Modulating the Precursor and Terpene Synthase Supply for the Whole-Cell Biocatalytic Production of the Sesquiterpene (+)-Zizaene in a Pathway Engineered E. coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Engineering of Vectors and Strains
2.2. Evaluation of the Engineered Strains for the Production (+)-zizaene
2.3. Optimization of the Fermentation Conditions for the Production (+)-zizaene
2.4. Analytical Measurements
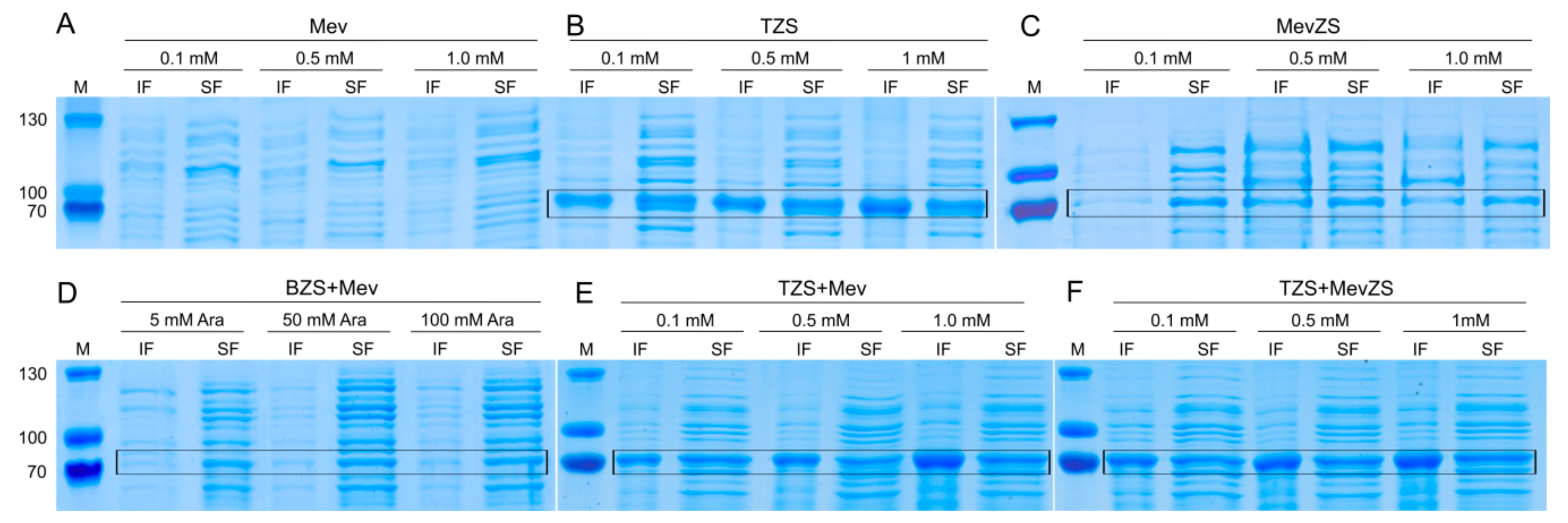
2.4.1. ZS Protein Gel Electrophoresis Analysis
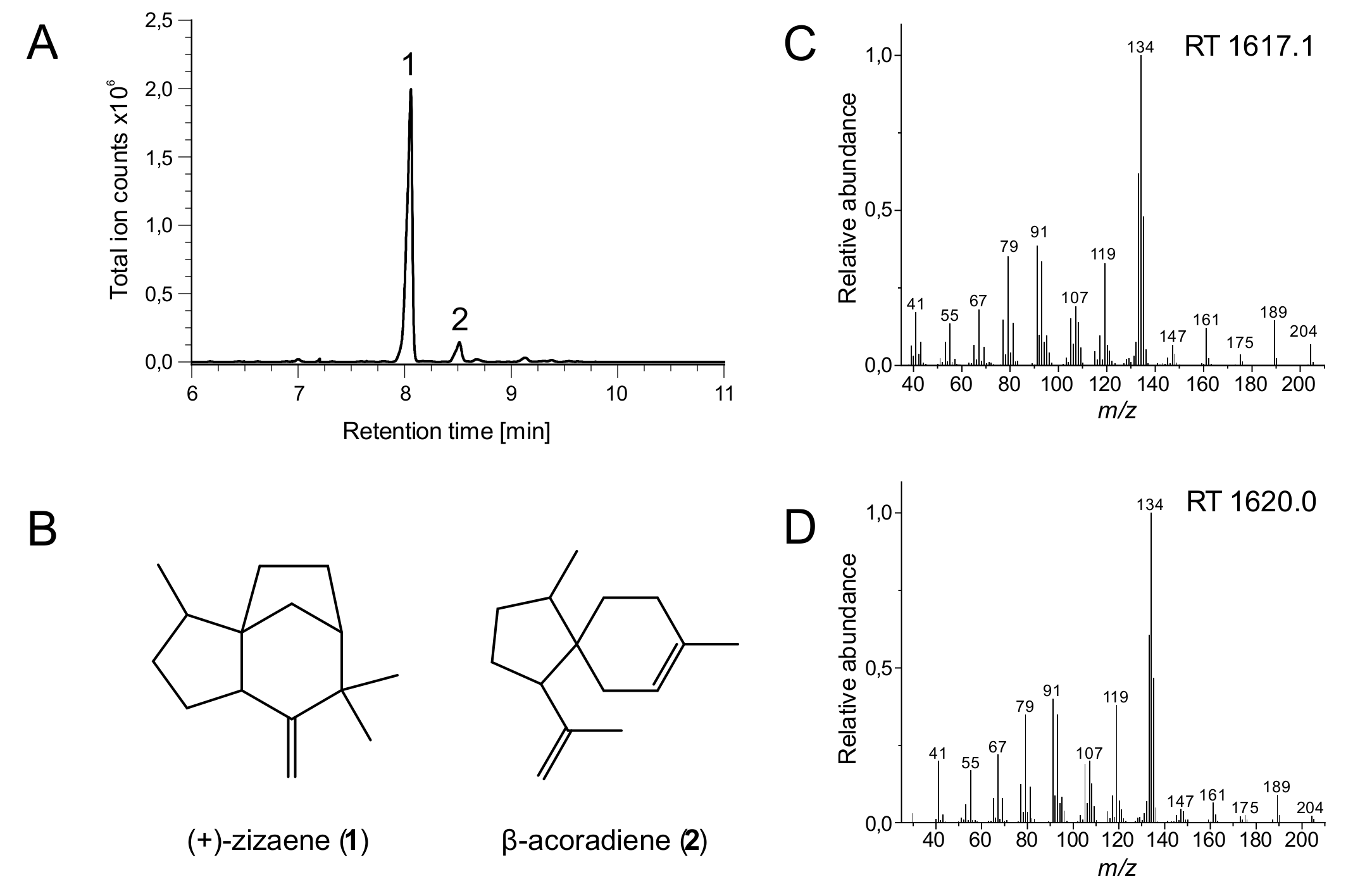
