–726. [Google Scholar] [CrossRef]
- Hertzberg, M.; Aspeborg, H.; Schrader, J.; Andersson, A.; Erlandsson, R.; Blomqvist, K.; Bhalerao, R.; Uhlén, M.; Teeri, T.T.; Lundeberg, J.; et al. A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. USA 2001, 98, 14732–14737. [Google Scholar] [CrossRef]
- Schrader, J.; Nilsson, J.; Mellerowicz, E.; Berglund, A.; Nilsson, P.; Hertzberg, M.; Sandberg, G. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 2004, 16, 2278–2292. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Beers, E.P.; Han, K.H. Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol. Genet. Genom. 2006, 276, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ye, C.Y.; Bisaria, A.; Tuskan, G.A.; Kalluri, U.C. Identification of candidate genes in Arabidopsis and Populus cell wall biosynthesis using text-mining, co-expression network analysis and comparative genomics. Plant Sci. 2011, 181, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Li, C.H.; Huang, J. Systematic identification of cell-wall related genes in Populus based on analysis of functional modules in co-expression network. PLoS ONE 2014, 9, e95176. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Wang, J.P.; Lin, Y.C.; Li, Q.; Sun, Y.H.; Chen, H.; Sederoff, R.R.; Chiang, V.L. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 2017, 245, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Sundell, D.; Street, N.R.; Kumar, M.; Mellerowicz, E.J.; Kucukoglu, M.; Johnsson, C.; Kumar, V.; Mannapperuma, C.; Delhomme, N.; Nilsson, O.; et al. AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 2017, 29, 1585–1604. [Google Scholar] [CrossRef] [PubMed]
- Chao, Q.; Gao, Z.F.; Zhang, D.; Zhao, B.G.; Dong, F.Q.; Fu, C.X.; Liu, L.J.; Wang, B.C. The developmental dynamics of the Populus stem transcriptome. Plant Biotechnol. J. 2019, 17, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S.; Douglas, C.J. Populus: A model system for plant biology. Annu. Rev. Plant Biol. 2007, 58, 435–458. [Google Scholar] [CrossRef] [PubMed]
- Porth, I.; EI-Kassaby, Y.A. Using Populus as a lignocellulosic feedstock for bioethanol. Biotechnol. J. 2015, 10, 510–524. [Google Scholar] [CrossRef]
- Biswal, A.K.; Atmodjo, M.A.; Pattathil, S.; Amos, R.A.; Yang, X.; Winkeler, K.; Collins, C.; Mohanty, S.S.; Ryno, D.; Tan, L.; et al. Working towards recalcitrance mechanisms: Increased xylan and homogalacturonan production by overexpression of GAlactUronosylTransferase12, GAUT12. causes increased recalcitrance and decreased growth in Populus. Biotechnol. Biofuels 2018, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Movahedi, A.; Sang, M.; Zhang, J.; Mohammadi, K.; Sun, W.; Yaghuti, A.A.Z.; Kadkhodaei, S.; Ruan, H.; Zhuge, Q. Functional analyses of PtROS1-RNAi in Poplars and evaluation of its effect on DNA methylation. J. Plant Biol. 2018, 61, 227–240. [Google Scholar] [CrossRef]
- Etchells, J.P.; Mishra, L.S.; Kumar, M.; Campbell, L.; Turner, S.R. Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 2015, 25, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Groover, A. Transcriptional regulation of secondary growth and wood formation. J. Integr. Plant Biol. 2010, 52, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Kim, H.T.; Hwang, I.; Han, K.H. Tissue-type-specific transcriptome analysis identifies developing xylem-specific promoters in poplar. Plant Biotechnol. J. 2012, 10, 587–596. [Google Scholar] [CrossRef]
