Trans-Species Polymorphism in Mitochondrial Genome of Camarodont Sea Urchins
Abstract
1. Introduction
2. Materials and Methods
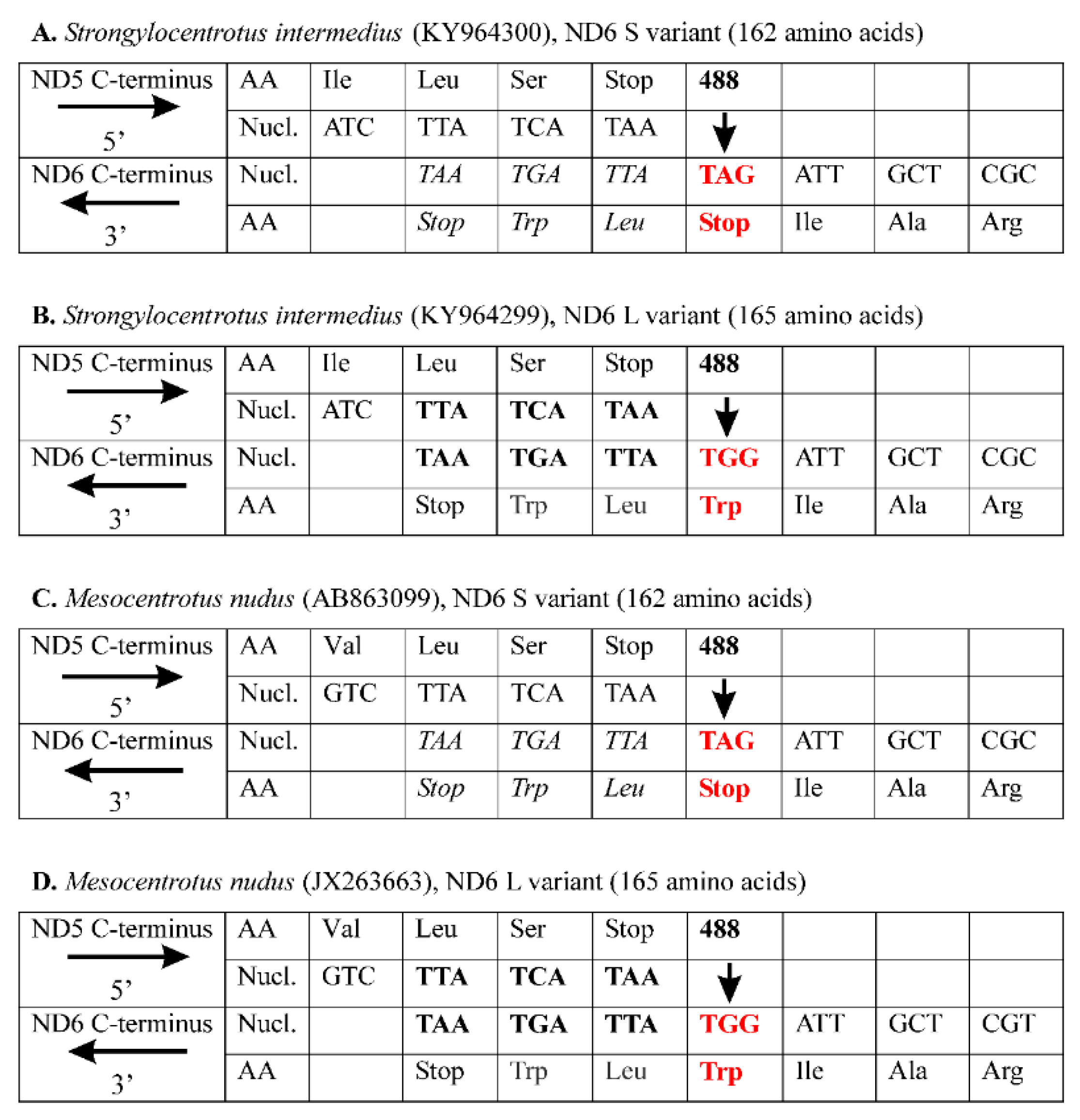
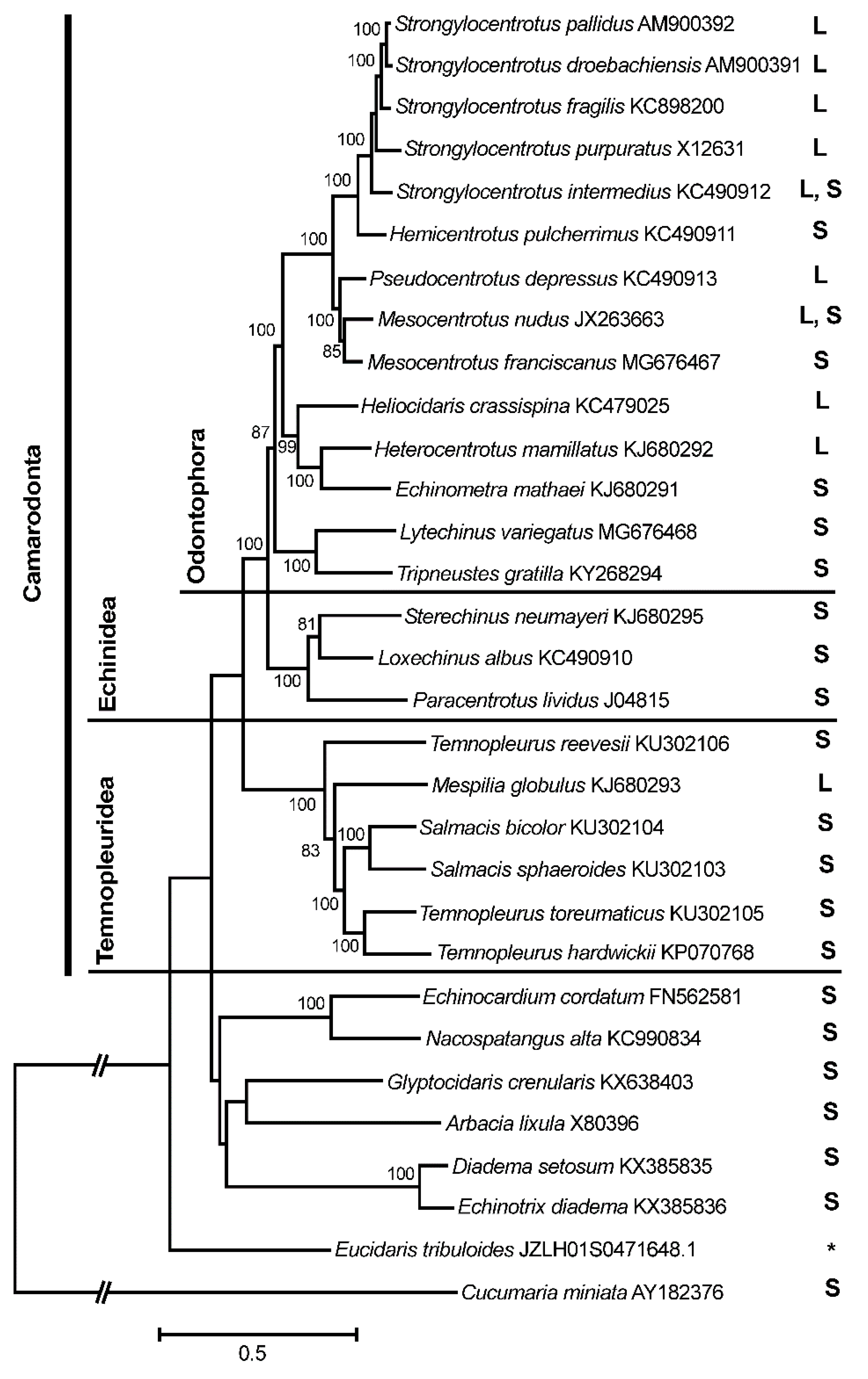
3. Results and Discussion
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
Disclosure Statement
References
- Jensen, M. The Strongylocentrotidae (Echinoidea), a morphologic and systematic study. Sarsia 1974, 57, 113–148. [Google Scholar] [CrossRef]
- Bazhin, A.G.; Stepanov, V.G. Sea Urchins Fam. Strongylocentrotidae of Seas of Russia; KamchatNIRO: Petropavlovsk-Kamchatsky, Russia, 2012; p. 196. [Google Scholar]
- Kafanov, A.I.; Pavlyuchkov, V.A. Ecology of the commercial sea urchins (genus Strongylocentrotus) of continental Japan Sea. Proc. TINRO Center (Vladivostok) 2001, 128, 349–373. [Google Scholar]
- Tatarenko, D.E.; Poltaraus, A.B. Assignment of the sea urchins Pseudocentrotus depressus to the family Strongylocentrotidae and description of a new genus Mesocentrotus belonging to this group based on the data of DNA-DNA hybridization and comparative morphology. Zool. Zhurn. 1993, 72, 61–72. [Google Scholar]
- Biermann, C.H.; Kessing, B.D.; Palumbi, S.R. Phylogeny and development of marine model species: Strongylocentrotid sea urchins. Evol. Dev. 2003, 5, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H. Molecular phylogenies and divergence times of sea urchin species of Strongylocentrotidae, Echinoida. Mol. Biol. Evol. 2003, 20, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Kober, K.M.; Bernardi, G. Phylogenomics of strongylocentrotid sea urchins. BMC Evol. Biol. 2013, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Láruson, Á.J. Rates and relations of mitochondrial genome evolution across the Echinoidea, with special focus on the superfamily Odontophora. Ecol. Evol. 2017, 7, 4543–4551. [Google Scholar] [CrossRef]