2.4.2. Identification and Measurement of (+)-zizaene by Gas Chromatography Analysis
3. Results
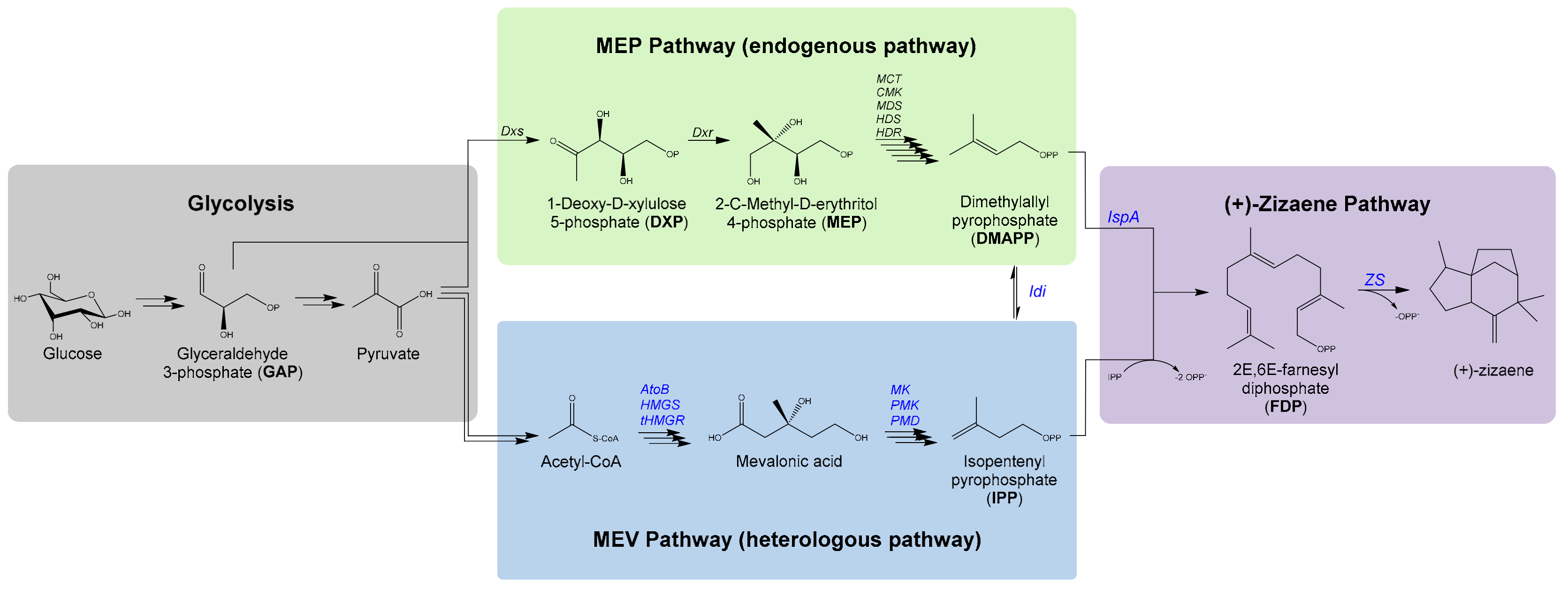
3.1. Engineering the Production of (+)-zizaene by the MEP Pathway
3.2. Improving the Titers of (+)-zizaene through the Mevalonate Pathway
3.3. Effect of Promoters on the Overexpression of the ZS Gene
3.4. Enhancing the ZS Supply by Engineering Multiple Copies of the ZS Gene
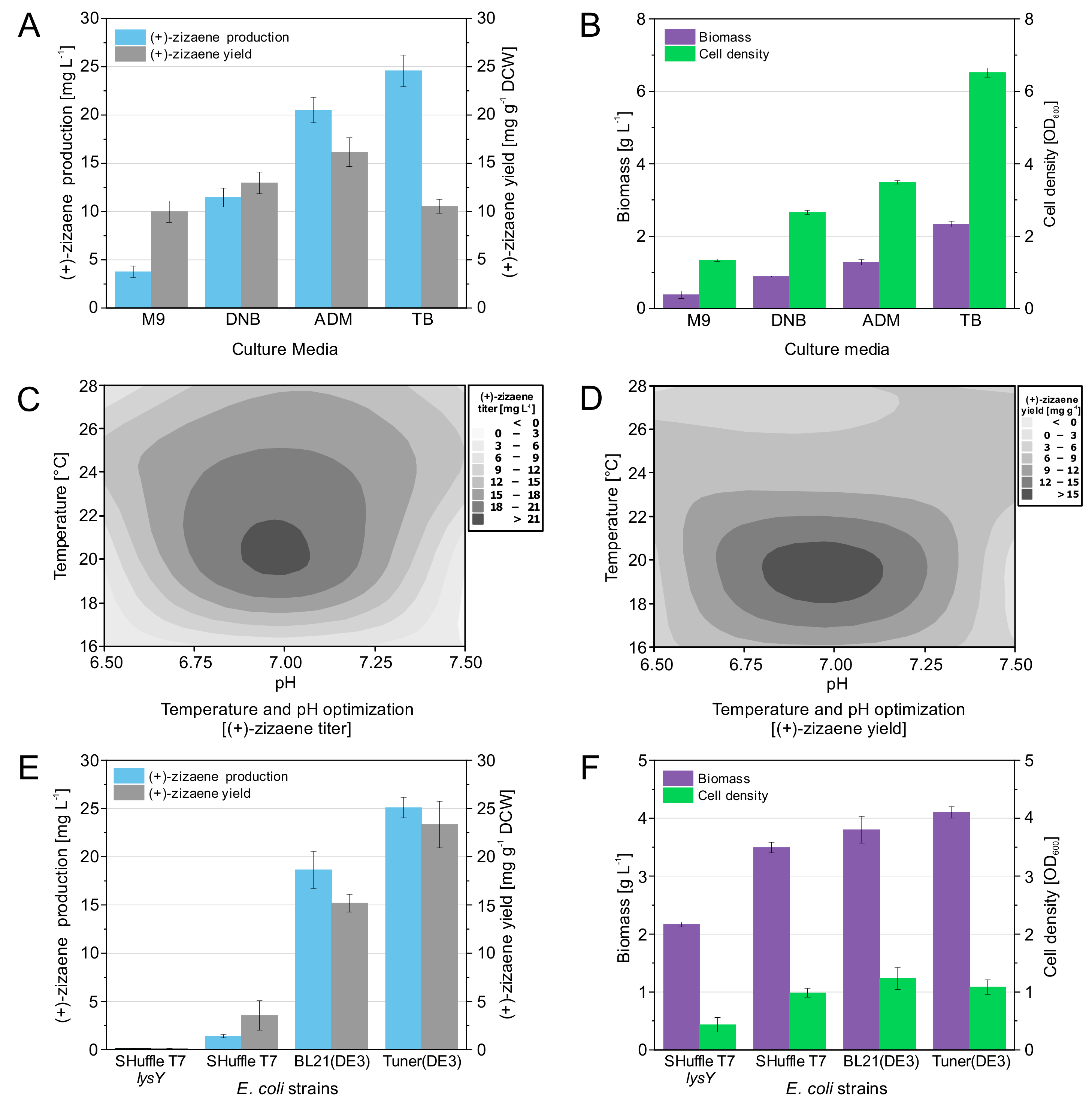
3.5. Optimization of Fermentation Conditions and Evaluation of