Assessing Anthocyanin Biosynthesis in Solanaceae as a Model Pathway for Secondary Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Flavonoid Biosynthesis Pathway Homologs
2.2. Comparison of Annotations from Different Sources
2.3. Phylogenetic Analysis of Flavonoid Biosynthesis Enzymes
2.4. Evaluation of Flower Color Modification Using Transgenic F3′5′H
3. Results
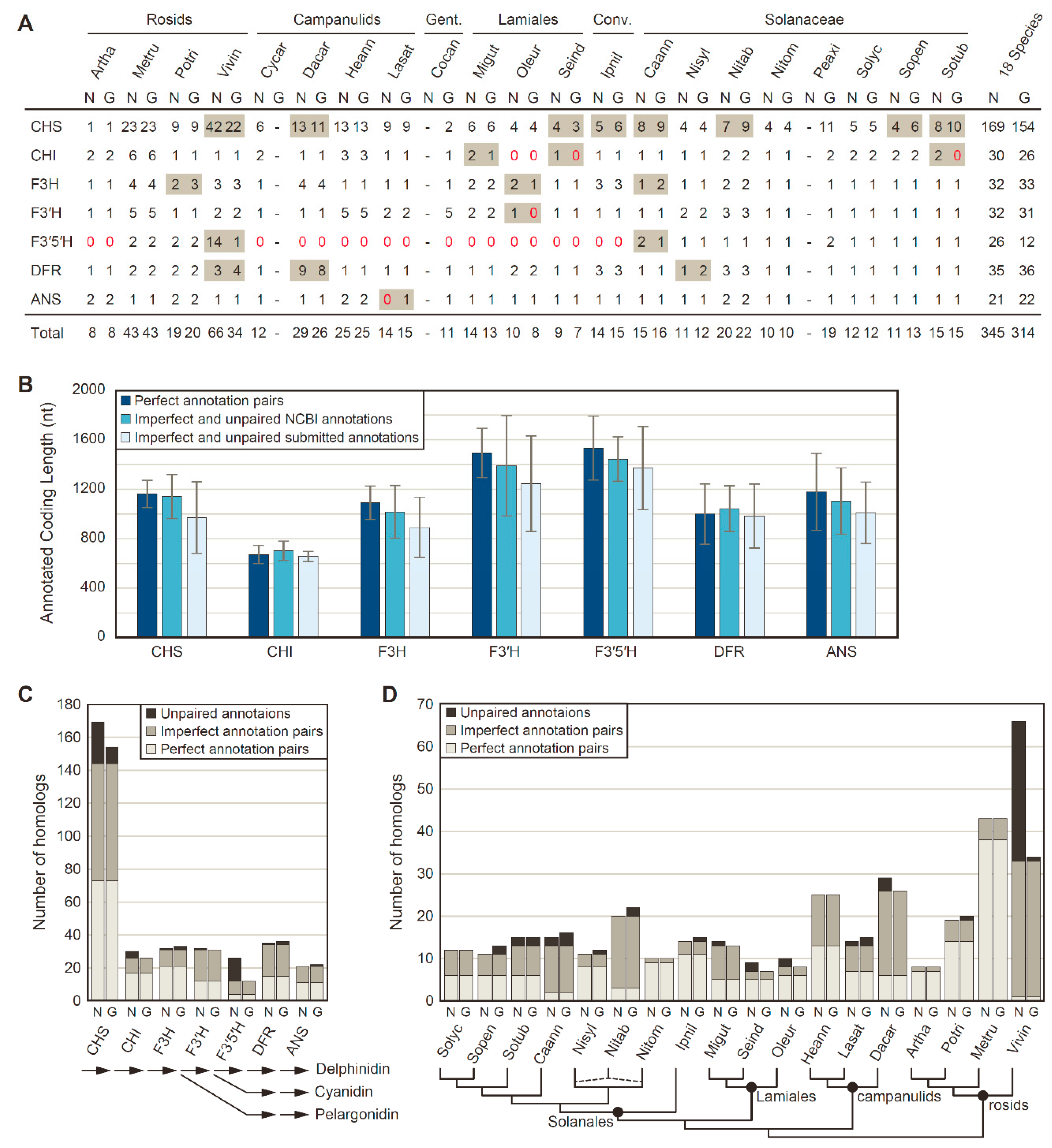
3.1. The NCBI Pipeline Annotates More and Longer Anthocyanin Biosynthesis Enzymes
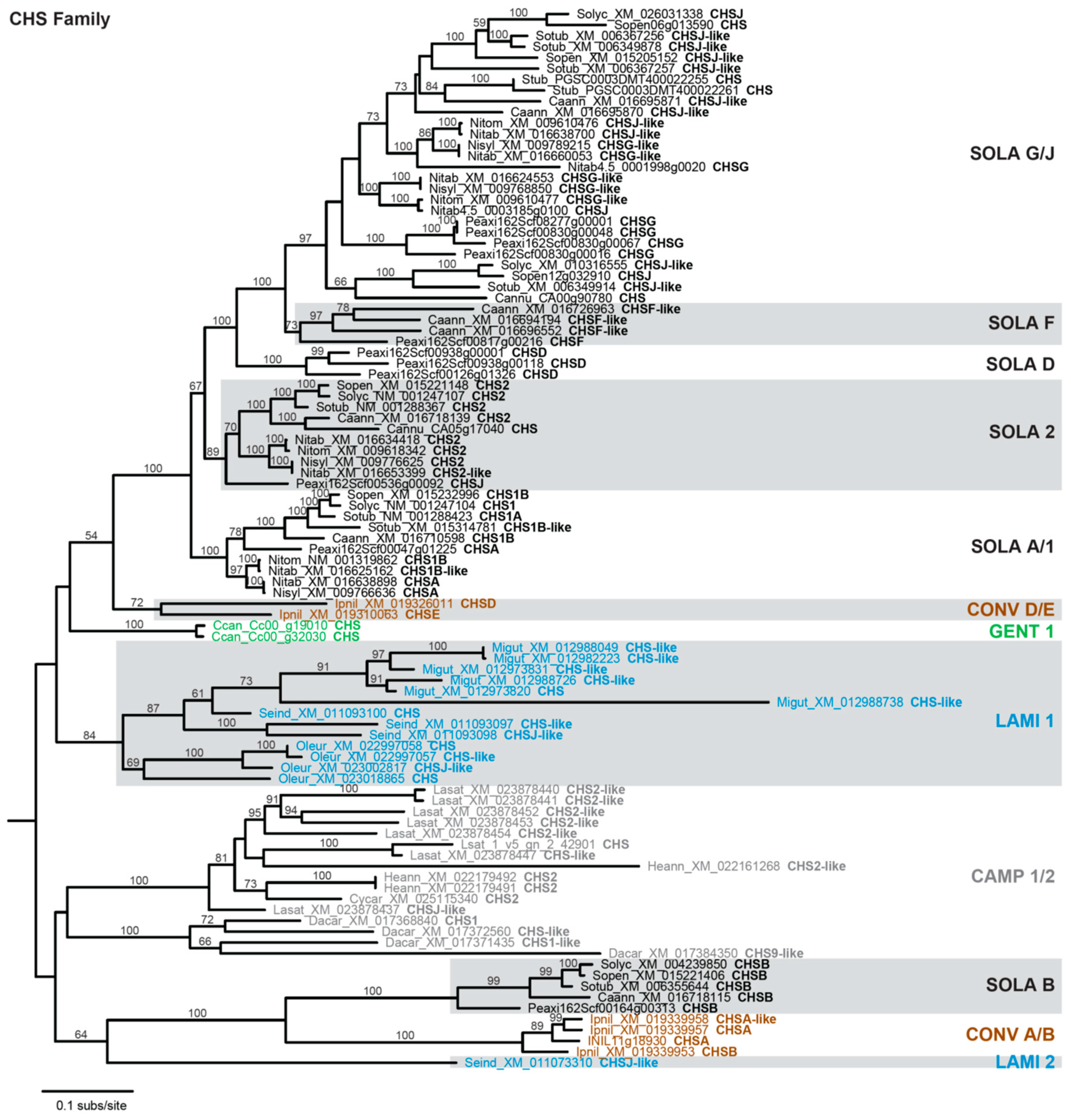
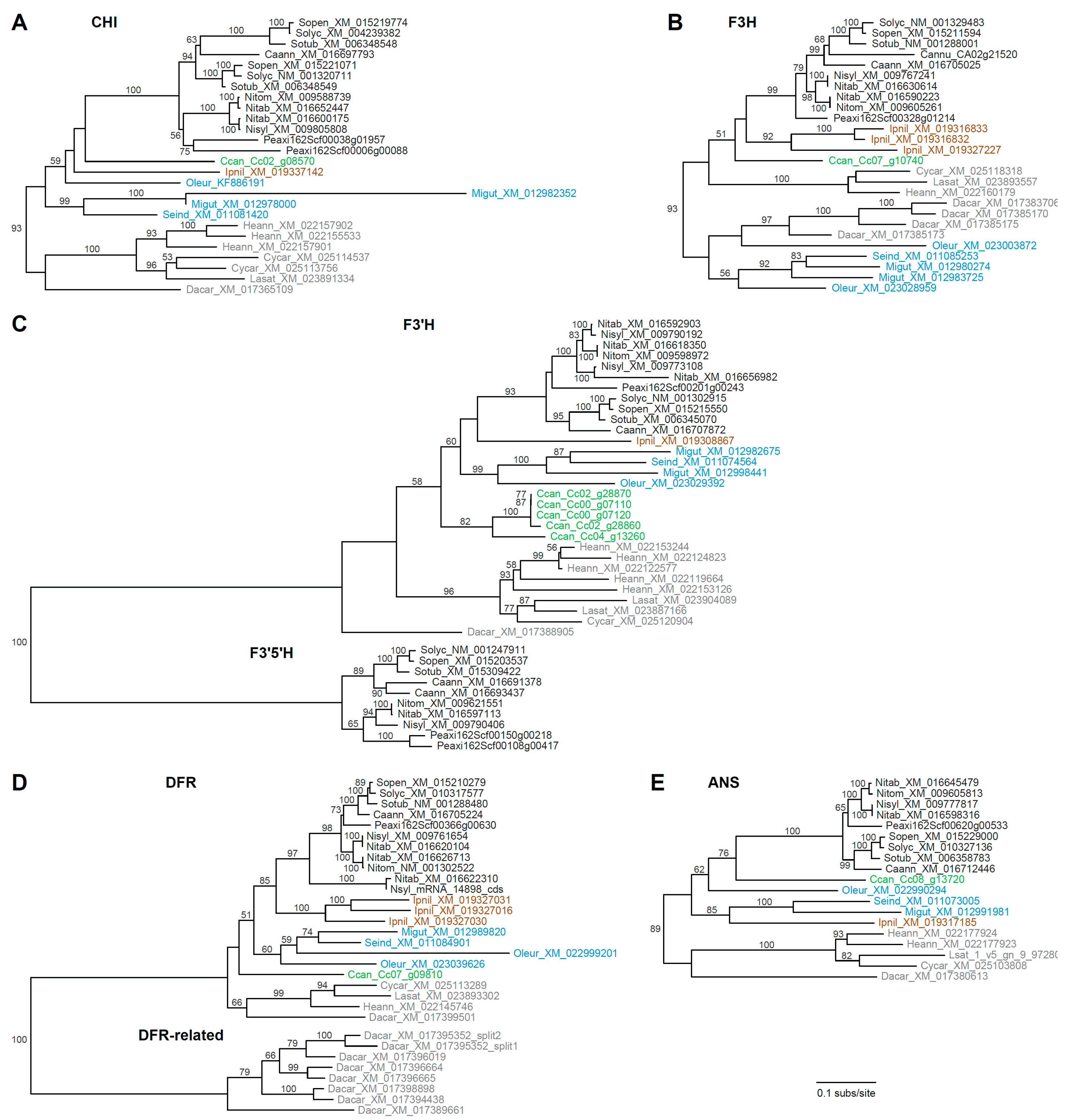
3.2. Rampant Lineage-Specific Duplication of Anthocyanin Biosynthesis Genes in Asterids
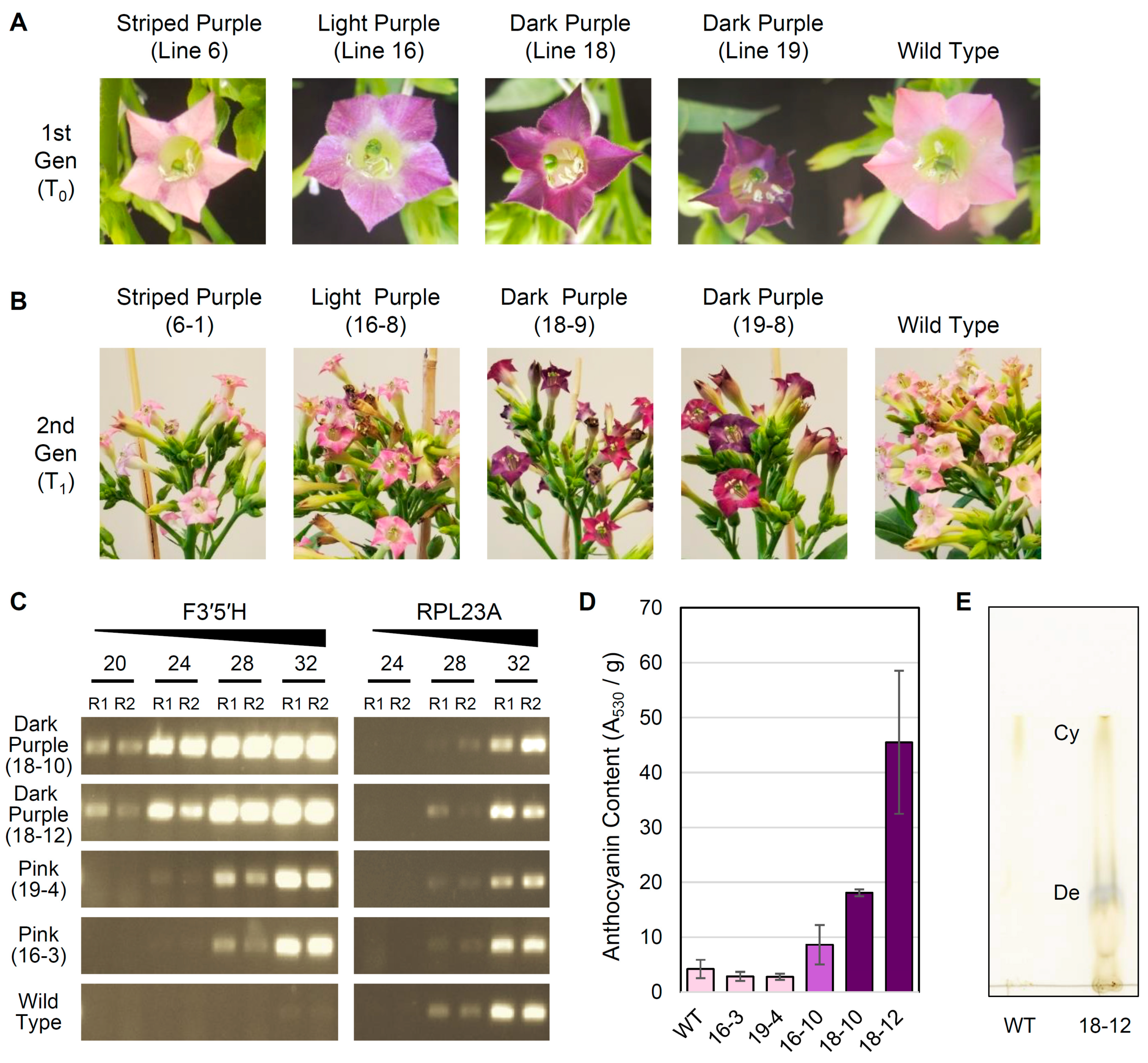
3.3. Overexpression of F3′5′H Produces Purple Flowers in Tobacco cv. Xanthi
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Olmstead, R.G.; Bohs, L. A summary of molecular systematic research in Solanaceae: 1982–2006. In Proceedings of the VI International Solanaceae Conference: Genomics Meets Biodiversity, Madison, WI, USA, 23–27 July 2006; Volune 745, pp. 255–268. [Google Scholar]
- Särkinen, T.; Bohs, L.; Olmstead, R.G.; Knapp, S. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): A dated 1000-tip tree. BMC Evol. Biol. 2013, 13, 214. [Google Scholar] [CrossRef]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; The Potato Genome Sequencing Consortium; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar]
- Tomato Genome, C. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Bombarely, A.; Rosli, H.G.; Vrebalov, J.; Moffett, P.; Mueller, L.A.; Martin, G.B. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 2012, 25, 1523–1530. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bovet, L.; Goepfert, S.; Bakaher, N.; Peitsch, M.C.; Ivanov, N.V. Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis. Genome Biol. 2013, 14, R60. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.; Scossa, F.; Bolger, M.E.; Lanz, C.; Maumus, F.; Tohge, T.; Quesneville, H.; Alseekh, S.; Sorensen, I.; Lichtenstein, G.; et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014, 46, 1034–1038. