Deep Splicing Code: Classifying Alternative Splicing Events Using Deep Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Preparation
- A set of exon skipping (ES) or cassette exon events: exons that can be either excluded or included with a frequency value known as inclLevel (inclLevel ∈ [0, 1]) and do not have any other alternative splice site.
- A set of exons having an alternative 3’ (ALT3) splice site used with a frequency value known as 3usageLevel (3usageLevel ∈ [0, 1]).
- A set of exons having an alternative 5’ (ALT5) splice site used with a frequency value known as 5usageLevel (5usageLevel ∈ [0, 1]).
- A set of constitutive exons (CON): internal exons with no ESTs evidence of any type of alternative splicing; the inclusion level and all the splice sites usage levels are equal to 1.
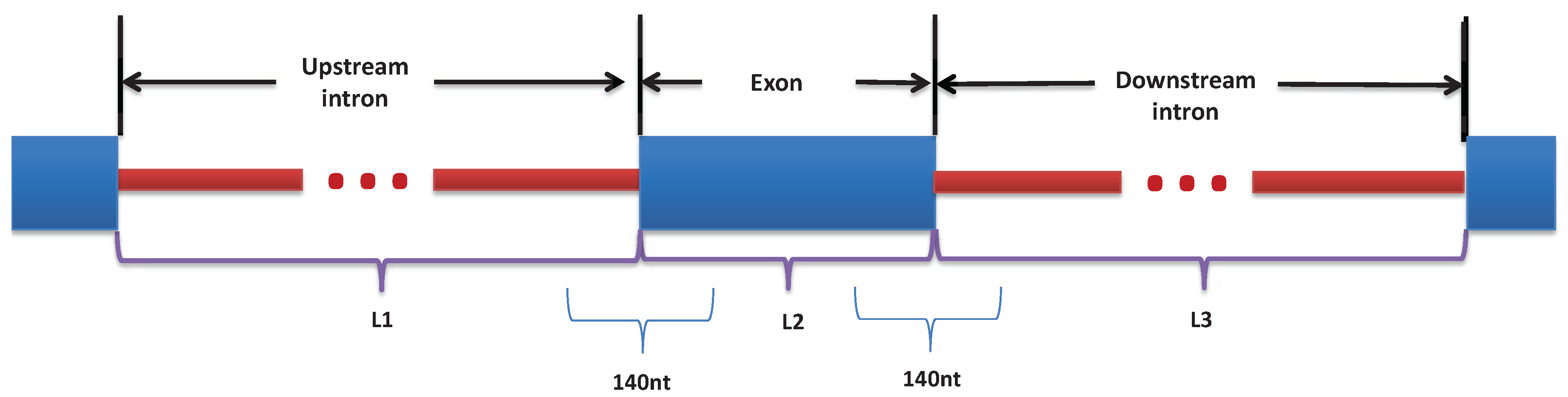
2.2. Features Selection
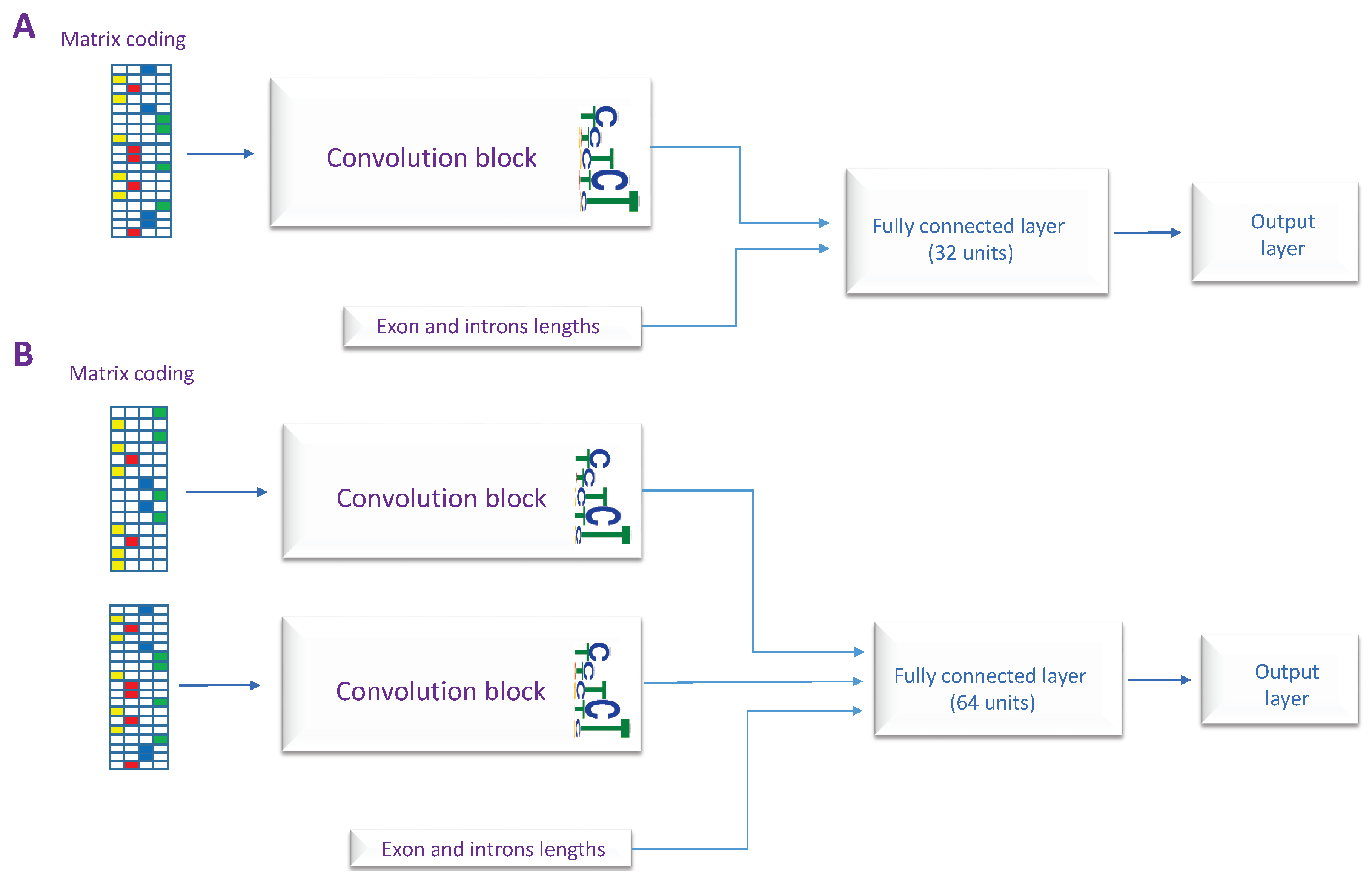
2.3. The Proposed Models
2.4. Implementation Details
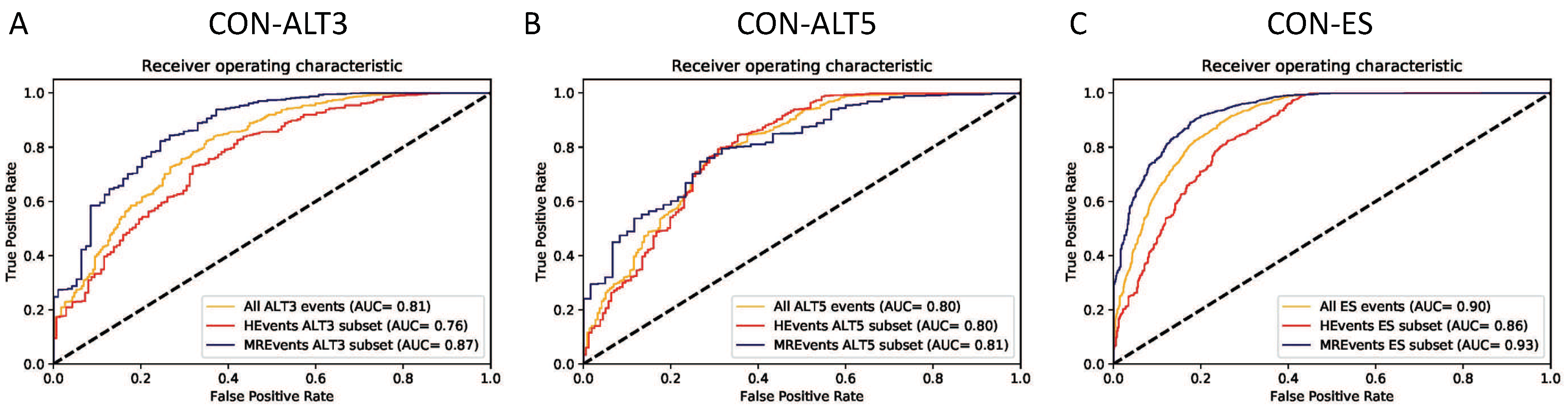
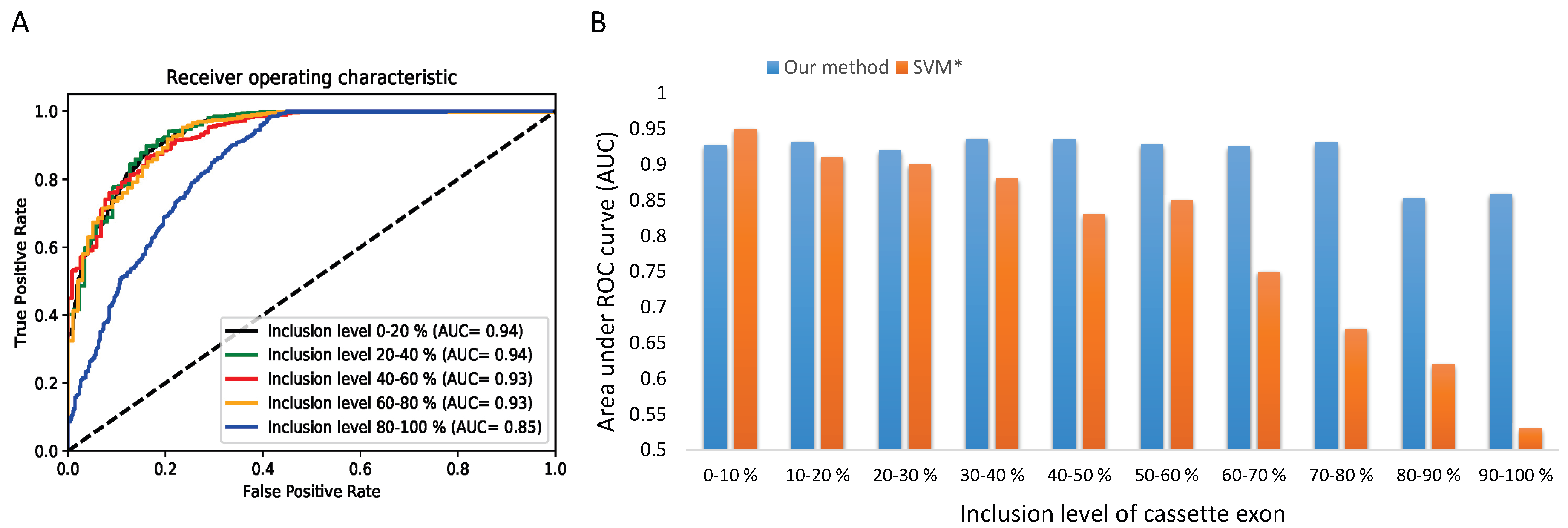
3. Results and Comparisons
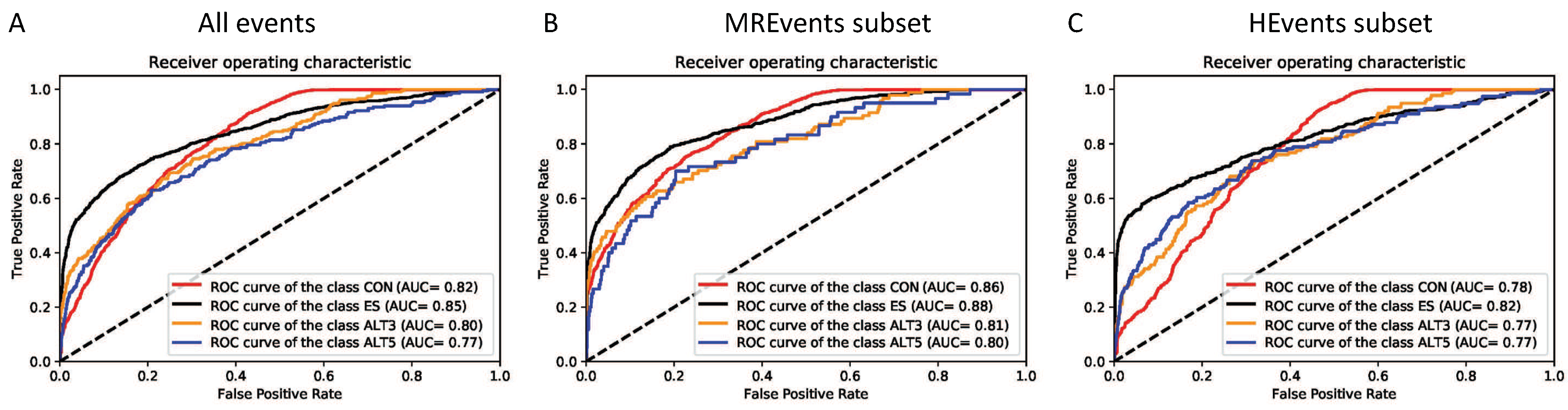
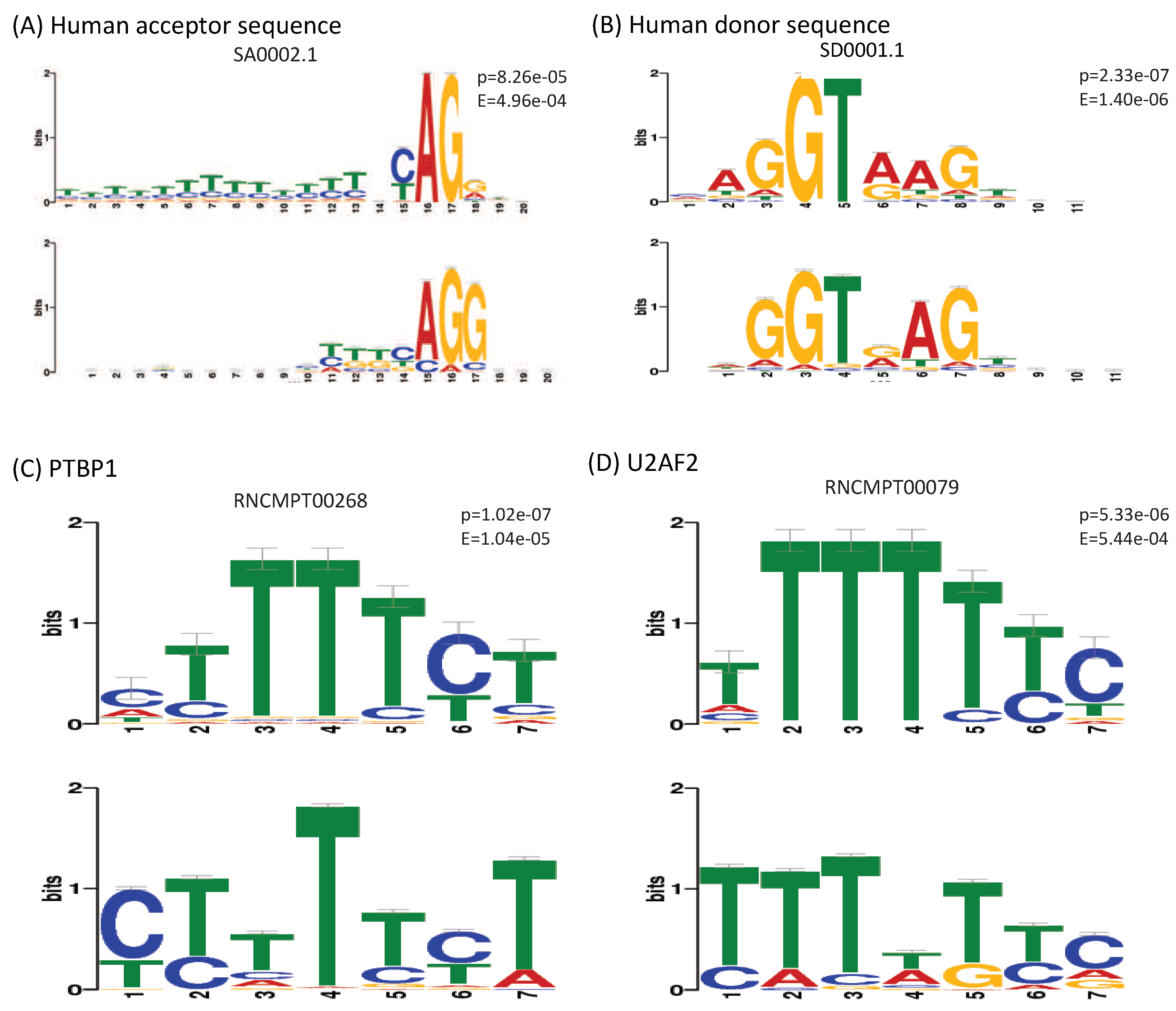
4. Models Interpretation and Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Barbosa-Morais, N.L.; Irimia, M.; Pan, Q.; Xiong, H.Y.; Gueroussov, S.; Lee, L.J.; Slobodeniuc, V.; Kutter, C.; Watt, S.; Çolak, R.; et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012, 338, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Sugnet, C.W.; Kent, W.J.; Ares, M., Jr.; Haussler, D. Transcriptome and genome conservation of alternative splicing events in humans and mice. In Biocomputing 2004; World Scientific: Singapore, 2003; pp. 66–77. [Google Scholar]
