Joint RNA-Seq and miRNA Profiling Analyses to Reveal Molecular Mechanisms in Regulating Thickness of Pod Canopy in Brassica napus
Abstract
1. Introduction
2. Materials and Methods
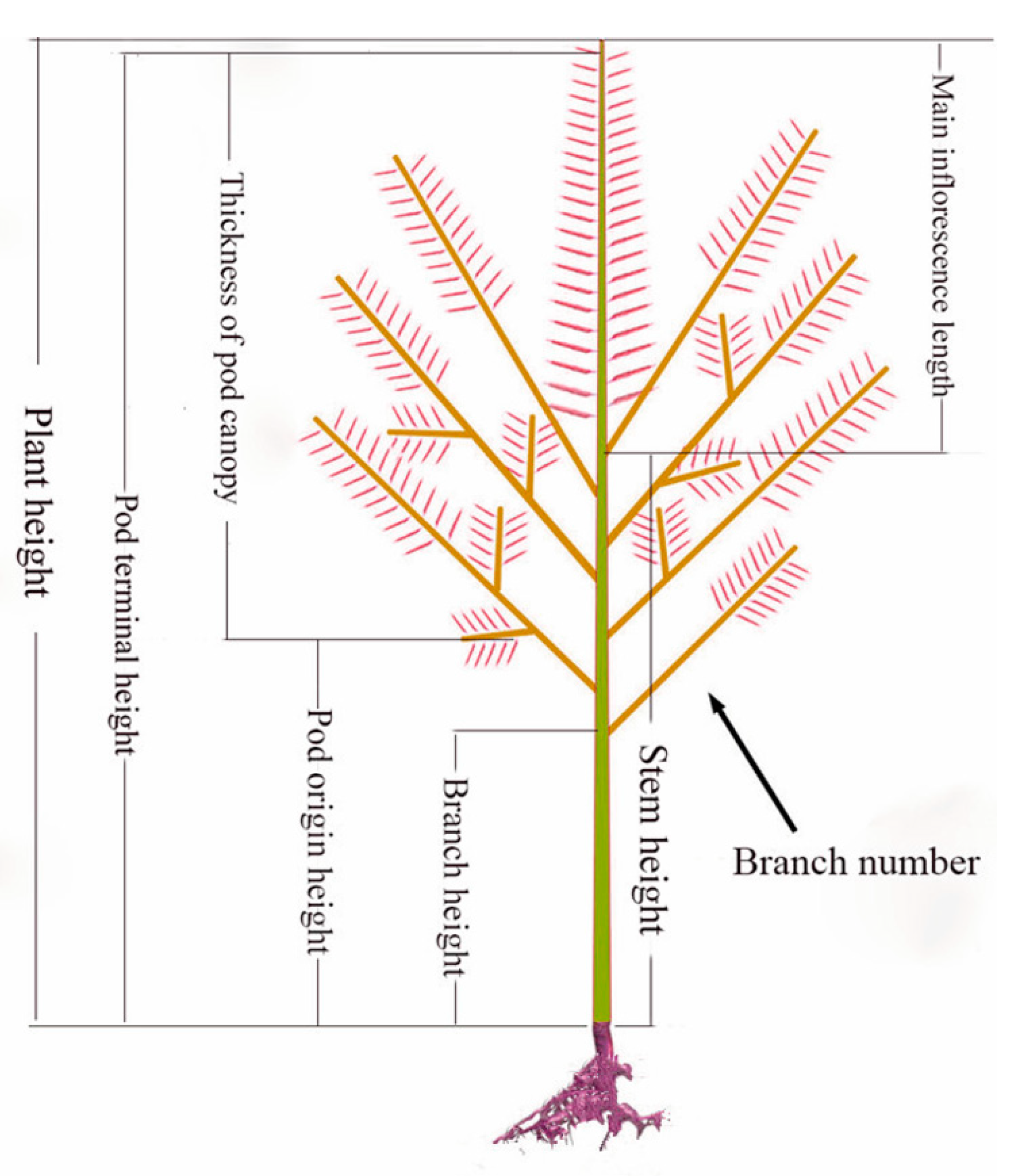
2.1. Plant Materials, RNA Sequencing, and Small RNA Sequencing
2.2. Transcriptome Analysis
2.3. Small RNA Data Analysis
2.4. qRT-PCR Validation
3. Results
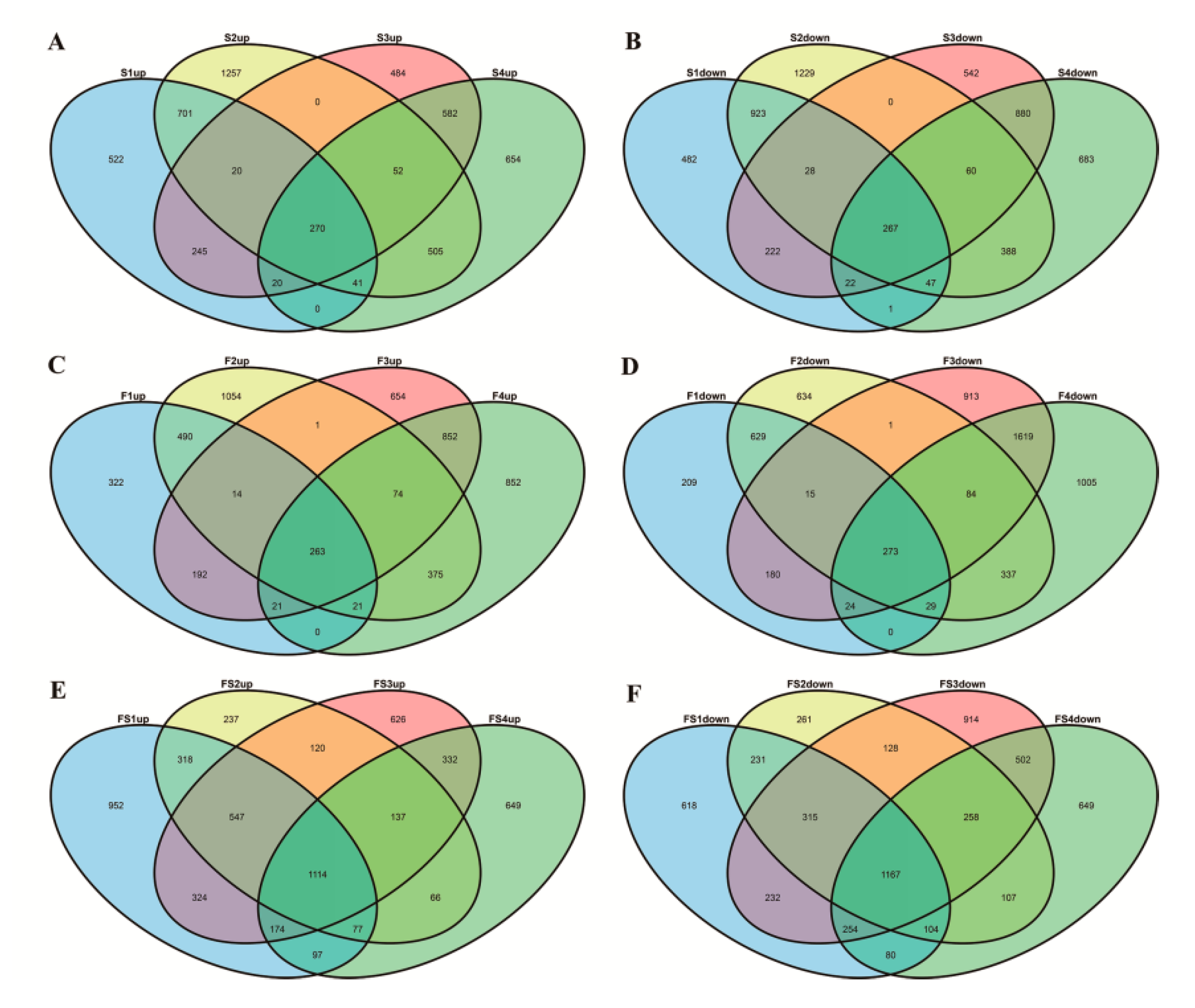
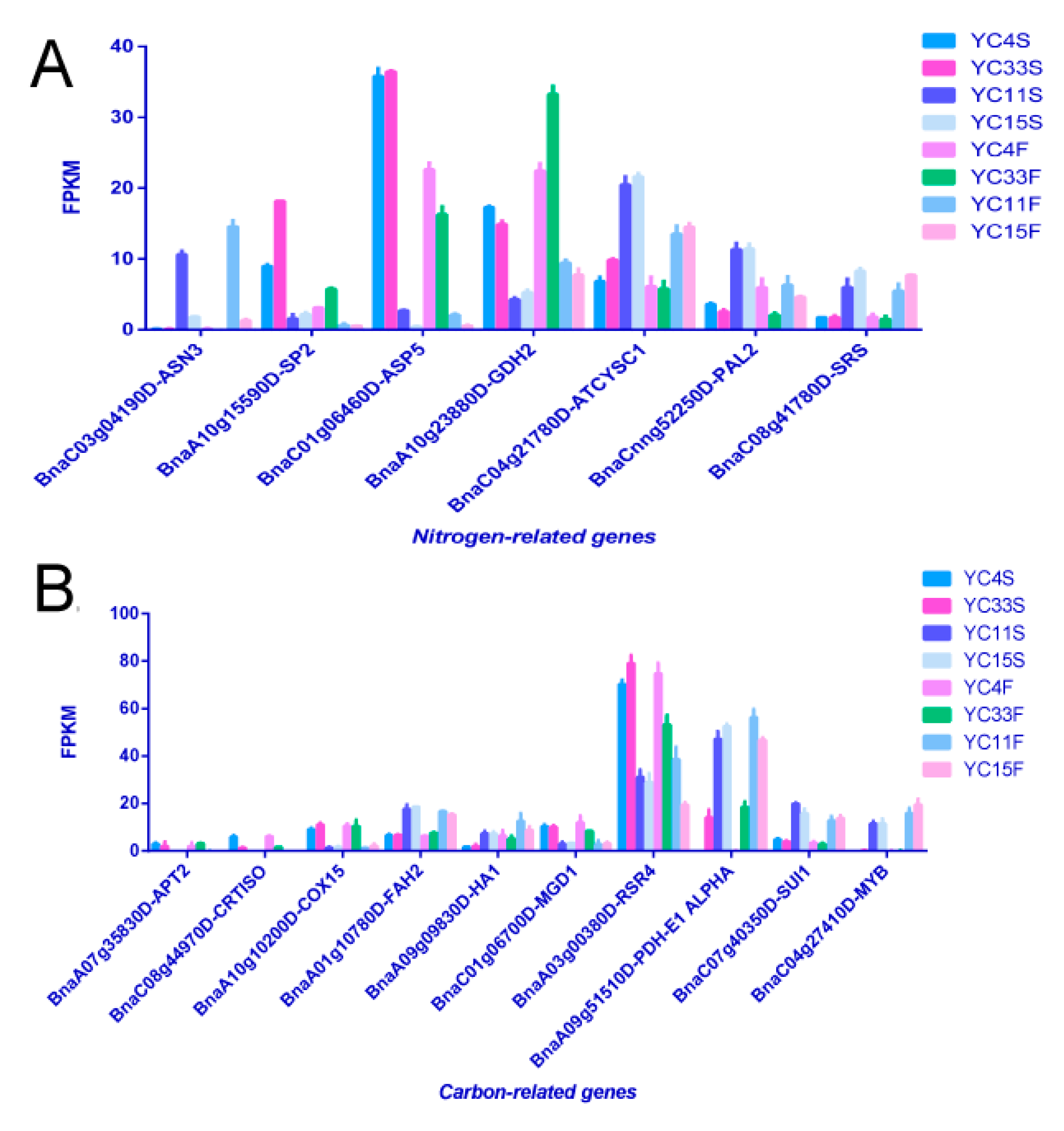
3.1. Identification of Differentially Expressed Genes by RNA-seq
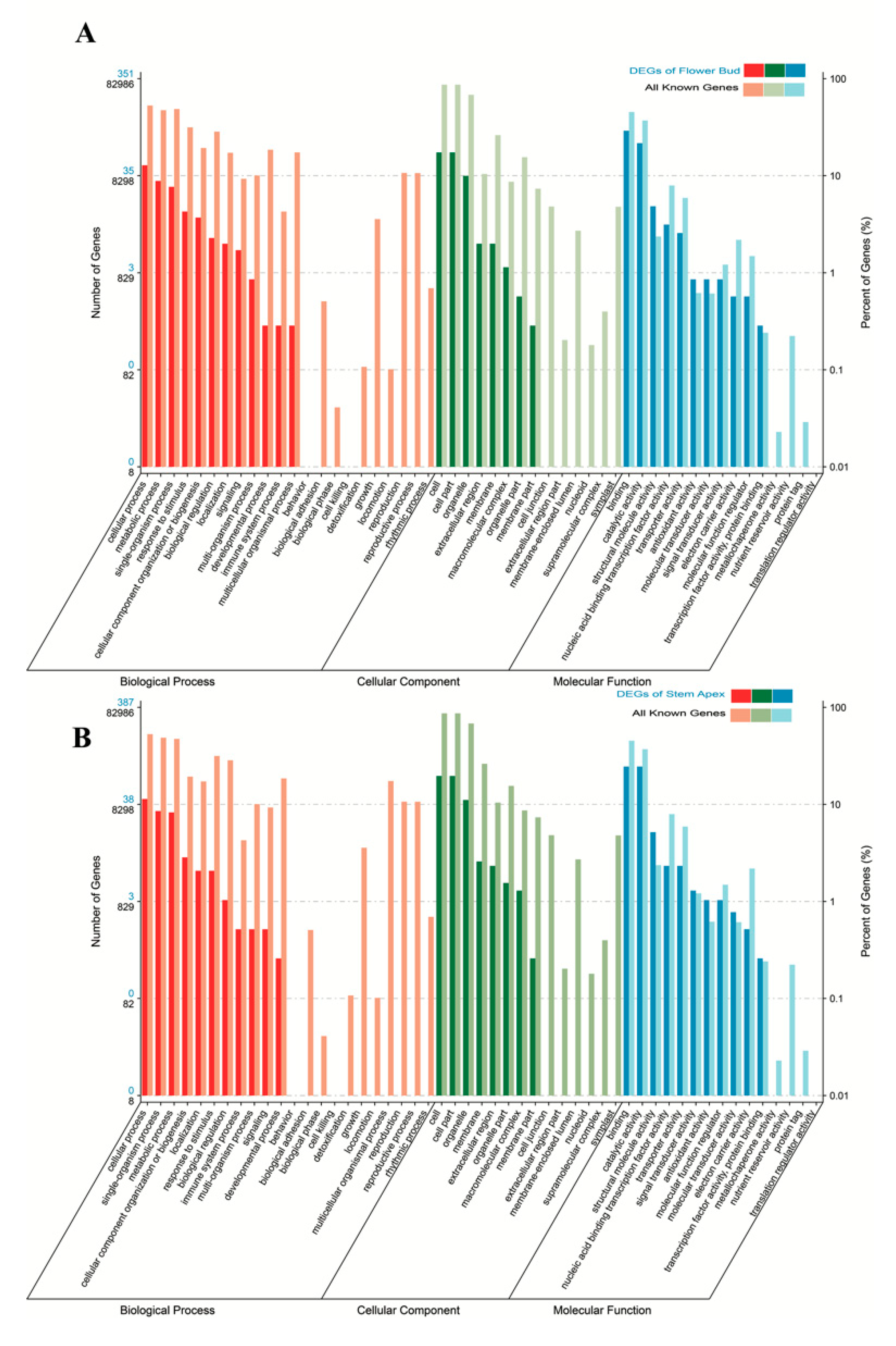
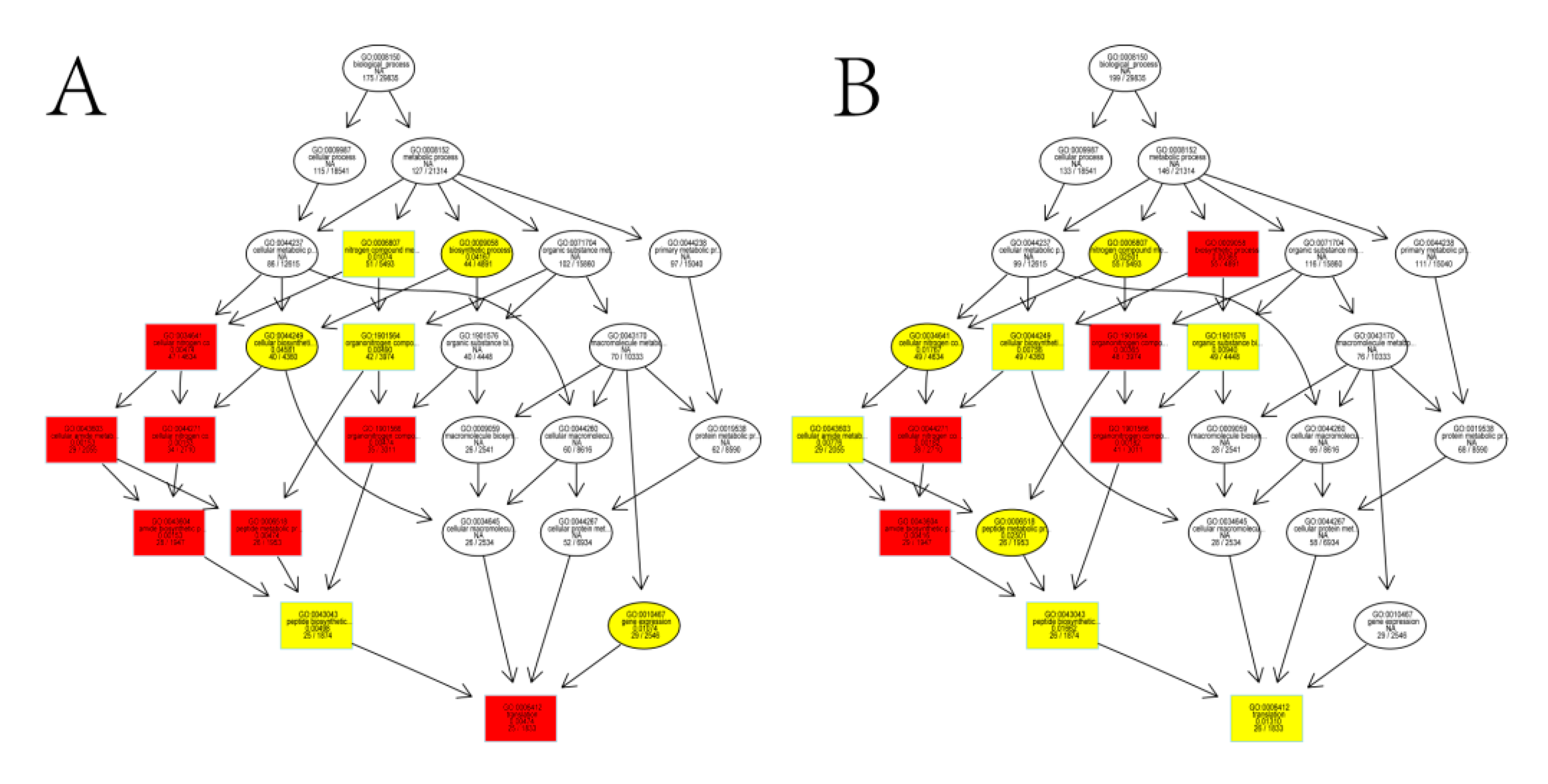
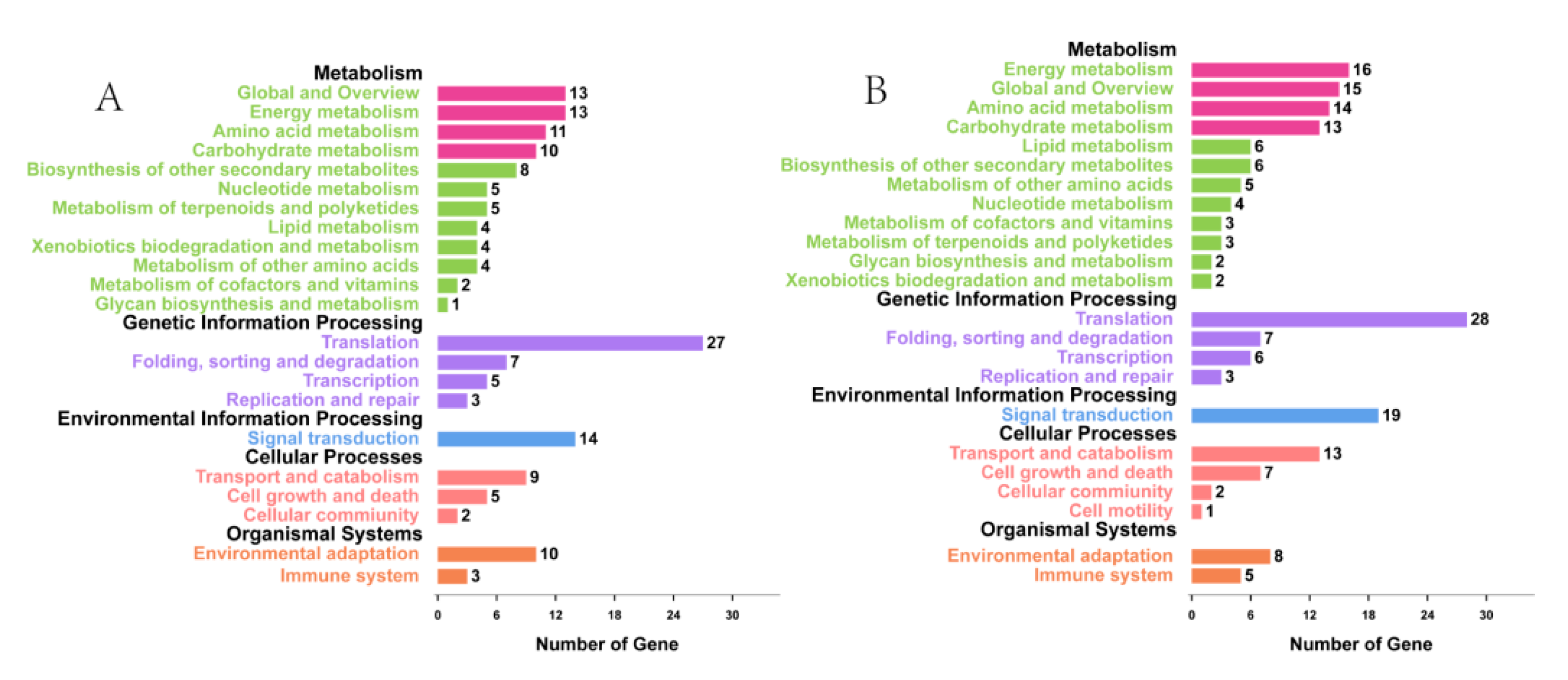
3.2. Functional Classification by Gene Ontology (GO) and Metabolic Pathway Analysis
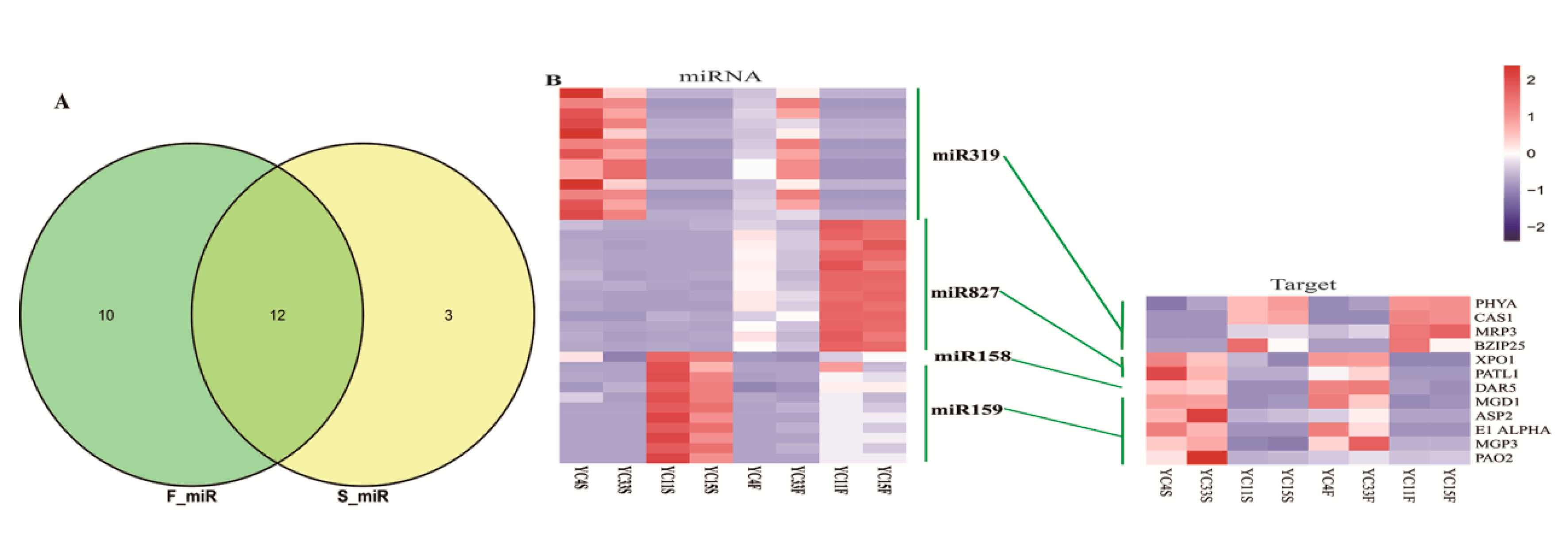
3.3. MiRNAs Differentially Expressed in High- and Low-TPC Samples
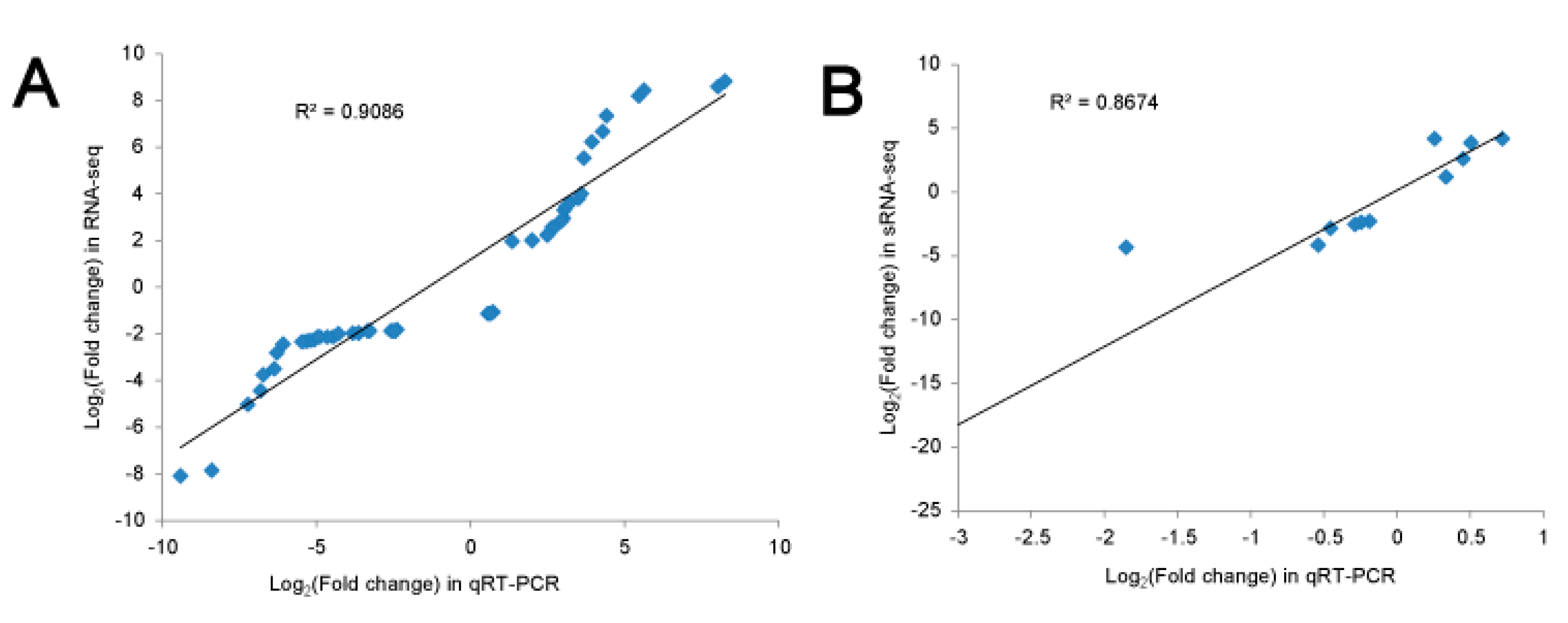
3.4. Validation of Differential Gene and miRNA Expression by qRT-PCR
3.5. MiRNA and Gene Network Analysis in High- and Low-TPC Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, S.; Xie, K.; Li, W.; Zou, T.; Ren, Y.; Wang, S.; Deng, Q.; Zheng, A.; Zhu, J.; Liu, H.; et al. Re-sequencing and genetic variation identification of a rice line with ideal plant architecture. Rice 2012, 5, 18. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Richards, R.A. Selectable traits to increase crop photosynthesis and yield of grain crops. J. Exp. Bot. 2000, 51, 447–458. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Romay, M.C.; Gore, M.A.; Flint-Garcia, S.A.; Zhang, Z.; Millard, M.J.; Gardner, C.A.C.; McMullen, M.D.; Holland, J.B.; Bradbury, P.J.; et al. The Genetic Architecture Of Maize Height. Genetics 2014, 196, 1337–1356. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zhang, T. Quantitative trait loci controlling plant architectural traits in cotton. Plant Sci. 2009, 177, 317–323. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Huyghe, C. Genetics and genetic modifications of plant architecture in grain legumes: A review. Agronomie 1998, 18, 383–411. [Google Scholar] [CrossRef]