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Shirasawa, K.; Miyatake, K.; Nunome, T.; Negoro, S.; Ohyama, A.; Yamaguchi, H.; Sato, S.; Isobe, S.; Tabata, S.; et al. Draft genome sequence of eggplant (Solanum melongena L.): The representative solanum species indigenous to the old world. DNA Res. 2014, 21, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [PubMed]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef] [PubMed]
- Bombarely, A.; Moser, M.; Amrad, A.; Bao, M.; Bapaume, L.; Barry, C.S.; Bliek, M.; Boersma, M.R.; Borghi, L.; Bruggmann, R.; et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2016, 2, 16074. [Google Scholar] [CrossRef]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom. 2017, 18, 448. [Google Scholar] [CrossRef]
- Xu, S.; Brockmoller, T.; Navarro-Quezada, A.; Kuhl, H.; Gase, K.; Ling, Z.; Zhou, W.; Kreitzer, C.; Stanke, M.; Tang, H.; et al. Wild tobacco genomes reveal the evolution of nicotine biosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, 6133–6138. [Google Scholar] [CrossRef]
- Hulse-Kemp, A.M.; Maheshwari, S.; Stoffel, K.; Hill, T.A.; Jaffe, D.; Williams, S.R.; Weisenfeld, N.; Ramakrishnan, S.; Kumar, V.; Shah, P.; et al. Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 2018, 5, 4. [Google Scholar] [CrossRef]
- Hoshino, A.; Jayakumar, V.; Nitasaka, E.; Toyoda, A.; Noguchi, H.; Itoh, T.; Shin, I.T.; Minakuchi, Y.; Koda, Y.; Nagano, A.J.; et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat. Commun. 2016, 7, 13295. [Google Scholar] [CrossRef] [PubMed]
- Hellsten, U.; Wright, K.M.; Jenkins, J.; Shu, S.; Yuan, Y.; Wessler, S.R.; Schmutz, J.; Willis, J.H.; Rokhsar, D.S. Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 19478–19482. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Han, X.; Zhang, Y.; Li, D.; Wei, X.; Ding, X.; Zhang, X. Deep resequencing reveals allelic variation in Sesamum indicum. BMC Plant Biol. 2014, 14, 225. [Google Scholar] [CrossRef] [PubMed]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef] [PubMed]