- Tazi, J.; Bakkour, N.; Stamm, S. Alternative splicing and disease. Biochim. Biophys. Acta Mol. Basis Dis. 2009, 1792, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Blanco, M.A.; Baraniak, A.P.; Lasda, E.L. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004, 22, 535. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, B.M. Splice variants as cancer biomarkers. Clin. Biochem. 2004, 37, 584–594. [Google Scholar] [CrossRef]
- Xiong, H.Y.; Alipanahi, B.; Lee, L.J.; Bretschneider, H.; Merico, D.; Yuen, R.K.; Hua, Y.; Gueroussov, S.; Najafabadi, H.S.; Hughes, T.R.; et al. The human splicing code reveals new insights into the genetic determinants of disease. Science 2015, 347, 1254806. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.K.; Xiong, H.Y.; Lee, L.J.; Frey, B.J. Deep learning of the tissue-regulated splicing code. Bioinformatics 2014, 30, i121–i129. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.; Gazzara, M.R.; Barash, Y. Integrative deep models for alternative splicing. Bioinformatics 2017, 33, i274–i282. [Google Scholar] [CrossRef]
- Oubounyt, M.; Louadi, Z.; Tayara, H.; Chong, K.T. Deep Learning Models Based on Distributed Feature Representations for Alternative Splicing Prediction. IEEE Access 2018, 6, 58826–58834. [Google Scholar] [CrossRef]