- Busov, V.B.; Brunner, A.M.; Strauss, S.H. Genes for control of plant stature and form. New Phytol. 2008, 177, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J. Molecular basis of plant architecture. Annu. Rev. Plant Biol. 2008, 59, 253–279. [Google Scholar] [CrossRef] [PubMed]
- Monna, L.; Kitazawa, N.; Yoshino, R.; Suzuki, J.; Masuda, H.; Maehara, Y.; Tanji, M.; Sato, M.; Nasu, S.; Minobe, Y. Positional cloning of rice semidwarfing gene, sd-1: Rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 2002, 9, 11–17. [Google Scholar] [CrossRef]
- Petrásek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertová, D.; Wisniewska, J.; Tadele, Z.; Kubes, M.; Covanová, M.; et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Maekawa, M.; Shimamoto, K.; Kyozuka, J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 2001, 231, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Wang, R.; Qian, Q.; Yan, M.; Meng, X.; Fu, Z.; Yan, C.; Jiang, B.; Su, Z.; Li, J.; et al. DWARF27, an Iron-Containing Protein Required for the Biosynthesis of Strigolactones, Regulates Rice Tiller Bud Outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef] [PubMed]
- Arite, T.; Umehara, M.; Ishikawa, S.; Hanada, A.; Maekawa, M.; Yamaguchi, S.; Kyozuka, J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009, 50, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- Wang, X.; Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 2006, 313, 1118. [Google Scholar] [CrossRef] [PubMed]
- Rupp, H.-M.; Frank, M.; Werner, T.; Strnad, M.; Schmülling, T. Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J. 1999, 18, 557–563. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Wu, M.; Li, L.; Jin, C.; Zhang, Q.; Chen, C.; Song, W.; Wang, C. Small RNA Sequencing Reveals Differential miRNA Expression in the Early Development of Broccoli (Brassica oleracea var. italica) Pollen. Front. Plant Sci. 2017, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Guo, Z.; Li, L. Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev. Biol. 2013, 380, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 2006, 18, 1121. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Song, Y.; Chen, Z.; Yu, D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 2009, 136, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Kutter, C.; Schöb, H.; Stadler, M.; Meins, F.; Si-Ammour, A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 2007, 19, 2417–2429. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jung, J.-H.; Reyes, J.L.; Kim, Y.-S.; Kim, S.-Y.; Chung, K.-S.; Kim, J.A.; Lee, M.; Lee, Y.; Narry Kim, V.; et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005, 42, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Wang, L.-J.; Mao, Y.-B.; Cai, W.-J.; Xue, H.-W.; Chen, X.-Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204. [Google Scholar] [CrossRef]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-Like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef]
- Alonso-Peral, M.M.; Sun, C.; Millar, A.A. MicroRNA159 can act as a switch or tuning microRNA independently of its abundance in Arabidopsis. PLoS ONE 2012, 7, e34751. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.-W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. A MicroRNA as a Translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Wollmann, H.; Mica, E.; Todesco, M.; Long, J.A.; Weigel, D. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 2010, 137, 3633–3642. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-T.; Chen, P.-W.; Chen, L.-C.; Yang, C.-C.; Chen, S.-Y.; Huang, G.; Lin, T.C.; Ku, H.-M.; Chen, J.J.W. Suppressive effect of microRNA319 expression on rice plant height. Theor. Appl. Genet. 2017, 130, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- An, W.; Gong, W.; He, S.; Pan, Z.; Sun, J.; Du, X. MicroRNA and mRNA expression profiling analysis revealed the regulation of plant height in Gossypium hirsutum. BMC Genom. 2015, 16, 886. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Mai, Y.-X.; Zhang, Y.-C.; Luo, Q.; Yang, H.-Q. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol. Plant 2010, 3, 794–806. [Google Scholar] [CrossRef]
- Wang, L.; Sun, S.; Jin, J.; Fu, D.; Yang, X.; Weng, X.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 15504–15509. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Hao, M.; Wang, W.; Mei, D.; Wells, R.; Liu, J.; Wang, H.; Sang, S.; Tang, M.; Zhou, R.; et al. Integrative RNA- and miRNA-profile analysis reveals a likely role of br and auxin signaling in branch angle regulation of B. napus. Int. J. Mol. Sci. 2017, 18, 887. [Google Scholar] [CrossRef]