- Motavkin, P.A.; Evdokimov, V.V. A hybrid kind of sea urchins Strongylocentrotus. DAN SSSR 1978, 241, 1451–1453. [Google Scholar]
- Wang, L.; Han, J.; Xu, W.; Wang, X.; Dong, Y.; Zhou, Z. The species hybridization between sea urchin Strongylocentrotus nudus and Strongylocentrotus intermedius and the seeding production. Fish. Sci. 2003, 22, 9–11. [Google Scholar]
- Lessios, H. Reproductive isolation between species of sea urchins. Bull. Mar. Sci. 2007, 81, 191–208. [Google Scholar]
- Pudovkin, A.I.; Serov, O.L.; Glazko, V.I. Allozyme variation in four echinoids from Peter the Great bay of the Sea of Japan. Genetika 1984, 20, 1139–1147. [Google Scholar]
- Manchenko, G.P. Genetic variability of proteins in sea urchin Strongylocentrotus intermedius. Genetika 1985, 21, 763–769. [Google Scholar]
- Manchenko, G.P.; Yakovlev, S.N. Genetic divergence between three sea urchin species of the genus Strongylocentrotus from the Sea of Japan. Biochem. Syst. Ecol. 2001, 29, 31–44. [Google Scholar] [CrossRef]
- Zaslavskaya, N.I.; Vashchenko, M.A.; Zhadan, P.M. The genetic structure of populations of the sea urchin Strongylocentrotus intermedius from the northwestern Sea of Japan in connection with a shift in spawning time. Russ. J. Mar. Biol. 2012, 38, 325–338. [Google Scholar]
- Balakirev, E.S.; Pavlyuchkov, V.A.; Ayala, F.J. Complete mitochondrial genome of the phenotypically-diverse sea urchin Strongylocentrotus intermedius (Strongylocentrotidae, Echinoidea). Mitochond. DNA Part B Res. 2017, 2, 613–614. [Google Scholar] [CrossRef]
- Nam, W.; Kanno, M.; Watanabe, M.; Ikeda, M.; Kijima, A. Genetic population structure of Strongylocentrotus nudus inferred from microsatellite DNA and mtDNA analyses. Nippon Suisan Gakkaishi 2014, 80, 726–740. [Google Scholar] [CrossRef][Green Version]