- Busch, A.; Hertel, K.J. Splicing predictions reliably classify different types of alternative splicing. RNA 2015, 21, 813–823. [Google Scholar] [CrossRef]
- Gazzara, M.R.; Vaquero-Garcia, J.; Lynch, K.W.; Barash, Y. In silico to in vivo splicing analysis using splicing code models. Methods 2014, 67, 3–12. [Google Scholar] [CrossRef][Green Version]
- Shepard, P.J.; Choi, E.A.; Busch, A.; Hertel, K.J. Efficient internal exon recognition depends on near equal contributions from the 3’ and 5’ splice sites. Nucleic Acids Res. 2011, 39, 8928–8937. [Google Scholar] [CrossRef] [PubMed]
- Arias, M.A.; Lubkin, A.; Chasin, L.A. Splicing of designer exons informs a biophysical model for exon definition. RNA 2015, 21, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Koren, E.; Lev-Maor, G.; Ast, G. The emergence of alternative 3’ and 5’ splice site exons from constitutive exons. PLoS Comput. Biol. 2007, 3, e95. [Google Scholar] [CrossRef] [PubMed]
- Fox-Walsh, K.L.; Dou, Y.; Lam, B.J.; Hung, S.p.; Baldi, P.F.; Hertel, K.J. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. USA 2005, 102, 16176–16181. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R.; Ast, G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003, 13, 1631–1637. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, S. Identify alternative splicing events based on position-specific evolutionary conservation. PLoS ONE 2008, 3, e2806. [Google Scholar] [CrossRef]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A primer on deep learning in genomics. Nat. Genet. 2018, 51, 12–18. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, Y.; Liao, X.; Shi, W.; Li, K.; Zou, Q.; Peng, S. Deep learning in omics: A survey and guideline. Brief. Funct. Genom. 2018, 18, 41–57. [Google Scholar] [CrossRef]
