Autophagy Related Gene (ATG3) is a Key Regulator for Cell Growth, Development, and Virulence of Fusarium oxysporum
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungus Isloation and Culturing
2.2. Fungal Transformastion
2.3. Generation of Deletion Mutants
2.4. Overexpression of FoATG3 Mutant Strains
2.5. Evaluation of Radial Growth, Conidiation, Formation, and Germination
2.6. RNA Extraction
2.7. Quantitative RT-qPCR
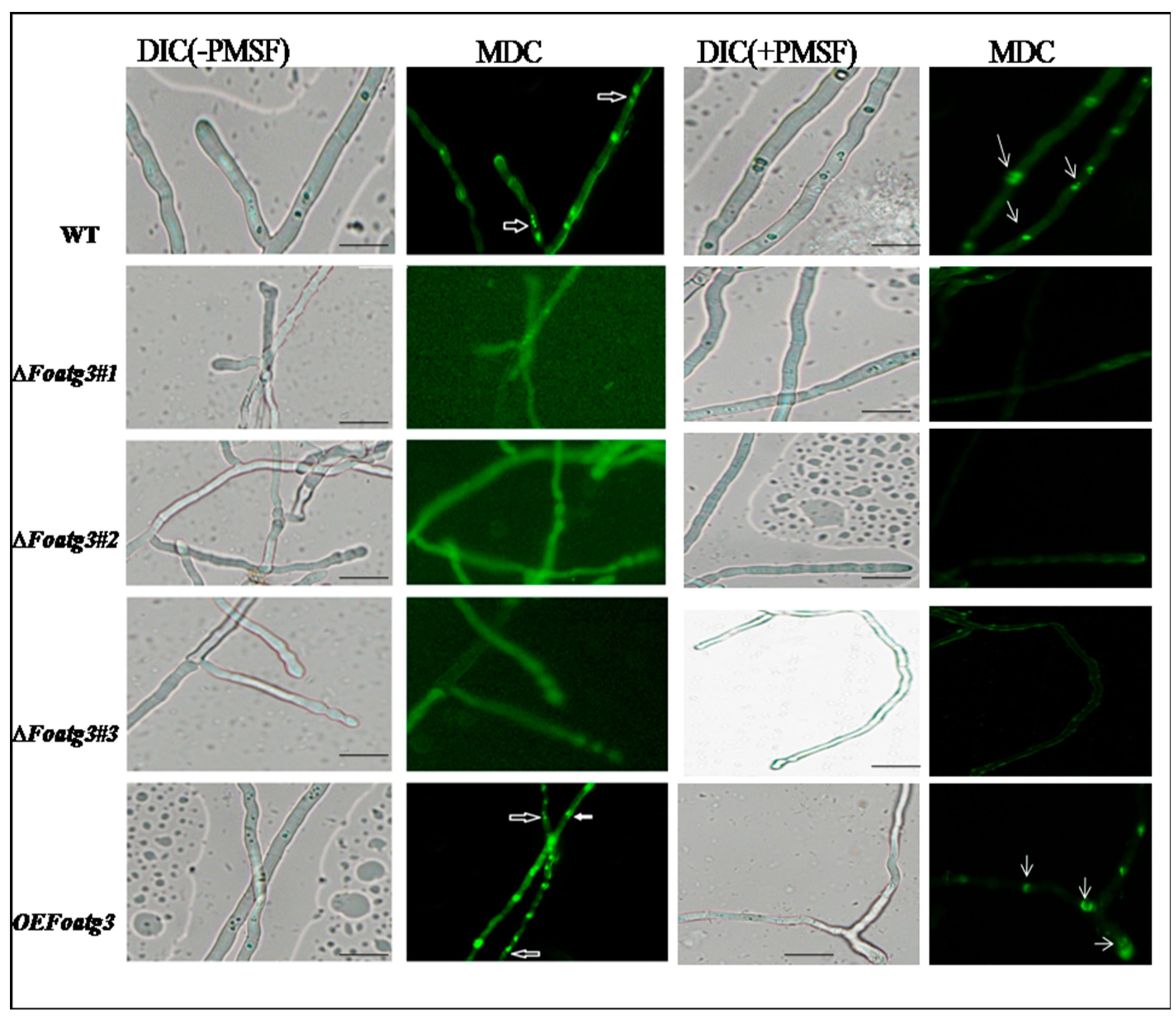
2.8. Analysis of Autophagy
2.9. Pathogenicity Test
2.10. Optical and Epifluorescence Microscopy
3. Results
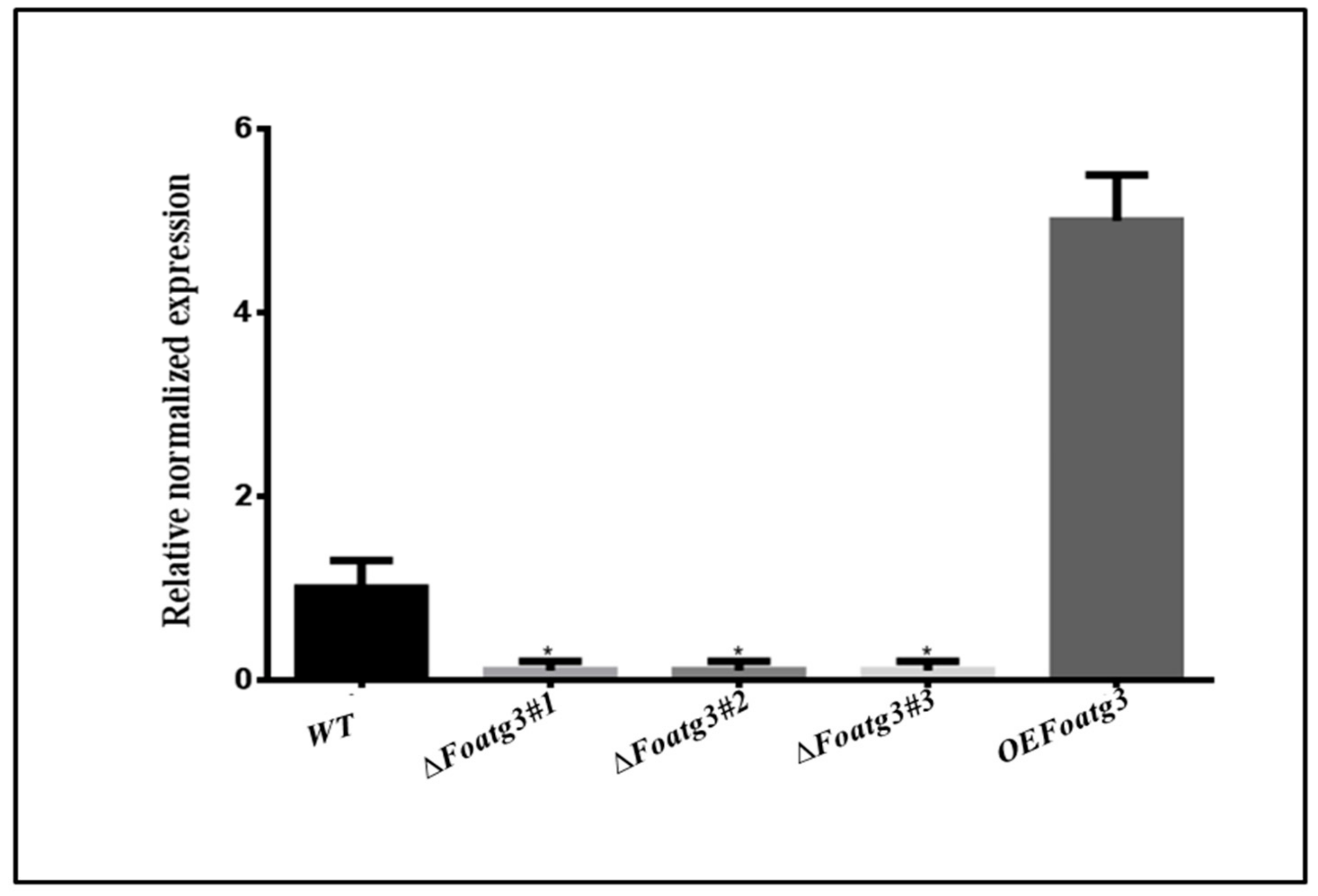