- Balakirev, E.S.; Pavlyuchkov, V.A.; Ayala, F.J. DNA variation and symbiotic associations in phenotypically-diverse sea urchin Strongylocentrotus intermedius. Proc. Natl. Acad. Sci. USA 2008, 105, 16218–16223. [Google Scholar] [CrossRef]
- Balakirev, E.S.; Pavlyuchkov, V.A.; Anisimova, M.; Ayala, F.J. DNA polymorphism and selection at the bindin locus in three Strongylocentrotus sp. (Echinoidea). BMC Genet. 2016, 17, 66. [Google Scholar] [CrossRef]
- The National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 19 June 2019).
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Filatov, D.A. PROSEQ: A software for preparation and evolutionary analysis of DNA sequence data sets. Mol. Ecol. Notes 2002, 2, 621–624. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G.E. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 1–4. [Google Scholar] [CrossRef]
- Gelman, A.; Rubin, D.B. Inference from iterative simulation using multiple sequences. Stat. Sci. 1992, 7, 457–511. [Google Scholar] [CrossRef]
- Russo, C.A.M.; Takezaki, N.; Nei, M. Efficiencies of different genes and different tree-building methods in recovering a known vertebrate phylogeny. Mol. Biol. Evol. 1996, 13, 525–536. [Google Scholar] [CrossRef]
- Douady, C.J.; Delsuc, F.; Boucher, Y.; Doolittle, W.F.; Douzery, E.J. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Mol. Biol. Evol. 2003, 20, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.R.; Lin, H.H.; Han, L.Y.; Jiang, L.; Chen, X.; Chen, Y.Z. PROFEAT: A web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence. Nucleic Acids Res. 2006, 34 (Suppl. S2), W32–W37. [Google Scholar] [CrossRef]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- The Sequence Read Archive (SRA). The NCBI Database. Available online: https://www.ncbi.nlm.nih.gov/sra/ (accessed on 19 June 2019).