- Quang, D.; Xie, X. DanQ: A hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016, 44, e107. [Google Scholar] [CrossRef]
- Quang, D.; Xie, X. FactorNet: A deep learning framework for predicting cell type specific transcription factor binding from nucleotide-resolution sequential data. Methods 2019. [Google Scholar] [CrossRef]
- Nazari, I.; Tayara, H.; Chong, K.T. Branch Point Selection in RNA Splicing Using Deep Learning. IEEE Access 2019, 7, 1800–1807. [Google Scholar] [CrossRef]
- Paggi, J.M.; Bejerano, G. A sequence-based, deep learning model accurately predicts RNA splicing branchpoints. RNA 2018, 24, 1647–1658. [Google Scholar] [CrossRef]
- Hill, S.T.; Kuintzle, R.; Teegarden, A.; Merrill, E., III; Danaee, P.; Hendrix, D.A. A deep recurrent neural network discovers complex biological rules to decipher RNA protein-coding potential. Nucleic Acids Res. 2018, 46, 8105–8113. [Google Scholar] [CrossRef]
- Angermueller, C.; Lee, H.J.; Reik, W.; Stegle, O. DeepCpG: Accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol. 2017, 18, 67. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; MacLeod, J.; Liu, J. Discerning novel splice junctions derived from RNA-seq alignment: A deep learning approach. BMC Genom. 2018, 19, 971. [Google Scholar] [CrossRef]