- Cruz, F.; Julca, I.; Gomez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galan, B.; Frias, L.; Ribeca, P.; Derdak, S.; et al. Genome sequence of the olive tree, Olea europaea. Gigascience 2016, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F.J.; et al. Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E9413–E9422. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Briere, C.; Owens, G.L.; Carrere, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef]
- Reyes-Chin-Wo, S.; Wang, Z.; Yang, X.; Kozik, A.; Arikit, S.; Song, C.; Xia, L.; Froenicke, L.; Lavelle, D.O.; Truco, M.J.; et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017, 8, 14953. [Google Scholar] [CrossRef]
- Scaglione, D.; Reyes-Chin-Wo, S.; Acquadro, A.; Froenicke, L.; Portis, E.; Beitel, C.; Tirone, M.; Mauro, R.; Lo Monaco, A.; Mauromicale, G.; et al. The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 2016, 6, 19427. [Google Scholar] [CrossRef]
- McCarthy, E.W.; Berardi, A.E.; Smith, S.D.; Litt, A. Related allopolyploids display distinct floral pigment profiles and transgressive pigments. Am. J. Bot. 2017, 104, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Azuma, K.; Ohyama, A.; Ippoushi, K.; Ichiyanagi, T.; Takeuchi, A.; Saito, T.; Fukuoka, H. Structures and antioxidant activity of anthocyanins in many accessions of eggplant and its related species. J. Agric. Food Chem. 2008, 56, 10154–10159. [Google Scholar] [CrossRef] [PubMed]
- Gates, D.J.; Olson, B.; Clemente, T.E.; Smith, S.D. A novel R3 MYB transcriptional repressor associated with the loss of floral pigmentation in Iochroma. New Phytol. 2018, 217, 1346–1356. [Google Scholar] [CrossRef]
- Rodriguez-Saona, L.E.; Giusti, M.M.; Wrolstad, R.E. Anthocyanin pigment composition of red-fleshed potatoes. J. Food Sci. 1998, 63, 458–465. [Google Scholar] [CrossRef]
- Lewis, C.E.; Walker, J.R.; Lancaster, J.E.; Sutton, K.H. Determination of anthocyanins, flavonoids and phenolic acids in potatoes. I: Coloured cultivars of Solanum tuberosum L. J. Sci. Food Agric. 1998, 77, 45–57. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P.; Heidmann, I.; Forkmann, G.; Saedler, H. A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature 1987, 330, 677–678. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Nakano-Shimada, R.; Ohbayashi, M.; Okinaka, Y.; Kiyokawa, S.; Kikuchi, Y. Expression of chimeric P450 genes encoding flavonoid-3’, 5’-hydroxylase in transgenic tobacco and petunia plants. FEBS Lett. 1999, 461, 241–245. [Google Scholar] [CrossRef]