3.1. Over Expression and Deletion of FoATG3 Mutants in F. Oxysporum
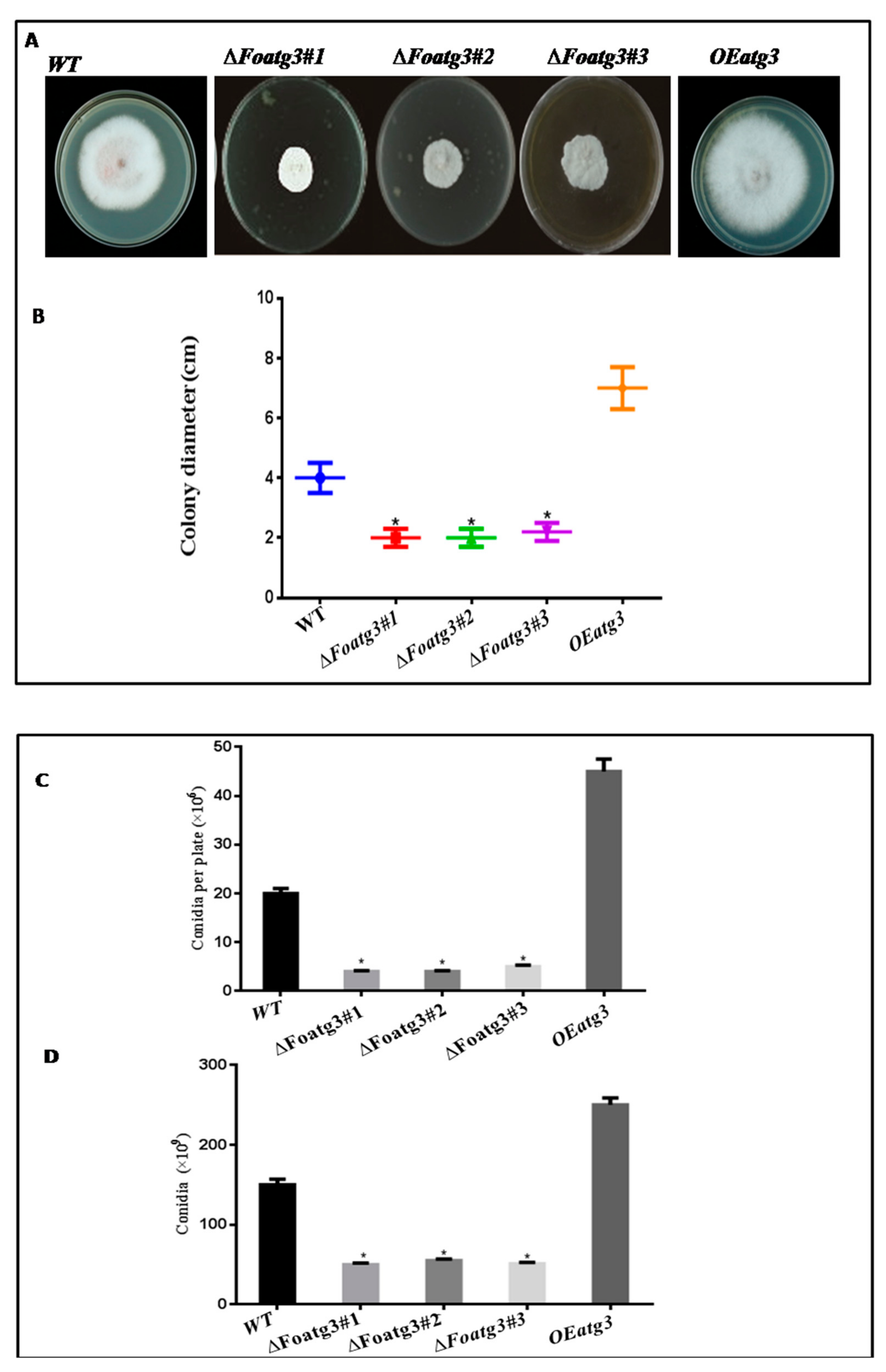
3.2. Role of Atg3 on Conidiation and Vegetative Growth of Fusarium Oxysporum
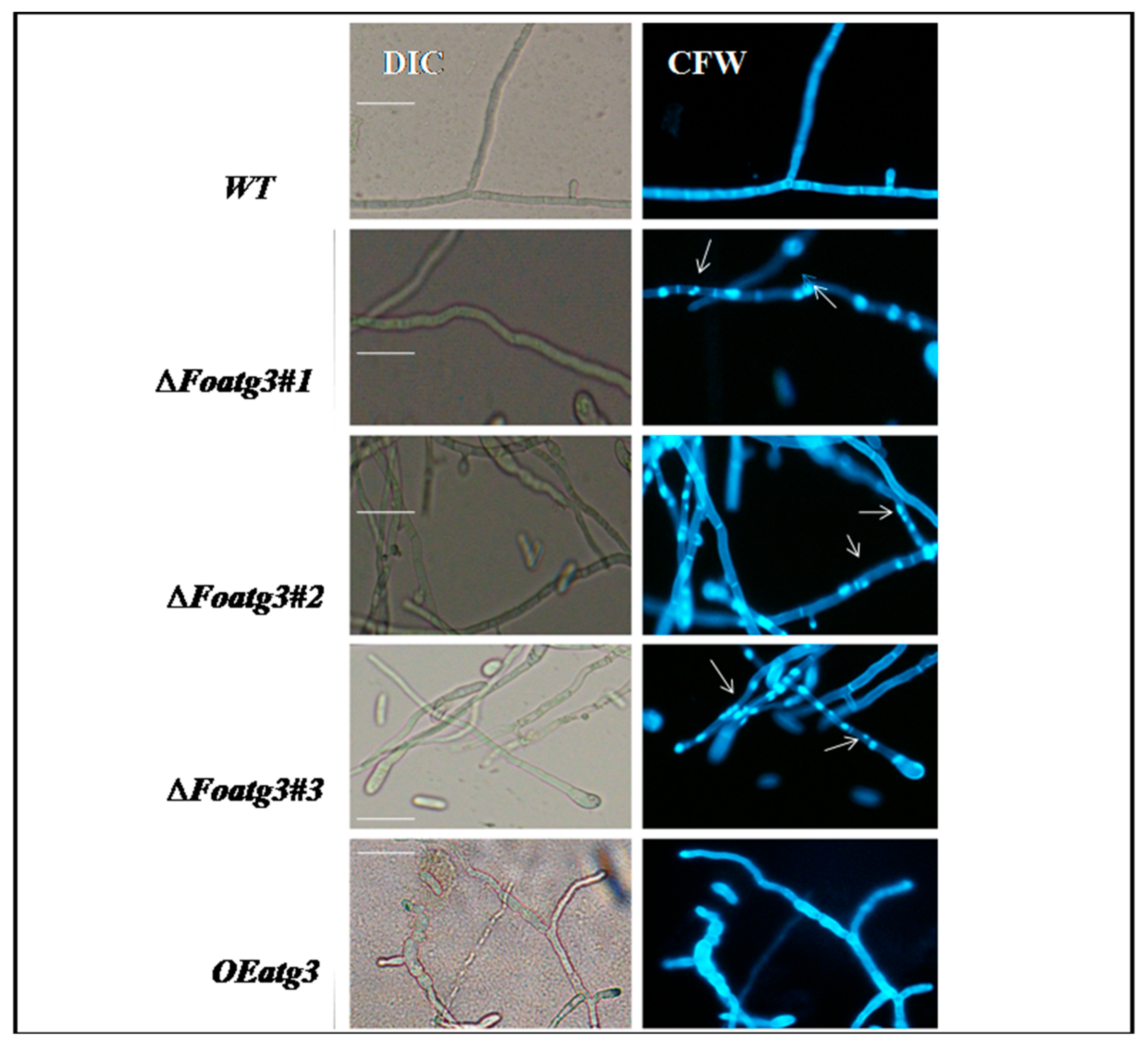
3.3. Role of Atg3 in Cellular Distribution of Nuclei in F. Oxysporum
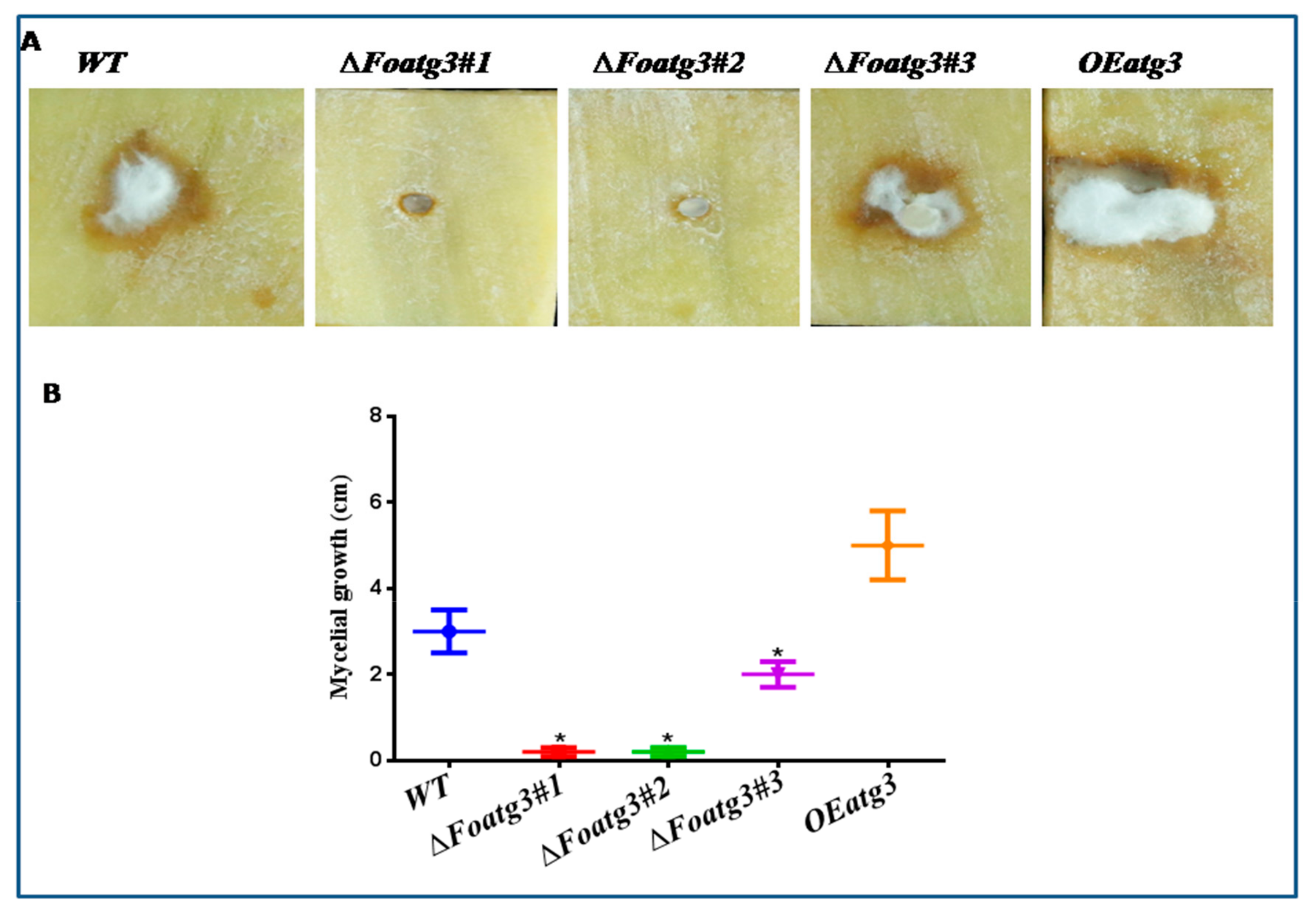
3.4. Autophagy Affects on Virulence of F. Oxysporum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ATG | Autophagy-related gene |
| WT | Wild type |
| OE | Overexpression |
| ∆ | Deletion mutant |
| MDC | Monodansylcadaverine |
| DIC | Differential interference contrast |
References
- El-Kassas, H.Y.; Khairy, H.M. A trial for biological control of a pathogenic fungus (Fusarium solani) by some marine microorganisms. Am. Eurasian J. Agric. Environ. Sci. 2009, 5, 434–440. [Google Scholar]
- Wharton, P.K.W. Fusarium Dry Rot. 2007. Available online: http://www.Potatodiseases.Org/contact.html (accessed on 23 May 2007).