- Nam, W.S.; Kanno, M.; Kijima, A. Strongylocentrotus nudus NADH:ubiquinone oxidoreductase 6 (ND6) gene, partial cds; mitochondrial gene for mitochondrial product. Unpublished. 2019. [Google Scholar]
- Kroh, A.; Smith, A.B. The phylogeny and classification of post-Palaeozoic echinoids. J. Syst. Palaeontol. 2010, 8, 147–212. [Google Scholar] [CrossRef]
- Smith, A.B.; Kroh, A. Phylogeny of sea urchins. In Sea Urchins: Biology and Ecology, 3rd ed.; Lawrence, J.M., Ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 1–14. [Google Scholar]
- Hopkins, M.J.; Smith, A.B. Dynamic evolutionary change in post-Paleozoic echinoids and the importance of scale when interpreting changes in rates of evolution. Proc. Natl. Acad. Sci. USA 2015, 112, 3758–3763. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.R.; Petsios, E.; Davidson, E.H.; Erkenbrack, E.M.; Gao, F.; Bottjer, D.J. Reorganization of sea urchin gene regulatory networks at least 268 million years ago as revealed by oldest fossil cidaroid echinoid. Sci. Rep. 2015, 5, 15541. [Google Scholar] [CrossRef]
- Thompson, J.R.; Erkenbrack, E.M.; Hinman, V.F.; McCauley, B.S.; Petsios, E.; Bottjer, D.J. Paleogenomics of echinoids reveals an ancient origin for the double-negative specification of micromeres in sea urchins. Proc. Natl. Acad. Sci. USA 2017, 14, 5870–5877. [Google Scholar] [CrossRef]
- Bronstein, O.; Kroh, A. The first mitochondrial genome of the model echinoid Lytechinus variegatus and insights into Odontophoran phylogenetics. Genomics 2019, 111, 710–718. [Google Scholar] [CrossRef]
- Koch, N.M.; Coppard, S.E.; Lessios, H.A.; Briggs, D.E.; Mooi, R.; Rouse, G.W. A phylogenomic resolution of the sea urchin tree of life. BMC Evol. Biol. 2018, 18, 189. [Google Scholar] [CrossRef]
- Roberti, M.; Polosa, P.L.; Musicco, C.; Milella, F.; Qureshi, S.A.; Gadaleta, M.N.; Jacobs, H.T.; Cantatore, P. In vivo mitochondrial DNA-protein interactions in sea urchin eggs and embryos. Curr. Genet. 1999, 34, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Roberti, M.; Polosa, P.L.; Bruni, F.; Deceglie, S.; Gadaleta, M.N.; Cantatore, P. MTERF factors: A multifunction protein family. Biomol. Concepts 2010, 1, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Hinman, V.F.; Nguyen, A.T.; Cameron, R.A.; Davidson, E.H. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13356–13361. [Google Scholar] [CrossRef] [PubMed]
- Gildor, T.; Ben-Tabou de-Leon, S. Comparative study of regulatory circuits in two sea urchin species reveals tight control of timing and high conservation of expression dynamics. PLoS Genet. 2015, 11, e1005435. [Google Scholar] [CrossRef] [PubMed]
- Klein, J. Origin of major histocompatibility complex polymorphism: The trans-species hypothesis. Hum. Immunol. 1987, 19, 155–162. [Google Scholar] [CrossRef]
- Klein, J.; Sato, A.; Nagl, S.; O’hUigín, C. Molecular trans-species polymorphism. Annu. Rev. Ecol. Syst. 1998, 29, 21. [Google Scholar] [CrossRef]
- Klein, J.; Sato, A.; Nikolaidis, N. MHC, TSP, and the origin of species: From immunogenetics to evolutionary genetics. Annu. Rev. Genet. 2007, 41, 281–304. [Google Scholar] [CrossRef]
- Takahata, N. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc. Natl. Acad. Sci. USA 1990, 87, 2419–2423. [Google Scholar] [CrossRef]
- Azevedo, L.; Serrano, C.; Amorim, A.; Cooper, D.N. Trans-species polymorphism in humans and the great apes is generally maintained by balancing selection that modulates the host immune response. Hum. Genomics 2015, 9, 21. [Google Scholar] [CrossRef]
- Těšický, M.; Vinkler, M. Trans-species polymorphism in immune genes: general pattern or MHC-restricted phenomenon? J. Immunol. Res. 2015, 838035. [Google Scholar] [CrossRef]
- Wu, Q.; Han, T.S.; Chen, X.; Chen, J.F.; Zou, Y.P.; Li, Z.W.; Xu, Y.C.; Guo, Y.L. Long-term balancing selection contributes to adaptation in Arabidopsis and its relatives. Genome Biol. 2017, 18, 217. [Google Scholar] [CrossRef]