- Okinaka, Y.; Shimada, Y.; Nakano-Shimada, R.; Ohbayashi, M.; Kiyokawa, S.; Kikuchi, Y. Selective accumulation of delphinidin derivatives in tobacco using a putative flavonoid 3’,5’-hydroxylase cDNA from Campanula medium. Biosci. Biotechnol. Biochem. 2003, 67, 161–165. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Nishihara, M.; Mishiba, K.; Yamamura, S. Heterologous expression of two gentian cytochrome P450 genes can modulate the intensity of flower pigmentation in transgenic tobacco plants. Mol. Breed. 2006, 17, 91–99. [Google Scholar] [CrossRef]
- Vanderkrol, A.R.; Lenting, P.E.; Veenstra, J.; Vandermeer, I.M.; Koes, R.E.; Gerats, A.G.M.; Mol, J.N.M.; Stuitje, A.R. An anti-sense chalcone synthase gene in transgenic plants inhibits flower pigmentation. Nature 1988, 333, 866–869. [Google Scholar] [CrossRef]
- Nishihara, M.; Nakatsuka, T.; Yamamura, S. Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. FEBS Lett. 2005, 579, 6074–6078. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, T.; Pitaksutheepong, C.; Yamamura, S.; Nishihara, M. Induction of differential flower pigmentation patterns by RNAi using promoters with distinct tissue-specific activity. Plant Biotechnol. Rep. 2007, 1, 251–257. [Google Scholar] [CrossRef]
- Holton, T.A.; Brugliera, F.; Tanaka, Y. Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 1993, 4, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Budde, I.; Hain, R. Stilbene synthase gene expression causes changes in flower colour and male sterility in tobacco. Plant J. 1997, 11, 489–498. [Google Scholar] [CrossRef]
- Davies, K.M.; Bloor, S.J.; Spiller, G.B.; Deroles, S.C. Production of yellow colour in flowers: Redirection of flavonoid biosynthesis in Petunia. Plant J. 1998, 13, 259–266. [Google Scholar] [CrossRef]