- Han, R.; Jian, C.; Lv, J.; Yan, Y.; Chi, Q.; Li, Z.; Wang, Q.; Zhang, J.; Liu, X.; Zhao, H. Identification and characterization of microRNAs in the flag leaf and developing seed of wheat (Triticum aestivum L.). BMC Genom. 2014, 15, 289. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.-W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Lu, K.; Li, T.; He, J.; Chang, W.; Zhang, R.; Liu, M.; Yu, M.; Fan, Y.; Ma, J.; Sun, W.; et al. qPrimerDB: A thermodynamics-based gene-specific qPCR primer database for 147 organisms. Nucleic Acids Res. 2018, 46, D1229–D1236. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Cai, G.; Yang, Q.; Chen, H.; Yang, Q.; Zhang, C.; Fan, C.; Zhou, Y. Genetic dissection of plant architecture and yield-related traits in Brassica napus. Sci. Rep. 2016, 6, 21625. [Google Scholar] [CrossRef]
- Zhang, J.; Song, Q.; Cregan, P.B.; Nelson, R.L.; Wang, X.; Wu, J.; Jiang, G.-L. Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genom. 2015, 16, 217. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, K.; Nie, H.; Zeng, Q.; Wu, B.; Qian, J.; Fang, Z. Rice nitrate transporter OsNPF7.2 positively regulates tiller number and grain yield. Rice 2018, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Anwar, S.; Kuai, J.; Ullah, S.; Fahad, S.; Zhou, G. Optimization of nitrogen rate and planting density for improving yield, nitrogen use efficiency, and lodging resistance in oilseed rape. Front. Plant Sci. 2017, 8, 532. [Google Scholar] [CrossRef] [PubMed]
- Kruger, N.J.; von Schaewen, A. The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant Biol. 2003, 6, 236–246. [Google Scholar] [CrossRef]
- Sahrawy, M.; Ávila, C.; Chueca, A.; Cánovas, F.M.; López-Gorgé, J. Increased sucrose level and altered nitrogen metabolism in Arabidopsis thaliana transgenic plants expressing antisense chloroplastic fructose-1,6-bisphosphatase. J. Exp. Bot. 2004, 55, 2495–2503. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Fernie, A.R.; Stitt, M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 2010, 3, 973–996. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Xin, Z.; Zheng, Z.-L. The OSU1/QUA2/TSD2-encoded putative methyltransferase is a critical modulator of carbon and nitrogen nutrient balance response in Arabidopsis. PLoS ONE 2008, 3, e1387. [Google Scholar] [CrossRef]