E. coli Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carvalho, I.T.; Estevinho, B.N.; Santos, L. Application of microencapsulated essential oils in cosmetic and personal healthcare products—A review. Int. J. Cosmet. Sci. 2016, 38, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Hüsnü, K.; Başer, C.; Demirci, F. Chemistry of essential oils. In Flavours and Fragrances; Berger, R.G., Ed.; Springer-Verlag: Berlin, Germany, 2007; pp. 43–86. [Google Scholar]
- Belhassen, E.; Filippi, J.J.; Brévard, H.; Joulain, D.; Baldovini, N. Volatile constituents of vetiver: A review. Flavour Fragr. J. 2015, 30, 26–82. [Google Scholar] [CrossRef]
- Gnansounou, E.; Alves, C.M.; Raman, J.K. Multiple applications of vetiver grass—A review. Int. J. Environ. Sci. 2017, 2, 125–141. [Google Scholar]
- Sanganeria, P. Essential Oils Market Report Summer 2018; Report summer 2018; Ultra International B.V: Spijkenisse, The Netherlands, 2018; pp. 36–39. [Google Scholar]
- Vetiver oil market expected to reach $169 Million By 2022. Available online: https://www.grandviewresearch.com/press-release/global-vetiver-oil-market (accessed on 22 April 2019).