- Davies, K.M.; Schwinn, K.E.; Deroles, S.C.; Manson, D.G.; Lewis, D.H.; Bloor, S.J.; Bradley, J.M. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 2003, 131, 259–268. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Katsumoto, Y.; Fukuchi-Mizutani, M.; Fukui, Y.; Brugliera, F.; Holton, T.A.; Karan, M.; Nakamura, N.; Yonekura-Sakakibara, K.; Togami, J.; Pigeaire, A.; et al. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 2007, 48, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Brugliera, F. Flower colour and cytochromes P450. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013, 368, 20120432. [Google Scholar] [CrossRef] [PubMed]
- Hajdukiewicz, P.; Svab, Z.; Maliga, P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994, 25, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Koncz, C.; Schell, J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986, 204, 383–396. [Google Scholar] [CrossRef]
- Clemente, T. Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). Methods Mol. Biol. 2006, 343, 143–154. [Google Scholar] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Sigmon, B.A.; Adams, R.P.; Mower, J.P. Complete chloroplast genome sequencing of vetiver grass (Chrysopogon zizanioides) identifies markers that distinguish the non-fertile ‘Sunshine’ cultivar from other accessions. Ind. Crops Prod. 2017, 108, 629–635. [Google Scholar] [CrossRef]
- Hepburn, N.J.; Schmidt, D.W.; Mower, J.P. Loss of two introns from the Magnolia tripetala mitochondrial cox2 gene implicates horizontal gene transfer and gene conversion as a novel mechanism of intron loss. Mol. Biol. Evol. 2012, 29, 3111–3120. [Google Scholar] [CrossRef]
- Jiang, C.; Kim, S.Y.; Suh, D.Y. Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol. Phylogenet. Evol. 2008, 49, 691–701. [Google Scholar] [CrossRef]
- Abe, I.; Morita, H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat. Prod. Rep. 2010, 27, 809–838. [Google Scholar] [CrossRef]
- Parage, C.; Tavares, R.; Rety, S.; Baltenweck-Guyot, R.; Poutaraud, A.; Renault, L.; Heintz, D.; Lugan, R.; Marais, G.A.; Aubourg, S.; et al. Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiol. 2012, 160, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Falginella, L.; Castellarin, S.D.; Testolin, R.; Gambetta, G.A.; Morgante, M.; Di Gaspero, G. Expansion and subfunctionalisation of flavonoid 3’,5’-hydroxylases in the grapevine lineage. BMC Genom. 2010, 11, 562. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.E.; Spelt, C.E.; Mol, J.N.M. The chalcone synthase multigene family of Petunia hybrida (V30)-differential, light-regulated expression during flower development and uv-light induction. Plant Mol. Biol. 1989, 12, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Durbin, M.L.; McCaig, B.; Clegg, M.T. Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Mol. Biol. 2000, 42, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Ayabe, S.-I.; Akashi, T. Cytochrome P450s in flavonoid metabolism. Phytochem. Rev. 2006, 5, 271–282. [Google Scholar] [CrossRef]
- Inagaki, Y.; Johzuka-Hisatomi, Y.; Mori, T.; Takahashi, S.; Hayakawa, Y.; Peyachoknagul, S.; Ozeki, Y.; Iida, S. Genomic organization of the genes encoding dihydroflavonol 4-reductase for flower pigmentation in the Japanese and common morning glories. Gene 1999, 226, 181–188. [Google Scholar] [CrossRef]
- Hirner, A.A.; Veit, S.; Seitz, H.U. Regulation of anthocyanin biosynthesis in UV-A-irradiated cell cultures of carrot and in organs of intact carrot plants. Plant Sci. 2001, 161, 315–322. [Google Scholar] [CrossRef]
- Xu, Z.S.; Huang, Y.; Wang, F.; Song, X.; Wang, G.L.; Xiong, A.S. Transcript profiling of structural genes involved in cyanidin-based anthocyanin biosynthesis between purple and non-purple carrot (Daucus carota L.) cultivars reveals distinct patterns. BMC Plant Biol. 2014, 14, 262. [Google Scholar] [CrossRef]
- Tropf, S.; Lanz, T.; Rensing, S.A.; Schroder, J.; Schroder, G. Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J. Mol. Evol. 1994, 38, 610–618. [Google Scholar] [CrossRef]
- Helariutta, Y.; Kotilainen, M.; Elomaa, P.; Kalkkinen, N.; Bremer, K.; Teeri, T.H.; Albert, V.A. Duplication and functional divergence in the chalcone synthase gene family of Asteraceae: Evolution with substrate change and catalytic simplification. Proc. Natl. Acad. Sci. USA 1996, 93, 9033–9038. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Huang, J.; Gu, H.; Zhong, Y.; Yang, Z. Duplication and adaptive evolution of the chalcone synthase genes of Dendranthema (Asteraceae). Mol. Biol. Evol. 2002, 19, 1752–1759. [Google Scholar] [CrossRef][Green Version]