- Yoneyama, K.; Xie, X.; Kisugi, T.; Nomura, T.; Yoneyama, K. Nitrogen and phosphorus fertilization negatively affects strigolactone production and exudation in sorghum. Planta 2013, 238, 885–894. [Google Scholar] [CrossRef]
- Razaq, M.; Zhang, P.; Shen, H.-L. Influence of nitrogen and phosphorous on the growth and root morphology of Acer mono. PLoS ONE 2017, 12, e0171321. [Google Scholar] [CrossRef]
- Enquist, B.J.; Niklas, K.J. Global allocation rules for patterns of biomass partitioning in seed plants. Science 2002, 295, 1517–1520. [Google Scholar] [CrossRef]
- Reinhardt, D.; Kuhlemeier, C. Plant architecture. EMBO Rep. 2002, 3, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G. local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 2002, 53, 203–224. [Google Scholar] [CrossRef] [PubMed]
- De Smet, I.; Signora, L.; Beeckman, T.; Inzé, D.; Foyer, C.H.; Zhang, H. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 2003, 33, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Signora, L.; De Smet, I.; Foyer, C.H.; Zhang, H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 2001, 28, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Vlieghe, K.; Vuylsteke, M.; Florquin, K.; Rombauts, S.; Maes, S.; Ormenese, S.; Van Hummelen, P.; Van de Peer, Y.; Inzé, D.; De Veylder, L. Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J. Cell Sci. 2003, 116, 4249–4259. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Wang, Y.; Li, X.; Wu, S.; Wang, Y.; Luo, J.; Mattson, N.; Xu, Y. Interactions between ethylene, gibberellin and abscisic acid in regulating submergence induced petiole elongation in Nelumbo nucifera. Aquat. Bot. 2017, 137, 9–15. [Google Scholar] [CrossRef]
- Pellny, T.K.; Van Aken, O.; Dutilleul, C.; Wolff, T.; Groten, K.; Bor, M.; De Paepe, R.; Reyss, A.; Van Breusegem, F.; Noctor, G.; et al. Mitochondrial respiratory pathways modulate nitrate sensing and nitrogen-dependent regulation of plant architecture in Nicotiana sylvestris. Plant J. 2008, 54, 976–992. [Google Scholar] [CrossRef]