- Zuallaert, J.; Godin, F.; Kim, M.; Soete, A.; Saeys, Y.; De Neve, W. SpliceRover: Interpretable convolutional neural networks for improved splice site prediction. Bioinformatics 2018, 34, 4180–4188. [Google Scholar] [CrossRef]
- Jaganathan, K.; Panagiotopoulou, S.K.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting splicing from primary sequence with deep learning. Cell 2019, 176, 535–548. [Google Scholar] [CrossRef]
- Bretschneider, H.; Gandhi, S.; Deshwar, A.G.; Zuberi, K.; Frey, B.J. COSSMO: Predicting competitive alternative splice site selection using deep learning. Bioinformatics 2018, 34, i429–i437. [Google Scholar] [CrossRef]
- Busch, A.; Hertel, K.J. HEXEvent: A database of Human EXon splicing Events. Nucleic Acids Res. 2012, 41, D118–D124. [Google Scholar] [CrossRef]
- Rosenbloom, K.R.; Armstrong, J.; Barber, G.P.; Casper, J.; Clawson, H.; Diekhans, M.; Dreszer, T.R.; Fujita, P.A.; Guruvadoo, L.; Haeussler, M.; et al. The UCSC genome browser database: 2015 update. Nucleic Acids Res. 2014, 43, D670–D681. [Google Scholar] [CrossRef]
- Piovesan, A.; Caracausi, M.; Ricci, M.; Strippoli, P.; Vitale, L.; Pelleri, M.C. Identification of minimal eukaryotic introns through GeneBase, a user-friendly tool for parsing the NCBI Gene databank. DNA Res. 2015, 22, 495–503. [Google Scholar] [CrossRef][Green Version]
- Castle, J.C.; Zhang, C.; Shah, J.K.; Kulkarni, A.V.; Kalsotra, A.; Cooper, T.A.; Johnson, J.M. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008, 40, 1416. [Google Scholar] [CrossRef]