- Koes, R.E.; Spelt, C.E.; Reif, H.J.; van den Elzen, P.J.; Veltkamp, E.; Mol, J.N. Floral tissue of Petunia hybrida (V30) expresses only one member of the chalcone synthase multigene family. Nucleic Acids Res. 1986, 14, 5229–5239. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ryder, T.B.; Hedrick, S.A.; Bell, J.N.; Liang, X.W.; Clouse, S.D.; Lamb, C.J. Organization and differential activation of a gene family encoding the plant defense enzyme chalcone synthase in Phaseolus vulgaris. Mol. Gen. Genet. 1987, 210, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.D.; Rausher, M.D. Gene loss and parallel evolution contribute to species difference in flower color. Mol. Biol. Evol. 2011, 28, 2799–2810. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, K.; Taniguchi, M.; Tanaka, Y. Functional analysis of Antirrhinum kelloggii flavonoid 3’-hydroxylase and flavonoid 3’,5’-hydroxylase genes; critical role in flower color and evolution in the genus Antirrhinum. J. Plant Res. 2012, 125, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.D.; Wang, S.; Rausher, M.D. Functional evolution of an anthocyanin pathway enzyme during a flower color transition. Mol. Biol. Evol. 2013, 30, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.A.; Streisfeld, M.A.; Nutter, L.I.; Cross, K.A. The genetic basis of a rare flower color polymorphism in Mimulus lewisii provides insight into the repeatability of evolution. PLoS ONE 2013, 8, e81173. [Google Scholar] [CrossRef] [PubMed]
- Heppel, S.C.; Jaffe, F.W.; Takos, A.M.; Schellmann, S.; Rausch, T.; Walker, A.R.; Bogs, J. Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 2013, 82, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.J.; Chuang, Y.N.; Chiou, C.Y.; Chin, D.C.; Shen, F.Q.; Yeh, K.W. Methylation effect on chalcone synthase gene expression determines anthocyanin pigmentation in floral tissues of two Oncidium orchid cultivars. Planta 2012, 236, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Huang, H.; Wang, L.; Sun, Y.; Dai, S. Transcriptomics and metabolite analysis reveals the molecular mechanism of anthocyanin biosynthesis branch pathway in different Senecio cruentus cultivars. Front. Plant Sci. 2016, 7, 1307. [Google Scholar] [CrossRef] [PubMed]