- Efremov, R.G.; Baradaran, R.; Sazanov, L.A. The architecture of respiratory complex I. Nature 2010, 465, 441–445. [Google Scholar] [CrossRef]
- Baradaran, R.; Berrisford, J.M.; Minhas, G.S.; Sazanov, L.A. Crystal structure of the entire respiratory complex I. Nature 2013, 494, 443–448. [Google Scholar] [CrossRef]
- Sazanov, L.A. A giant molecular proton pump: Structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 2015, 16, 375–388. [Google Scholar] [CrossRef]
- Fiedorczuk, K.; Letts, J.A.; Degliesposti, G.; Kaszuba, K.; Skehel, M.; Sazanov, L.A. Atomic structure of the entire mammalian mitochondrial complex I. Nature 2016, 538, 406–410. [Google Scholar] [CrossRef]
- Bai, Y.; Attardi, G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998, 17, 4848–4858. [Google Scholar] [CrossRef]
- Cardol, P.; Matagne, R.F.; Remacle, C. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas. Implication for the structural organization of the enzyme. J. Mol. Biol. 2002, 319, 1211–1221. [Google Scholar] [CrossRef]
- Kirby, D.M.; Kahler, S.G.; Freckmann, M.L.; Reddihough, D.; Thorburn, D.R. Leigh disease caused by the mitochondrial DNA G14459A mutation in unrelated families. Ann. Neurol. 2000, 48, 102–104. [Google Scholar] [CrossRef]
- Chinnery, P.F.; Brown, D.T.; Andrews, R.M.; Singh-Kler, R.; Riordan-Eva, P.; Lindley, J.; Applegarth, D.A.; Turnbull, D.M.; Howell, N. The mitochondrial ND6 gene is a hot spot for mutations that cause Leber’s hereditary optic neuropathy. Brain 2001, 124, 209–218. [Google Scholar] [CrossRef]
- Ravn, K.; Wibrand, F.; Hansen, F.J.; Horn, N.; Rosenberg, T.; Schwartz, M. An mtDNA mutation, 14453G–>A, in the NADH dehydrogenase subunit 6 associated with severe MELAS syndrome. Eur. J. Hum. Genet. 2001, 9, 805–809. [Google Scholar] [CrossRef]
- Ugalde, C.; Triepels, R.H.; Coenen, M.J.; Van Den Heuvel, L.P.; Smeets, R.; Uusimaa, J.; Briones, P.; Campistol, J.; Majamaa, K.; Nijtmans, L.G.; et al. Impaired complex I assembly in a Leigh syndrome patient with a novel missense mutation in the ND6 gene. Ann. Neurol. 2003, 54, 665–669. [Google Scholar] [CrossRef]
- DeHaan, C.; Habibi-Nazhad, B.; Yan, E.; Salloum, N.; Parliament, M.; Allalunis-Turner, J. Mutation in mitochondrial complex I ND6 subunit is associated with defective response to hypoxia in human glioma cells. BMC Mol. Cancer 2004, 3, 19. [Google Scholar]
- Uehara, N.; Mori, M.; Tokuzawa, Y.; Mizuno, Y.; Tamaru, S.; Kohda, M.; Moriyama, Y.; Nakachi, Y.; Matoba, N.; Yamazaki, T.; et al. New MT-ND6 and NDUFA1 mutations in mitochondrial respiratory chain disorders. Ann. Clin. Transl. Neurol. 2014, 5, 361–369. [Google Scholar] [CrossRef]
- Fiedorczuk, K.; Sazanov, L.A. Mammalian mitochondrial complex I structure and disease-causing mutations. Trends Cell Biol. 2018, 28, 835–867. [Google Scholar] [CrossRef]
- Da Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Ramos, M.J.; Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008, 9, 119. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, S.; Xu, J.; Guo, Y.; Yang, G. Adaptive Evolution of Mitochondrial Energy Metabolism Genes Associated with Increased Energy Demand in Flying Insects. PLoS ONE 2014, 9, e99120. [Google Scholar] [CrossRef]
- Almeida, D.; Maldonado, E.; Vasconcelos, V.; Antunes, A. Adaptation of the mitochondrial genome in Cephalopods: Enhancing proton translocation channels and the subunit interactions. PLoS ONE 2015, 10, e0135405. [Google Scholar] [CrossRef]
- Wang, Z.; Shi, X.; Sun, L.; Bai, Y.; Zhang, D.; Tang, B. Evolution of mitochondrial energy metabolism genes associated with hydrothermal vent adaption of Alvinocaridid shrimps. Genes Genom. 2017, 39, 1367–1376. [Google Scholar] [CrossRef]
- Hao, Y.J.; Zou, Y.L.; Ding, Y.R.; Xu, W.Y.; Yan, Z.T.; Li, X.D.; Fu, W.B.; Li, T.J.; Chen, B. Complete mitochondrial genomes of Anopheles stephensi and An. dirus and comparative evolutionary mitochondriomics of 50 mosquitoes. Sci. Rep. 2017, 7, 7666. [Google Scholar] [CrossRef]