- Logemann, J.; Schell, J.; Willmitzer, L. Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 1987, 163, 16–20. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Zhu, C.; Zhang, C.; Cui, S. Detection of porcine parvovirus using a taqman-based real-time PCR with primers and probe designed for the NS1 gene. Virol. J. 2010, 7, 353. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Kim, M.D.; Choi, Y.I.; Park, S.D.; Yun, D.J.; Noh, E.W.; Lee, H.S.; Kwak, S.S. Transgenic poplar expressing Arabidopsis NDPK2 enhances growth as well as oxidative stress tolerance. Plant Biotechnol. J. 2010, 9, 334–347. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Jeon, H.W.; Cho, J.S.; Park, E.J.; Han, K.H.; Choi, Y.I.; Ko, J.H. Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 2016, 14, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Wilton, O.C. Correlation of cambial activity with flowering and regeneration. Bot. Gaz. 1938, 99, 854–864. [Google Scholar] [CrossRef]
- Melzer, S.; Lens, F.; Gennen, J.; Vanneste, S.; Rohde, A.; Beeckman, T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 2008, 40, 1489–1492. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Geisler, M.; Johansson, H.; Harholt, J.; Scheller, H.V.; Mellerowicz, E.J.; Kleczkowski, L.A. UDP-glucose pyrophosphorylase is not rate limiting, but is essential in Arabidopsis. Plant Cell Physiol. 2009, 50, 998–1011. [Google Scholar] [CrossRef]
- Barratt, D.H.; Derbyshire, P.; Findlay, K.; Pike, M.; Wellner, N.; Lunn, J.; Feil, R.; Simpson, C.; Maule, A.J.; Smith, A.M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 13124–13129. [Google Scholar] [CrossRef]
- Rende, U.; Wang, W.; Gandla, M.L.; Jönsson, L.J.; Niittylä, T. Cytosolic invertase contributes to the supply of substrate for cellulose biosynthesis in developing wood. New Phytol. 2017, 214, 796–807. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, H.E.; Döring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.G.; Howells, R.M.; Huttly, A.K.; Vickers, K.; Turner, S.R. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 2003, 100, 1450–1455. [Google Scholar] [CrossRef] [PubMed]
- Seifert, G.J. Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr. Opin. Plant Biol. 2004, 7, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Zhong, R.; Ye, Z.H. Biochemical characterization of xylan xylosyltransferases involved in wood formation in poplar. Plant Signal. Behav. 2012, 7, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Saleme, M.L.S.; Cesarino, I.; Vargas, L.; Kim, H.; Vanholme, R.; Goeminne, G.; Van Acker, R.; Fonseca, F.C.A.; Pallidis, A.; Voorend, W.; et al. Silencing CAFFEOYL SHIKIMATE EXTERASE affects lignification and improves saccharification in Poplar. Plant Physiol. 2017, 175, 1040–1057. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Matthews, M.L.; Williams, C.M.; Shi, R.; Yang, C.; Tunlaya-Anukit, S.; Chen, H.C.; Li, Q.; Liu, J.; Lin, C.Y.; et al. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 2018, 9, 1579. [Google Scholar] [CrossRef] [PubMed]
- Pineau, C.; Freydier, A.; Ranocha, P.; Jauneau, A.; Turner, S.; Lemonnier, G.; Renou, J.P.; Tarkowski, P.; Sandberg, G.; Jouanin, L.; et al. hca: An Arabidopsis mutant exhibiting unusual cambial activity and altered vascular patterning. Plant J. 2015, 44, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgenes, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 11, 805–815. [Google Scholar] [CrossRef]
- Suer, S.; Agusti, J.; Sanchez, P.; Schwarz, M.; Greb, T. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 2011, 23, 3247–3259. [Google Scholar] [CrossRef] [PubMed]
- Kucukoglu, M.; Nilsson, J.; Zheng, B.; Chaabouni, S.; Nilsson, O. WUSCHEL-RELATED HOMEOBOX4, WOX4.-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 2017, 215, 642–657. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.J.; Li, X.; Huang, J.; Gropp, G.M.; Gjetvaj, B.; Lindsay, D.L.; Wei, S.; Coutu, C.; Chen, Z.; Wan, X.C.; et al. SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme. Nat. Commun. 2015, 6, 7243. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).