- Roy, M.; Kim, N.; Xing, Y.; Lee, C. The effect of intron length on exon creation ratios during the evolution of mammalian genomes. RNA 2008, 14, 2261–2273. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; Volume 16, pp. 265–283. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Reddi, S.J.; Kale, S.; Kumar, S. On the Convergence of Adam and Beyond. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Fox-Walsh, K.L.; Hertel, K.J. Splice-site pairing is an intrinsically high fidelity process. Proc. Natl. Acad. Sci. USA 2009, 106, 1766–1771. [Google Scholar] [CrossRef]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef]
- Sugnet, C.W.; Srinivasan, K.; Clark, T.A.; O’Brien, G.; Cline, M.S.; Wang, H.; Williams, A.; Kulp, D.; Blume, J.E.; Haussler, D.; et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006, 2, e4. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Ray, D.; Kazan, H.; Cook, K.B.; Weirauch, M.T.; Najafabadi, H.S.; Li, X.; Gueroussov, S.; Albu, M.; Zheng, H.; Yang, A.; et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 2013, 499, 172. [Google Scholar] [CrossRef]
- Chong, A.; Zhang, G.; Bajic, V.B. Information for the Coordinates of Exons (ICE): A human splice sites database. Genomics 2004, 84, 762–766. [Google Scholar] [CrossRef]
- Lin, J.C.; Tarn, W.Y. Exon selection in α-tropomyosin mRNA is regulated by the antagonistic action of RBM4 and PTB. Mol. Cell. Biol. 2005, 25, 10111–10121. [Google Scholar] [CrossRef]