- Li, X.D.; Jiang, G.F.; Yan, L.Y.; Li, R.; Mu, Y.; Deng, W.A. Positive Selection Drove the Adaptation of Mitochondrial Genes to the Demands of Flight and High-Altitude Environments in Grasshoppers. Front. Genet. 2018, 9, 605. [Google Scholar] [CrossRef]
- Bbole, I.; Zhao, J.L.; Tang, S.J.; Katongo, C. Mitochondrial genome annotation and phylogenetic placement of Oreochromis andersonii and O. macrochir among the cichlids of southern Africa. PLoS ONE 2018, 13, e0203095. [Google Scholar] [CrossRef]
- Zhuang, X.; Cheng, C.H. ND6 gene “lost” and found: Evolution of mitochondrial gene rearrangement in Antarctic notothenioids. Mol. Biol. Evol. 2010, 27, 1391–1403. [Google Scholar] [CrossRef]
- Jin, Y.; Wo, Y.; Tong, H.; Song, S.; Zhang, L.; Brown, R.P. Evolutionary analysis of mitochondrially encoded proteins of toad-headed lizards, Phrynocephalus, along an altitudinal gradient. BMC Genom. 2018, 19, 185. [Google Scholar] [CrossRef]
- Lamb, A.M.; Gan, H.M.; Greening, C.; Joseph, L.; Lee, Y.P.; Morán-Ordóñez, A.; Sunnucks, P.; Pavlova, A. Climate-driven mitochondrial selection: A test in Australian songbirds. Mol. Ecol. 2018, 27, 898–918. [Google Scholar] [CrossRef]
- Gu, P.; Liu, W.; Yao, Y.F.; Ni, Q.Y.; Zhang, M.W.; Li, D.Y.; Xu, H.L. Evidence of adaptive evolution of alpine pheasants to high-altitude environment from mitogenomic perspective. DNA Mapp. Seq. Anal. 2016, 27, 455–462. [Google Scholar] [CrossRef]
- Finch, T.M.; Zhao, N.; Korkin, D.; Frederick, K.H.; Eggert, L.S. Evidence of positive selection in mitochondrial complexes I and V of the african elephant. PLoS ONE 2014, 9, e92587. [Google Scholar] [CrossRef]
- Xu, S.; Luosang, J.; Hua, S.; He, J.; Ciren, A.; Wang, W.; Tong, X.; Liang, Y.; Wang, J.; Zheng, X. High altitude adaptation and phylogenetic analysis of Tibetan horse based on the mitochondrial genome. J. Genet. Genom. 2007, 34, 720–729. [Google Scholar] [CrossRef]
- Ning, T.; Xiao, H.; Li, J.; Hua, S.; Zhang, Y.P. Adaptive evolution of the mitochondrial ND6 gene in the domestic horse. Genet. Mol. Res. 2010, 9, 144–150. [Google Scholar] [CrossRef]
- Ning, T.; Li, J.; Lin, K.; Xiao, H.; Wylie, S.; Hua, S.; Li, H.; Zhang, Y.-P. Complex Evolutionary Patterns Revealed by Mitochondrial Genomes of the Domestic Horse. Curr. Mol. Med. 2014, 14, 1286–1298. [Google Scholar] [CrossRef]
- Yu, L.; Wang, X.; Ting, N.; Zhang, Y. Mitogenomic analysis of Chinese snub-nosed monkeys: Evidence of positive selection in NADH dehydrogenase genes in high-altitude adaptation. Mitochondrion 2011, 11, 497–503. [Google Scholar] [CrossRef]
- Elson, J.L.; Turnbull, D.M.; Howell, N. Comparative genomics and the evolution of human mitochondrial DNA: Assessing the effects of selection. Am. J. Hum. Genet. 2004, 74, 229–238. [Google Scholar] [CrossRef]
- Grantham, R. Amino acid difference formula to help explain protein evolution. Science 1974, 185, 862–864. [Google Scholar] [CrossRef]
- Schneider, G.; Wrede, P. The rational design of amino acid sequences by artificial neural networks and simulated molecular evolution: De novo design of an idealized leader peptidase cleavage site. Biophys. J. 1994, 66, 335–344. [Google Scholar] [CrossRef]
- Gott, J.M.; Emeson, R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000, 34, 499–531. [Google Scholar] [CrossRef]
- Knoop, V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell. Mol. Life Sci. 2011, 68, 567–586. [Google Scholar] [CrossRef]
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Levanon, E.Y.; et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014, 24, 365–376. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Pett, W. Animal Mitochondrial DNA as We Do Not Know It: Mt-Genome Organization and Evolution in Nonbilaterian Lineages. Genome Biol. Evol. 2016, 8, 2896–2913. [Google Scholar] [CrossRef]
- Liscovitch-Brauer, N.; Alon, S.; Porath, H.T.; Elstein, B.; Unger, R.; Ziv, T.; Admon, A.; Levanon, E.Y.; Rosenthal, J.J.C. Eisenberg, E.; et al. Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods. Cell 2017, 169, 191–202. [Google Scholar] [CrossRef]