- Chełkowski, J. Toxinogenicity of Fusarium Species Causing Dry Rot of Potato Tubers; Elsevier, B.V.: New York, NY, USA, 1989; Chapter 25; pp. 435–440. [Google Scholar]
- Slininger, P.J.; Burkhead, K.D.; Schisler, D.A. Antifungal and sprout regulatory bioactivities of phenylacetic acid, indole-3 acetic acid, and tyrosol isolated from the potato dry rot suppressive bacterium Enterobacter cloacae S11:T:07. J. Ind. Microbiol. Biotechnol. 2004, 31, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Secor, G.A.; Gudmestad, N.C. Managing fungal diseases of potato. Can. J. Plant Pathol. 1999, 21, 213–221. [Google Scholar] [CrossRef]
- Shattock, R. Compendium of Potato Diseases, Second Edition. Plant Pathol. 2002, 51, 520. [Google Scholar] [CrossRef]
- Saccardo, P.A. Sylloge Fungorum Omnium Hucusque Cognitorum; Edwards Brothers Malloy: Ann Arbor, MI, USA, 1882. [Google Scholar]
- Secor, G.A.; Salas, B. Fusarium dry rot and fusarium wilt. In Compendium of Potato Diseases; Stevenson, W.R., Loria, F., Franc, G.D., Weingartner, D.P., Eds.; APS Press: St. Paul, MN, USA, 2001; pp. 23–25. [Google Scholar]
- Yangui, T.; Sayadi, S.; Dhouib, A. Sensitivity of pectobacterium carotovorum to hydroxytyrosol-rich extracts and their effect on the development of soft rot in potato tubers during storage. Crop Prot. 2013, 53, 52–57. [Google Scholar] [CrossRef]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. BMC Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.R.; Vierstra, R.D. Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 2005, 8, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cai, S.Y.; Yin, L.L.; Shi, K. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 2015, 11, 2033–2047. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, J.Q.; Chen, Z.X. The perplexing role of autophagy in plant innate immune responses. Mol. Plant Pathol. 2014, 15, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.M.; Zhao, P.; Wang, W.; Zou, J. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 2015, 22, 245–257. [Google Scholar] [CrossRef]
- Bartoszewska, M.; Kiel, J.A. The role of macroautophagy in development of filamentous fungi. Antioxid. Redox Signal. 2011, 14, 2271–2287. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.A.; Lu, J.P.; Liu, X.H.; Rehman, A. Multifunction of autophagy-related genes in filamentous fungi. Microbiol. Res. 2012, 167, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.F.; Guo, M.; Wang, H.; Lu, J.P.; Liu, J.; Zhang, C.; Gong, Z.; Lu, M. Autophagy, a conserved mechanism for protein degradation, responds to Heat, and other abiotic stresses in Capsicum annuum L. Front. Plant Sci. 2016, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.; Suttangkakul, A.; Vierstra, R.D. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-Lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009, 149, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.I.; Cho, H.J.; Kim, S.R.; Park, O.K. The rab GTPase rabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in arabidopsis. Plant Physiol. 2013, 161, 1722–1736. [Google Scholar] [CrossRef] [PubMed]
- Maruthachalam, K.; Klosterman, S.J.; Kang, S.; Hayes, R.J. Identification of pathogenicity related genes in the Vascular Wilt Fungus Verticillium dahliae by agrobacterium tumefaciens-mediated T-DNA Insertional mutagenesis. Mol. Biotechnol. 2011, 49, 209–221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Y.G.; Chen, Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTechniques 2007, 43, 649–650. [Google Scholar] [CrossRef] [PubMed]
- Klosterman, S.J.; Subbarao, K.V.; Kang, S.C.; Veronese, P. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 2011, 7, e1002137. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Prot. 2006, 1, 3111–3120. [Google Scholar] [CrossRef]
- Di Pietro, A.; Roncero, M.I.G. Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant Microbe Interact. 1998, 11, 91–98. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdalla, F.C.; Abeliovich, H.; Abraham, R.T.; Acevedo-Arozena, A.; Adeli, K.; Agholme, L.; Agnello, M.; Agostinis, P.; Aguirre-Ghiso, J.A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012, 8, 445–544. [Google Scholar] [CrossRef] [PubMed]
- Biederbick, A.; Kern, H.F.; Elsasser, H.P. Monodansylcadaverine (Mdc) is a specific in-vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 1995, 66, 3–14. [Google Scholar] [PubMed]