- Zamore, P.D.; Green, M.R. Biochemical characterization of U2 snRNP auxiliary factor: An essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991, 10, 207–214. [Google Scholar] [CrossRef]
- Makeyev, A.V.; Liebhaber, S.A. The poly (C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 2002, 8, 265–278. [Google Scholar] [CrossRef]
- Förch, P.; Puig, O.; Kedersha, N.; Martínez, C.; Granneman, S.; Séraphin, B.; Anderson, P.; Valcárcel, J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 2000, 6, 1089–1098. [Google Scholar] [CrossRef]
- Cereda, M.; Pozzoli, U.; Rot, G.; Juvan, P.; Schweitzer, A.; Clark, T.; Ule, J. RNAmotifs: Prediction of multivalent RNA motifs that control alternative splicing. Genome Biol. 2014, 15, R20. [Google Scholar] [CrossRef]
- Bao, S.; Moakley, D.F.; Zhang, C. The Splicing Code Goes Deep. Cell 2019, 176, 414–416. [Google Scholar] [CrossRef]
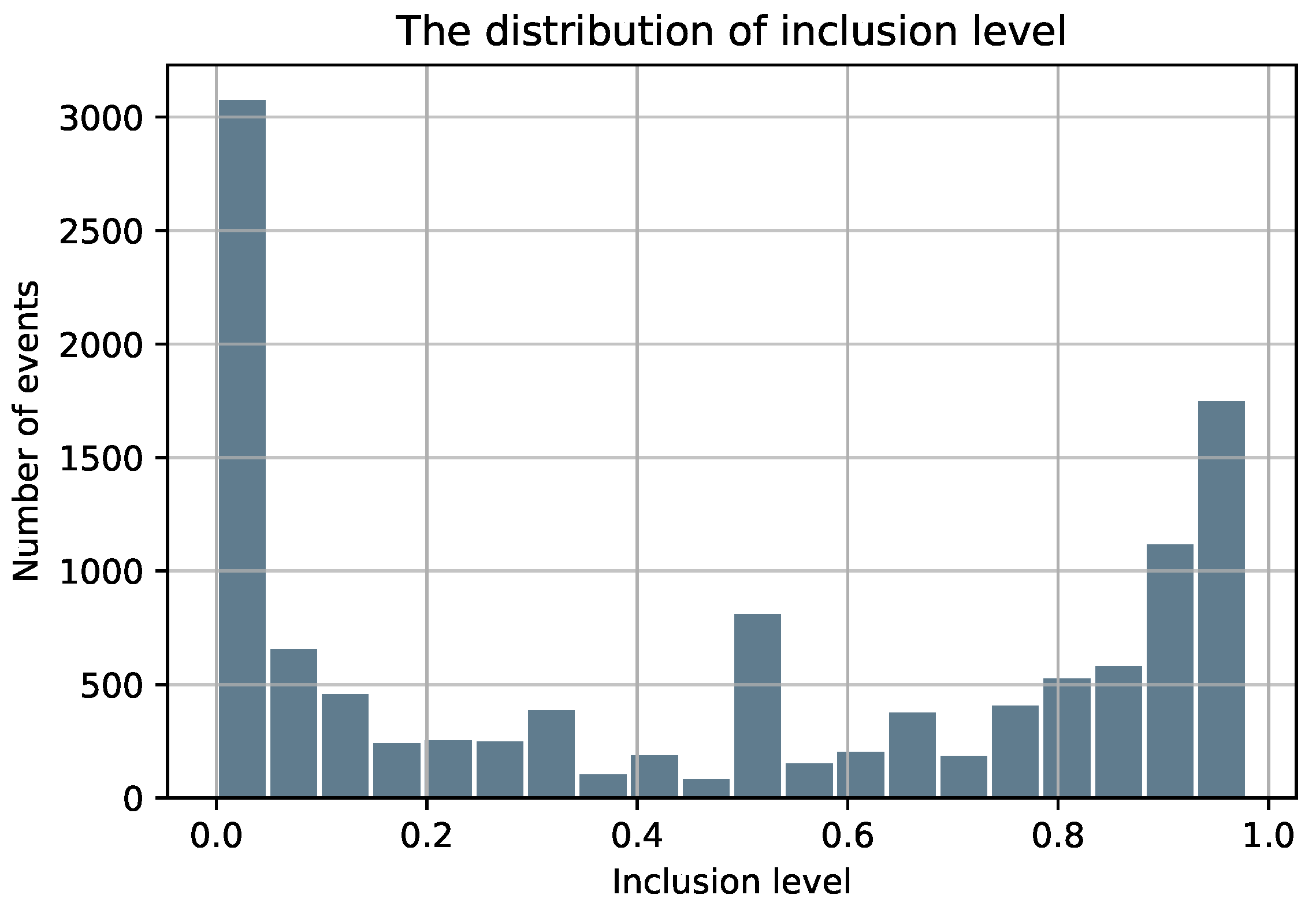


| AS Type | Cons | ES | Alt3 | Alt5 |
|---|---|---|---|---|
| Number of events | 39,128 | 11,590 | 2332 | 2172 |
| The Model | Architecture Used | Inputs | The Output Layer |
|---|---|---|---|
| CON-ES | Architecture B | Upstream and downstream sequences | Sigmoid |
| CON-ALT3 | Architecture A | Upstream sequence | Sigmoid |
| CON-ALT5 | Architecture A | Downstream sequence | Sigmoid |
| DSC | Architecture B | Upstream and downstream sequences | Softmax |
| AS Type | HEevents | MREvents |
|---|---|---|
| ES | 4952 | 6638 |
| ALT3 | 1388 | 944 |
| ALT5 | 1568 | 604 |
| Model | All Events | MREvents Subset | HEvents Subset |
|---|---|---|---|