- Talansier, E.; Braga, M.E.M.; Rosa, P.T.V.; Paolucci-Jeanjean, D.; Meireles, M.A.A. Supercritical fluid extraction of vetiver roots: A study of SFE kinetics. J. Supercrit. Fluids 2008, 47, 200–208. [Google Scholar] [CrossRef]
- Akhila, A.; Rani, M. Chemical constituents and essential oil biogenesis in Vetiveria zizanioides. In Vetiveria: The Genus Vetiveria; Maffei, M., Ed.; Taylor & Francis: London, UK, 2002; pp. 80–117. ISBN 0-415-27586-5. [Google Scholar]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, S.; Frister, T.; Alemdar, S.; Li, Z.; Scheper, T.; Beutel, S. SUMO-fusion, purification, and characterization of a (+)-zizaene synthase from Chrysopogon zizanioides. Biochem. Biophys. Res. Commun. 2015, 458, 883–889. [Google Scholar] [CrossRef]
- Martin, V.J.J.; Yoshikuni, Y.; Keasling, J.D. The in vivo synthesis of plant sesquiterpenes by Escherichia coli. Biotechnol. Bioeng. 2001, 75, 497–503. [Google Scholar] [CrossRef]
- Nybo, S.E.; Saunder, J.; McCormick, S. Metabolic engineering of Escherichia coli for production of valerenadiene. J. Biotechnol. 2017, 262, 60–66. [Google Scholar] [CrossRef]
- Guan, Z.; Breazeale, S.D.; Raetz, C.R.H. Extraction and identification by mass spectrometry of undecaprenyl diphosphate-MurNAc-pentapeptide-GlcNAc from Escherichia coli. Anal. Biochem. 2005, 345, 336–339. [Google Scholar] [CrossRef]
- Okada, K.; Minehira, M.; Zhu, X.; Suzuki, K.; Nakagawa, T.; Matsuda, H.; Kawamukai, M. The ispB gene encoding octaprenyl diphosphate synthase is essential for growth of Escherichia coli. J. Bacteriol. 1997, 179, 3058–3060. [Google Scholar] [CrossRef]
- Kim, S.; Keasling, J.D. Nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 2001, 72, 408–415. [Google Scholar] [CrossRef]
- Sørensen, H.P.; Mortensen, K.K. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 2005, 115, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.; Pitera, D.; Withers, S.; Newman, J.; Keasling, J. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003, 21, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Yahya, P.P.; Ouellet, M.; Chan, R.; Mukhopadhyay, A.; Keasling, J.D.; Lee, T.S. Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun. 2011, 2, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Christianson, D.W. Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 2006, 106, 3412–3442. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, F.; Hartwig, S.; Scheper, T.; Beutel, S. Catalytical specificity, reaction mechanisms, and conformational changes during catalysis of the recombinant SUMO (+)-zizaene synthase from Chrysopogon zizanioides. ACS Omega 2019, 4, 6199–6209. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012; ISBN 978-1-936113-42-2. [Google Scholar]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Alonso-Gutierrez, J.; Chan, R.; Batt, T.S.; Adams, P.D.; Keasling, J.D.; Petzold, C.J.; Lee, T.S.; Adams, P.D. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab. Eng. 2013, 19, 33–41. [Google Scholar] [CrossRef]
- Li, Z.; Carstensen, B.; Rinas, U. Smart sustainable bottle (SSB) system for E. coli based recombinant protein production. Microb. Cell Fact. 2014, 13, 2–9. [Google Scholar] [CrossRef]