- Zorenc, Z.; Veberic, R.; Slatnar, A.; Koron, D.; Miosic, S.; Chen, M.H.; Haselmair-Gosch, C.; Halbwirth, H.; Mikulic-Petkovsek, M. A wild ‘albino’ bilberry (Vaccinium myrtillus L.) from Slovenia shows three bottlenecks in the anthocyanin pathway and significant differences in the expression of several regulatory genes compared to the common blue berry type. PLoS ONE 2017, 12, e0190246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, X.; Zhan, Z.; Cao, L.; Zeng, A.; Chang, G.; Liang, Y. Transcriptome sequencing and metabolism analysis reveals the role of cyanidin metabolism in dark-red onion (Allium cepa L.) bulbs. Sci. Rep. 2018, 8, 14109. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, A.; Ohno, S.; Hosokawa, M.; Tatsuzawa, F.; Doi, M. Endogenous post-transcriptional gene silencing of flavone synthase resulting in high accumulation of anthocyanins in black dahlia cultivars. Planta 2013, 237, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Lou, Q.; Liu, Y.; Qi, Y.; Jiao, S.; Tian, F.; Jiang, L.; Wang, Y. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J. Exp. Bot. 2014, 65, 3157–3164. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Vickrey, T.L.; McNally, M.G.; Sato, S.J.; Clemente, T.E.; Mower, J.P. Assessing Anthocyanin Biosynthesis in Solanaceae as a Model Pathway for Secondary Metabolism. Genes 2019, 10, 559. https://doi.org/10.3390/genes10080559
Li Z, Vickrey TL, McNally MG, Sato SJ, Clemente TE, Mower JP. Assessing Anthocyanin Biosynthesis in Solanaceae as a Model Pathway for Secondary Metabolism. Genes. 2019; 10(8):559. https://doi.org/10.3390/genes10080559
Chicago/Turabian StyleLi, Zuo, Trisha L. Vickrey, Moira G. McNally, Shirley J. Sato, Tom Elmo Clemente, and Jeffrey P. Mower. 2019. "Assessing Anthocyanin Biosynthesis in Solanaceae as a Model Pathway for Secondary Metabolism" Genes 10, no. 8: 559. https://doi.org/10.3390/genes10080559
APA StyleLi, Z., Vickrey, T. L., McNally, M. G., Sato, S. J., Clemente, T. E., & Mower, J. P. (2019). Assessing Anthocyanin Biosynthesis in Solanaceae as a Model Pathway for Secondary Metabolism. Genes, 10(8), 559. https://doi.org/10.3390/genes10080559