- Gommans, W.M.; Mullen, S.P.; Maas, S. RNA editing: A driving force for adaptive evolution? Bioessays 2009, 31, 1137–1145. [Google Scholar] [CrossRef]
- Duan, Y.; Dou, S.; Luo, S.; Zhang, H.; Lu, J. Adaptation of A-to-I RNA editing in Drosophila. PLoS Genet. 2017, 13, e1006648. [Google Scholar] [CrossRef]
- Buckley, K.M.; Terwilliger, D.P.; Smith, L.C. Sequence variations in 185/333 Messages from the purple Sea Urchin suggest posttranscriptional modifications to increase immune diversity. J. Immunol. 2008, 181, 8585–8594. [Google Scholar] [CrossRef]
- Smith, L.C. Innate immune complexity in the purple sea urchin: Diversity of the sp185/333 system. Front. Immunol. 2012, 3, 70. [Google Scholar] [CrossRef]
- Smith, L.C.; Lun, C.M. The SpTransformer gene family (formerly Sp185/333) in the purple Sea Urchin and the functional diversity of the anti-pathogen rSpTransformer-E1 protein. Front. Immunol. 2017, 8, 725. [Google Scholar] [CrossRef]
- Roth, M.O.; Wilkins, A.G.; Cooke, G.M.; Raftos, D.A.; Nair, S.V. Characterization of the highly variable immune response gene family, He185/333, in the sea urchin, Heliocidaris erythrogramma. PLoS ONE 2014, 9, e62079. [Google Scholar] [CrossRef][Green Version]
- Fučíková, K.; Lewis, P.O.; González-Halphen, D.; Lewis, L.A. Gene arrangement convergence, diverse intron content, and genetic code modifications in mitochondrial genomes of Sphaeropleales (Chlorophyta). Genome Biol. Evol. 2014, 6, 2170–2180. [Google Scholar] [CrossRef]
- Hayashi-Ishimaru, Y.; Ohama, T.; Kawatsu, Y.; Nakamura, K.; Osawa, S. UAG is a sense codon in several chlorophycean mitochondria. Curr. Genet. 1996, 30, 29–33. [Google Scholar] [CrossRef]
- Laforest, M.J.; Roewer, I.; Lang, B.F. Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG ‘Stop’ codons recognized as leucine. Nucleic Acids Res. 1997, 25, 626–632. [Google Scholar] [CrossRef]
- Clegg, J.B.; Weatherall, D.J.; Milner, P.F. Haemoglobin Constant Spring—A chain termination mutant? Nature 1971, 234, 337–340. [Google Scholar] [CrossRef]
- Clegg, J.B.; Weatherall, D.J.; Contopolou-Griva, I.; Caroutsos, K.; Poungouras, P.; Tsevrenis, H. Haemoglobin Icaria, a new chain-termination mutant with causes α thalassaemia. Nature 1974, 251, 245–247. [Google Scholar] [CrossRef]
- Hashimoto, S.; Nobuta, R.; Izawa, T.; Inada, T. Translation arrest as a protein quality control system for aberrant translation of the 3’-UTR in mammalian cells. FEBS Lett. 2019, 593, 777–787. [Google Scholar] [CrossRef]
- Balakirev, E.S.; Krupnova, T.N.; Ayala, F.J. DNA variation in the phenotypically-diverse brown alga Saccharina japonica. BMC Plant Biol. 2012, 12. [Google Scholar] [CrossRef]
- Balakirev, E.S.; Romanov, N.S.; Mikheev, P.B.; Ayala, F.J. Mitochondrial DNA variation and introgression in Siberian taimen Hucho taimen. PLoS ONE 2013, 8, e71147. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Tu, Q.; Cameron, R.A.; Worley, K.C.; Gibbs, R.A.; Davidson, E.H. Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis. Genome Res. 2012, 22, 2079–2087. [Google Scholar] [CrossRef]
- Tu, Q.; Cameron, R.A.; Davidson, E.H. Quantitative developmental transcriptomes of the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 2014, 385, 160–167. [Google Scholar] [CrossRef]
- Chen, Y.; Chang, Y.; Wang, X.; Qiu, X.; Liu, Y. De novo assembly and analysis of tissue-specific transcriptomes revealed the tissue-specific genes and profile of immunity from Strongylocentrotus intermedius. Fish Shellfish. Immunol. 2015, 46, 723–736. [Google Scholar] [CrossRef]
- Jia, Z.; Wang, Q.; Wu, K.; Wei, Z.; Zhou, Z.; Liu, X. De novo transcriptome sequencing and comparative analysis to discover genes involved in ovarian maturity in Strongylocentrotus nudus. Comp. Biochem. Physiol. D Genom. Proteom. 2017, 23, 27–38. [Google Scholar] [CrossRef]
- Ding, J.; Yang, D.; Chang, Y.; Wang, Y.; Zhang, W.; Chen, T. Comparative transcriptome analysis of tube feet of different colors in the sea urchin Strongylocentrotus intermedius. Genes Genom. 2017, 39, 1215–1225. [Google Scholar] [CrossRef]