- Song, J.M.; Wei, D.Z. Production and characterization of cellulases and xylanases of Cellulosimicrobium cellulans grown in pretreated and extracted bagasse and minimal nutrient medium M9. Biomass Bioenergy 2010, 34, 1930–1934. [Google Scholar] [CrossRef]
- Rodríguez-Aparicio, L.B.; Reglero, A.; Ortiz, A.I.; Luengo, J.M. Effect of physical and chemical conditions on the production of colominic acid by E. coli in defined medium. Appl. Microbiol. Biotechnol. 1988, 27, 474–483. [Google Scholar] [CrossRef]
- Van Del Dool, H.; Kratz, P. A generalization of the retention index system including linear temperature programmed gas-liquid partition chromatography. J. Chromatogr. A 1963, 11, 463–471. [Google Scholar] [CrossRef]
- Wang, C.; Yoon, S.H.; Jang, H.J.; Chung, Y.R.; Kim, J.Y.; Choi, E.S.; Kim, S.W. Metabolic engineering of Escherichia coli for α-farnesene production. Metab. Eng. 2011, 13, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, S.; Frister, T.; Alemdar, S.; Li, Z.; Krings, U.; Berger, R.G.; Scheper, T.; Beutel, S. Expression, purification and activity assay of a patchoulol synthase cDNA variant fused to thioredoxin in Escherichia coli. Protein Expr. Purif. 2014, 97, 61–71. [Google Scholar] [CrossRef]
- Alemdar, S.; Hartwig, S.; Frister, T.; König, J.C.; Scheper, T.; Beutel, S. Heterologous expression, purification, and biochemical characterization of α-humulene synthase from Zingiber zerumbet Smith. Appl. Biochem. Biotechnol. 2016, 178, 474–489. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.D.; Marshall, J.; Chang, M.; Nowroozi, F.; Paradise, E.; Pitera, D.; Newman, K.L.; Keasling, J.D. High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol. Bioeng. 2006, 95, 684–691. [Google Scholar] [CrossRef]
- Wang, C.; Yoon, S.H.; Shah, A.A.; Chung, Y.R.; Kim, J.Y.; Choi, E.S.; Keasling, J.D.; Kim, S.W. Farnesol production from Escherichia coli by harnessing the exogenous mevalonate pathway. Biotechnol. Bioeng. 2010, 107, 421–429. [Google Scholar] [CrossRef]
- Korz, D.; Rinas, U.; Hellmuth, K.; Sanders, E.; Deckwer, W. Simple fed-batch technique for high cell density cultivation of Escherichia coli. J. Biotechnol. 1995, 39, 59–65. [Google Scholar] [CrossRef]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. In Advances in Biochemical Engineering/Biotechnology; Schrader, J., Bohlmann, J., Eds.; Springer International: Basel, Switzerland, 2015; pp. 63–106. [Google Scholar]
- Cao, Y.; Zhang, R.; Liu, W.; Zhao, G.; Niu, W.; Guo, J.; Xian, M.; Liu, H. Manipulation of the precursor supply for high-level production of longifolene by metabolically engineered Escherichia coli. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Kumar, S.; Jain, K.K.; Bhardwaj, K.N.; Chakraborty, S.; Kuhad, R.C. Multiple genes in a single host: Cost-effective production of bacterial laccase (cotA), pectate lyase (pel), and endoxylanase (xyl) by simultaneous expression and cloning in single vector in E. coli. PLoS ONE 2015, 10, 1–15. [Google Scholar] [CrossRef]
- Samuelson, J. Bacterial Systems. In Production of Membrane Proteins: Strategies for Expression and Isolation; Robinson, A.S., Ed.; Wiley-VCH Verlag GmbH & Co: Weinheim, Germany, 2011; pp. 11–35. ISBN 9780470871942. [Google Scholar]