- Hu, M.; Pu, H.; Gao, J.; Long, W.; Chen, F.; Zhang, W.; Zhou, X.; Peng, Q.; Chen, S.; Zhang, J. Comparative analysis of miRNAs of two rapeseed genotypes in response to acetohydroxyacid synthase-inhibiting herbicides by high-throughput sequencing. PLoS ONE 2017, 12, e0184917. [Google Scholar] [CrossRef]
- Guoxing, H.A.U. Wuhan Chen. Study of Overexpressing miRNA 167a to Regulate the Architecture in Oryza sativa. Mol. Plant Breed. 2011, 9, 390–396. [Google Scholar]
- Zhao, Y.-T.; Wang, M.; Fu, S.-X.; Yang, W.-C.; Qi, C.-K.; Wang, X.-J. Small RNA Profiling in Two Brassica napus Cultivars Identifies MicroRNAs with Oil Production- and Development-Correlated Expression and New Small RNA Classes. Plant Physiol. 2012, 158, 813–823. [Google Scholar] [CrossRef]
- Wang, J.; Jian, H.; Wang, T.; Wei, L.; Li, J.; Li, C.; Liu, L. Identification of microRNAs actively involved in fatty acid biosynthesis in developing Brassica napus seeds using high-throughput sequencing. Front. Plant Sci. 2016, 7, 1570. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Jiang, J.; Hu, Z.; Lyu, T.; Yang, Y.; Jiang, J.; Cao, J. Over-expression of miR158 causes pollen abortion in Brassica campestris ssp. chinensis. Plant Mol. Biol. 2017, 93, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wen, H.; Teotia, S.; Du, Y.; Zhang, J.; Li, J.; Sun, H.; Tang, G.; Peng, T.; Zhao, Q. Suppression of microRNA159 impacts multiple agronomic traits in rice (Oryza sativa L.). BMC Plant Biol. 2017, 17, 215. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, Y.; Liu, Z.; Kong, D.; Duan, M.; Luo, L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 2010, 61, 4157–4168. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Shu, J.; Jin, W. Identification and validation of miRNAs associated with the resistance of maize (Zea mays L.) to Exserohilum turcicum. PLoS ONE 2014, 9, e87251. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.N.; Rothstein, S.J.; Spangenberg, G.; Kant, S. Role of microRNAs involved in plant response to nitrogen and phosphorous limiting conditions. Front. Plant Sci. 2015, 6, 629. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Gong, S.; Wang, Y.; Wang, F.; Bao, H.; Sun, J.; Cai, C.; Yi, K.; Chen, Z.; Zhu, C. MicroRNA166 modulates cadmium tolerance and accumulation in rice. Plant Physiol. 