| CON-ES | 0.899 (±0.008) | 0.928 (±0.007) | 0.858 (±0.015) |
| CON-ALT3 | 0.812 (±0.016) | 0.870 (±0.016) | 0.772 (±0.022) |
| CON-ALT5 | 0.821 (±0.014) | 0.838 (±0.028) | 0.813 (±0.017) |
| Exon Type | All Events | MREvents Subset | HEvents Subset |
|---|---|---|---|
| Constitutive exons | 0.835 (±0.007) | 0.870 (±0.005) | 0.798 (±0.001) |
| Cassette exon | 0.858 (±0.007) | 0.878 (±0.008) | 0.844 (±0.012) |
| Exon with an alternative 3-splice site | 0.819 (±0.014) | 0.832 (±0.025) | 0.795 (±0.020) |
| Exon with an alternative 5’-splice site | 0.772 (±0.017) | 0.788 (±0.029) | 0.774 (± 0020) |
| Average of the four classes | 0.821 | 0.842 | 0.803 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Louadi, Z.; Oubounyt, M.; Tayara, H.; Chong, K.T. Deep Splicing Code: Classifying Alternative Splicing Events Using Deep Learning. Genes 2019, 10, 587. https://doi.org/10.3390/genes10080587
Louadi Z, Oubounyt M, Tayara H, Chong KT. Deep Splicing Code: Classifying Alternative Splicing Events Using Deep Learning. Genes. 2019; 10(8):587. https://doi.org/10.3390/genes10080587
Chicago/Turabian StyleLouadi, Zakaria, Mhaned Oubounyt, Hilal Tayara, and Kil To Chong. 2019. "Deep Splicing Code: Classifying Alternative Splicing Events Using Deep Learning" Genes 10, no. 8: 587. https://doi.org/10.3390/genes10080587
APA StyleLouadi, Z., Oubounyt, M., Tayara, H., & Chong, K. T. (2019). Deep Splicing Code: Classifying Alternative Splicing Events Using Deep Learning. Genes, 10(8), 587. https://doi.org/10.3390/genes10080587