- Tegel, H.; Ottosson, J.; Hober, S. Enhancing the protein production levels in Escherichia coli with a strong promoter. FEBS J. 2011, 278, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Xu, H.; Yu, H. Significantly enhanced production of isoprene by ordered coexpression of genes dxs, dxr, and idi in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 2357–2365. [Google Scholar] [CrossRef] [PubMed]
- Du, F.L.; Yu, H.L.; Xu, J.H.; Li, C.X. Enhanced limonene production by optimizing the expression of limonene biosynthesis and MEP pathway genes in E. coli. Bioresour. Bioprocess. 2014, 1, 1–10. [Google Scholar] [CrossRef]
- Tan, S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 2001, 21, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Perrakis, A.; Romier, C. Assembly of protein complexes by coexpression in prokaryotic and eukaryotic hosts: An overview. In Structural Proteomics High-Throughput Methods; Kobe, B., Guss, M., Huber, T., Eds.; Humana Press: New York, NY, USA, 2008; ISBN 9781588298096. [Google Scholar]
- Jones, K.L.; Kim, S.W.; Keasling, J.D. Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab. Eng. 2000, 2, 328–338. [Google Scholar] [CrossRef]
- Mathis, J.R.; Back, K.; Starks, C.; Noel, J.; Poulter, C.D.; Chappel, J. Pre-steady-state study of recombinant sesquiterpene cyclases. Biochemistry 1997, 36, 8340–8348. [Google Scholar] [CrossRef]
- Wu, W.; Liu, F.; Davis, R.W. Engineering Escherichia coli for the production of terpene mixture enriched in caryophyllene and caryophyllene alcohol as potential aviation fuel compounds. Metab. Eng. Commun. 2018, 6, 13–21. [Google Scholar] [CrossRef]
- Huang, X.; Xiao, Y.; Köllner, T.G.; Zhang, W.; Wu, J.; Wu, J.; Guo, Y.; Zhang, Y. Identification and characterization of (E)-β-caryophyllene synthase and α/β-pinene synthase potentially involved in constitutive and herbivore-induced terpene formation in cotton. Plant Physiol. Biochem. 2013, 73, 302–308. [Google Scholar] [CrossRef]
- Chubukov, V.; Desmarais, J.J.; Wang, G.; Chan, L.J.G.; Baidoo, E.E.K.; Petzold, C.J.; Keasling, J.D.; Mukhopadhyay, A. Engineering glucose metabolism of Escherichia coli under nitrogen starvation. npj Syst. Biol. Appl. 2017, 3, 1–7. [Google Scholar] [CrossRef]
- Sørensen, H.P.; Mortensen, K.K. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Fact. 2005, 8, 1–8. [Google Scholar] [CrossRef]
- Strandberg, L.; Enfors, S.O. Factors influencing inclusion body formation in the production of a fused protein in Escherichia coli. Appl. Environ. Microbiol. 1991, 57, 1669–1674. [Google Scholar] [PubMed]


| Plasmid Reference | Plasmid Name | Description (Origin of Replication, Promoter, Antibiotic Resistance and Genes) | Reference |
|---|---|---|---|
| pETZS | pETSUMO::ZIZ(co) | pBR322, PT7, Kan, harboring the codon-optimized SUMO-fused ZS gene from Ch. zizanioides | [10] |
| pMev | pBbA5c-MevT(CO)-MBIS (CO, ispA) | p15A, PlacUV5, Cam, harboring the mevalonate pathway genes: AtoB, HMGS, tHMGR, MK, PMK, PMD, Idi and IspA*. | [18] |
| pJbei-6411 | pJbei-6411 | pBBR1, PBAD, Kan, harboring the arabinose operon with the cytochrome P450 (CYP153A6) from Sphingomonas sp. | [23] |
| pMevZS | pBbA5c-MevT(CO)-MBIS (CO, ispA)-SUMO::ZIZ(co) | p15A, PlacUV5, Cam, harboring the mevalonate pathway genes* and the codon-optimized SUMO-fused ZS gene from Ch. zizanioides | This study |
| pJbeiZS | pJbei-6411-SUMO::ZIZ(co) | pBBR1, PBAD, Kan, harboring the arabinose operon and the codon-optimized SUMO-fused ZS gene from Ch. zizanioides | This study |