2018, 177, 1691–1703. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, M.; Mafra, A.C.P.; Dias, S.M.G.; Vasilescu, C.; Calin, G.A. Using microRNA Networks to Understand Cancer. Int. J. Mol. Sci. 2018, 19, 1871. [Google Scholar] [CrossRef]
- Roy, A.; Sahoo, D.; Tripathy, B.C. Involvement of phytochrome A in suppression of photomorphogenesis in rice seedling grown in red light. Plant Cell Environ. 2013, 36, 2120–2134. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X.; Huang, H.; Wang, J.; Li, Z.; Wu, L.; Zhang, G.; Ma, Z. Efficiency of phosphorus utilization in phyA-expressing cotton lines. Indian J. Biochem. Biophys. 2012, 49, 250–256. [Google Scholar] [CrossRef]
- Donohue, K.; Heschel, M.S.; Butler, C.M.; Barua, D.; Sharrock, R.A.; Whitelam, G.C.; Chiang, G.C. Diversification of phytochrome contributions to germination as a function of seed-maturation environment. New Phytol. 2008, 177, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Shah, K.; Sharma, N.; Kaur, R.; Singh, J.; Vinson, C.; Rishi, V. A-ZIP53, a dominant negative reveals the molecular mechanism of heterodimerization between bZIP53, bZIP10 and bZIP25 involved in Arabidopsis seed maturation. Sci. Rep. 2017, 7, 14343. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Namasivayam, P.; Abdullah, M.P. The PAL2 promoter activities in relation to structural development and adaptation in Arabidopsis thaliana. Planta 2012, 235, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Simon, N.M.L.; Sawkins, E.; Dodd, A.N. Involvement of the SnRK1 subunit KIN10 in sucrose-induced hypocotyl elongation. Plant Signal. Behav. 2018, e1457913. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Lehneck, R.; Pöggeler, S.; Supuran, C.T. Sulfonamide inhibition studies of two β-carbonic anhydrases from the ascomycete fungus Sordaria macrospora, CAS1 and CAS2. J. Enzym. Inhib. Med. Chem. 2018, 33, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Truernit, E.; Haseloff, J. A Role for KNAT class II genes in root development. Plant Signal. Behav. 2007, 2, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Blanvillain, R.; Boavida, L.C.; McCormick, S.; Ow, D.W. EXPORTIN1 genes are essential for development and function of the gametophytes in Arabidopsis thaliana. Genetics 2008, 180, 1493–1500. [Google Scholar] [CrossRef]