- Josefsen, L.; Droce, A.; Sondergaard, T.E.; Sorensen, J.L. Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium graminearum. Autophagy 2012, 8, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Husaini, A.M.; Sakina, A.; Cambay, S.R. Host-pathogen interaction in Fusarium oxysporum infections: Where do we stand? Mol. Plant Microbe Interact. 2018, 31, 889–898. [Google Scholar] [CrossRef] [PubMed]
- De Sain, M.; Rep, M. The role of pathogen secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 2015, 16, 23970–23993. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M.; Manners, J.M. On the trail of a cereal killer: Recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 2012, 13, 399–413. [Google Scholar] [CrossRef]
- Gordon, T.R. Fusarium oxysporum and the fusarium wilt syndrome. Annu. Rev. Phytopathol. 2017, 55, 23–39. [Google Scholar] [CrossRef]
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef]
- Levine, B.; Klionsky, D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, S.; Zhang, J.; Russel, R.J.; Wang, Y.; Qiu, D.; Luo, X.; Khalid, A.R.; Wang, H.; Feng, L.; et al. Synergistic anti-oomycetes effect of melatonin with a biofungicides against oomycetic black shank diseses. J. Pineal Res. 2018, 65, e12492. [Google Scholar] [CrossRef]
- Klionsky, D.J. The molecular machinery of autophagy: Unanswered questions. J. Cell Sci. 2005, 118, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Roldan, M.C.; Kohli, M.; Roncero, M.I.G.; Philippsen, P. Nuclear dynamics during germination, conidiation, and hyphal fusion of Fusarium oxysporum. Eukaryot. Cell 2010, 9, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Corral-Ramos, C.; Roca, M.G.; Di Pietro, A.; Roncero, M.I.G. Autophagy contributes to regulation of nuclear dynamics during vegetative growth and hyphal fusion in Fusarium oxysporum. Autophagy 2015, 11, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, H.; Noda, T.; Shirano, Y.; Kato, T. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002, 129, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.F.; Liu, T.; Ouyang, J.; Wang, R. Genome-wide identification, classification, and expression analysis of autophagy associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 15, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.P.; Nair, U.; Klionsky, D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 2008, 19, 3290–3298. [Google Scholar] [CrossRef] [PubMed]
- Richie, D.L.; Fuller, K.K.; Fortwendel, J.; Miley, M.D.; McCarthy, J.W.; Feldmesser, M.; Rhodes, J.C.; Askew, D.S. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot. Cell 2007, 6, 2437–2447. [Google Scholar] [CrossRef]
- Shoji, J.Y.; Kikuma, T.; Arioka, M.; Kitamoto, K. Macroautophagy mediated degradation of whole nuclei in the filamentous fungus aspergillus oryzae. PLoS ONE 2010, 5, 220–234. [Google Scholar] [CrossRef]
- Nitsche, B.M.; Burggraaf-van Welzen, A.M.; Lamers, G.; Meyer, V. Autophagy promotes survival in aging submerged cultures of the filamentous fungus Aspergillus niger. Appl. Microbiol. Biotechnol. 2013, 97, 8205–8218. [Google Scholar] [CrossRef]
- Veneault-Fourrey, C.; Barooah, M.; Egan, M.; Wakley, G.; Talbot, N.J. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006, 312, 580–583. [Google Scholar] [CrossRef]
- Khalid, A.R.; Zhang, S.; Luo, X.; Mehmood, K.; Rahim, J.; Shaheen, H.; Dong, P.; Qiu, D.; Ren, M. Role of autophagy-related gene atg22 in developmental process and virulence of Fusarium oxysporum. Genes 2019, 10, 365. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid, A.R.; Lv, X.; Naeem, M.; Mehmood, K.; Shaheen, H.; Dong, P.; Qiu, D.; Ren, M. Autophagy Related Gene (ATG3) is a Key Regulator for Cell Growth, Development, and Virulence of Fusarium oxysporum. Genes 2019, 10, 658. https://doi.org/10.3390/genes10090658
Khalid AR, Lv X, Naeem M, Mehmood K, Shaheen H, Dong P, Qiu D, Ren M. Autophagy Related Gene (ATG3) is a Key Regulator for Cell Growth, Development, and Virulence of Fusarium oxysporum. Genes. 2019; 10(9):658. https://doi.org/10.3390/genes10090658
Chicago/Turabian StyleKhalid, A. Rehman, Xiulan Lv, Muhammad Naeem, Khalid Mehmood, Hamayun Shaheen, Pan Dong, Dan Qiu, and Maozhi Ren. 2019. "Autophagy Related Gene (ATG3) is a Key Regulator for Cell Growth, Development, and Virulence of Fusarium oxysporum" Genes 10, no. 9: 658. https://doi.org/10.3390/genes10090658
APA StyleKhalid, A. R., Lv, X., Naeem, M., Mehmood, K., Shaheen, H., Dong, P., Qiu, D., & Ren, M. (2019). Autophagy Related Gene (ATG3) is a Key Regulator for Cell Growth, Development, and Virulence of Fusarium oxysporum. Genes, 10(9), 658. https://doi.org/10.3390/genes10090658