| Strains | Genotype/Description | Reference | |
| E. coli TOP10 | F- mcrA (mrr-hsdRMS-mcrBC) ϕ80lacZ ∆M15 ∆lacX74 recA1 ara∆139 (ara-leu)7697 galU galK rpsL (StrR) endA1 nupG | IBA | |
| E. coli Bl21(DE3) | F- ompT hsdSB (rB-mB-) gal dcm (DE3) | Novagen | |
| E. coli Tuner(DE3) | F- ompT hsdSB (rB-mB-) gal dcm lacY1(DE3) | Novagen | |
| E. coli NEB 10-beta | Δ(ara-leu) 7697 araD139 fhuA ΔlacX74 galK16 galE15 e14- ϕ80dlacZΔM15 recA1 relA1 endA1 nupG rpsL (StrR) rph spoT1 Δ(mrr-hsdRMS-mcrBC) | NEB | |
| E. coli SHuffle T7 | fhuA2 lacZ::T7 gene1 (lon) ompT ahpC gal λatt::pNEB3-r1-cDsbC (SpecR, lacIq) ΔtrxB sulA11 R(mcr-73::miniTn10--TetS)2 (dcm) R(zgb-210::Tn10 --TetS) endA1 ∆gor ∆(mcrC-mrr)114::IS10 | NEB | |
| E. coli SHuffle T7 lysY | MiniF lysY (CamR)/fhuA2 lacZ::T7 gene1 (lon) ompT ahpC gal λatt::pNEB3-r1-cDsbC (SpecR, lacIq) ΔtrxB sulA11 R(mcr-73::miniTn10--TetS)2 (dcm) R(zgb-210::Tn10 --TetS) endA1 ∆gor ∆(mcrC-mrr)114::IS10 | NEB | |
| Mev | E. coli Bl21(DE3) harboring pMev | This study | |
| ZS | E. coli Bl21(DE3) harboring pETZS | This study | |
| MevZS | E. coli Bl21(DE3) harboring pMevZS | This study | |
| BZS+Mev | E. coli TOP10 harboring pMev and pJbeiZS | This study | |
| TZS+Mev | E. coli Bl21(DE3) harboring pMev and pETZS | This study | |
| TZS+MevZS | E. coli Bl21(DE3) harboring pMevZS and pETZS | This study | |
| T-TZS+MevZS | E. coli Tuner(DE3) harboring pMevZS and pETZS | This study | |
| SH-TZS+MevZS | E. coli SHuffle T7 harboring pMevZS and pETZS | This study | |
| SHL-TZS+MevZS | E. coli SHuffle T7 lysY harboring pMevZS and pETZS | This study | |
| Primers | Description | Reference |
|---|---|---|
| ZS-F-Mev | catccagcgtaataaataagGATCTAGGAGGTAATGGGCAGCAGCCATCATC | This study |
| ZS-R-Mev | gagatccttactcgagtttgTCACACCGGAATCAGATTTACATAC | This study |
| pJ6411-F-ZS | cccaagattacgtacattg | This study |
| pJ6411-R-ZS | ttctttatcctcctagatcttttgaattcccaaaaaaacg | This study |
| ZS-F-pJ6411 | ccgtttttttgggaattcaaaagatctaggaggataaagaaATGGGCAGCAGCCATCATC | This study |
| ZS-R-pJ6411 | tcaatgtacgtaatcttgggTCACACCGGAATCAGATTTACATAC | This study |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilar, F.; Scheper, T.; Beutel, S. Modulating the Precursor and Terpene Synthase Supply for the Whole-Cell Biocatalytic Production of the Sesquiterpene (+)-Zizaene in a Pathway Engineered E. coli. Genes 2019, 10, 478. https://doi.org/10.3390/genes10060478
Aguilar F, Scheper T, Beutel S. Modulating the Precursor and Terpene Synthase Supply for the Whole-Cell Biocatalytic Production of the Sesquiterpene (+)-Zizaene in a Pathway Engineered E. coli. Genes. 2019; 10(6):478. https://doi.org/10.3390/genes10060478
Chicago/Turabian StyleAguilar, Francisco, Thomas Scheper, and Sascha Beutel. 2019. "Modulating the Precursor and Terpene Synthase Supply for the Whole-Cell Biocatalytic Production of the Sesquiterpene (+)-Zizaene in a Pathway Engineered E. coli" Genes 10, no. 6: 478. https://doi.org/10.3390/genes10060478
APA StyleAguilar, F., Scheper, T., & Beutel, S. (2019). Modulating the Precursor and Terpene Synthase Supply for the Whole-Cell Biocatalytic Production of the Sesquiterpene (+)-Zizaene in a Pathway Engineered E. coli. Genes, 10(6), 478. https://doi.org/10.3390/genes10060478