- Lange, P.R.; Geserick, C.; Tischendorf, G.; Zrenner, R. Functions of chloroplastic adenylate kinases in Arabidopsis. Plant Physiol. 2008, 146, 492–504. [Google Scholar] [CrossRef]
- Song, H.X.; Ping, A.M.; Sun, M.X.; Qi, X.H.; Gao, M.Y.; Xu, X.Y.; Zhu, Z.J.; Li, M.L.; Hou, L.P. Identification of genes related to floral organ development in pak choi by expression profiling. Genet. Mol. Res. 2017, 16, 1–14. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, X.; Zhang, J. Alteration of osa-miR156e expression affects rice plant architecture and strigolactones (SLs) pathway. Plant Cell Rep. 2015, 34, 767–781. [Google Scholar] [CrossRef]
- Shen, Y.; Sun, S.; Hua, S.; Shen, E.; Ye, C.-Y.; Cai, D.; Timko, M.P.; Zhu, Q.-H.; Fan, L. Analysis of transcriptional and epigenetic changes in hybrid vigor of allopolyploid Brassica napus uncovers key roles for small RNAs. Plant J. 2017, 91, 874–893. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Huo, Q.; Yang, H.; Jian, H.; Qu, C.; Lu, K.; Li, J. Joint RNA-Seq and miRNA Profiling Analyses to Reveal Molecular Mechanisms in Regulating Thickness of Pod Canopy in Brassica napus. Genes 2019, 10, 591. https://doi.org/10.3390/genes10080591
Chen Z, Huo Q, Yang H, Jian H, Qu C, Lu K, Li J. Joint RNA-Seq and miRNA Profiling Analyses to Reveal Molecular Mechanisms in Regulating Thickness of Pod Canopy in Brassica napus. Genes. 2019; 10(8):591. https://doi.org/10.3390/genes10080591
Chicago/Turabian StyleChen, Zhiyou, Qiang Huo, Hong Yang, Hongju Jian, Cunmin Qu, Kun Lu, and Jiana Li. 2019. "Joint RNA-Seq and miRNA Profiling Analyses to Reveal Molecular Mechanisms in Regulating Thickness of Pod Canopy in Brassica napus" Genes 10, no. 8: 591. https://doi.org/10.3390/genes10080591
APA StyleChen, Z., Huo, Q., Yang, H., Jian, H., Qu, C., Lu, K., & Li, J. (2019). Joint RNA-Seq and miRNA Profiling Analyses to Reveal Molecular Mechanisms in Regulating Thickness of Pod Canopy in Brassica napus. Genes, 10(8), 591. https://doi.org/10.3390/genes10080591