- Sun, Z.H.; Zhang, J.; Zhang, W.J.; Chang, Y.Q. Gonadal transcriptomic analysis and identification of candidate sex-related genes in Mesocentrotus nudus. Gene 2019, 698, 72–81. [Google Scholar] [CrossRef]
- Zhan, Y.; Li, J.; Sun, J.; Zhang, W.; Li, Y.; Cui, D.; Hu, W.; Chang, Y. The Impact of Chronic Heat Stress on the Growth, Survival, Feeding, and Differential Gene Expression in the Sea Urchin Strongylocentrotus intermedius. Front. Genet. 2019, 10, 301. [Google Scholar] [CrossRef]
- Zhang, W.; Lv, Z.; Li, C.; Sun, Y.; Jiang, H.; Zhao, M.; Zhao, X.; Shao, Y.; Chang, Y. Transcriptome profiling reveals key roles of phagosome and NOD-like receptor pathway in spotting diseased Strongylocentrotus intermedius. Fish Shellfish. Immunol. 2019, 84, 521–531. [Google Scholar] [CrossRef]
- Zhao, C.; Ding, J.; Yang, M.; Shi, D.; Yin, D.; Hu, F.; Sun, J.; Chi, X.; Zhang, L.; Chang, Y. Transcriptomes reveal genes involved in covering and sheltering behaviors of the sea urchin Strongylocentrotus intermedius exposed to UV-B radiation. Ecotoxicol. Environ. Saf. 2019, 167, 236–241. [Google Scholar] [CrossRef]
- Liu, Z.; Pagani, M.; Zinniker, D.; DeConto, R.; Huber, M.; Brinkhuis, H.; Pearson, A. Global cooling during the Eocene-Oligocene climate transition. Science 2009, 323, 1187–1190. [Google Scholar] [CrossRef]
- Raup, D.M.; Sepkoski, J.J. Periodic extinction of families and genera. Science 1986, 231, 833–836. [Google Scholar] [CrossRef]
- Prothero, D.R. The late Eocene—Oligocene extinctions. Annu. Rev. Earth Planet. Sci. 1994, 22, 145–165. [Google Scholar] [CrossRef]
- Sun, J.; Ni, X.; Bi, S.; Wu, W.; Ye, J.; Meng, J.; Windley, B.F. Synchronous turnover of flora, fauna, and climate at the Eocene-Oligocene Boundary in Asia. Sci. Rep. 2014, 4, 7463. [Google Scholar] [CrossRef]
- Ducasse, O.; Lété, C.; Rousselle, L. Polymorphism and speciation medoc Ostracods at the Eocene/Oligocene boundary (Aquitaine, France). Develop. Palaeontol. Stratigr. 1988, 11, 939–947. [Google Scholar]
- Suto, I. The explosive diversification of the diatom genus Chaetoceros across the Eocene/Oligocene and Oligocene/Miocene boundaries in the Norwegian Sea. Mar. Micropaleont. 2006, 58, 259–269. [Google Scholar] [CrossRef]
- Steeman, M.E.; Hebsgaard, M.B.; Fordyce, R.E.; Ho, S.Y.W.; Rabosky, D.L.; Nielsen, R.; Willerslev, E. Radiation of extant cetaceans driven by restructuring of the oceans. Syst. Biol. 2009, 58, 573–585. [Google Scholar] [CrossRef]
- Ballard, J.W.O.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef]
- Dowling, D.K.; Friberg, U.; Lindell, J. Evolutionary implications of nonneutral mitochondrial genetic variation. Trends Ecol. Evol. 2008, 23, 546–554. [Google Scholar] [CrossRef]
- Lajbner, Z.; Pnini, R.; Camus, M.F.; Miller, J.; Dowling, D.K. Experimental evidence that thermal selection shapes mitochondrial genome evolution. Sci. Rep. 2018, 8, 9500. [Google Scholar] [CrossRef]
- Camus, M.F.; Wolff, J.N.; Sgrò, C.M.; Dowling, D.K. Experimental evidence that thermal selection has shaped the latitudinal distribution of mitochondrial haplotypes in Australian fruit flies. Mol. Biol. Evol. 2017, 34, 2600–2612. [Google Scholar] [CrossRef]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balakirev, E.S. Trans-Species Polymorphism in Mitochondrial Genome of Camarodont Sea Urchins. Genes 2019, 10, 592. https://doi.org/10.3390/genes10080592
Balakirev ES. Trans-Species Polymorphism in Mitochondrial Genome of Camarodont Sea Urchins. Genes. 2019; 10(8):592. https://doi.org/10.3390/genes10080592
Chicago/Turabian StyleBalakirev, Evgeniy S. 2019. "Trans-Species Polymorphism in Mitochondrial Genome of Camarodont Sea Urchins" Genes 10, no. 8: 592. https://doi.org/10.3390/genes10080592
APA StyleBalakirev, E. S. (2019). Trans-Species Polymorphism in Mitochondrial Genome of Camarodont Sea Urchins. Genes, 10(8), 592. https://doi.org/10.3390/genes10080592



