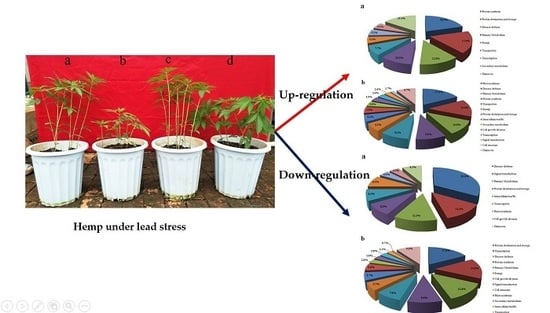
Protein Changes in Response to Lead Stress of Lead-Tolerant and Lead-Sensitive Industrial Hemp Using SWATH Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation
2.2. Determination of Physiological and Biochemical Indexes
2.3. Protein Preparation
2.4. Fractionation by High pH Reverse Phase Chromatography
2.5. LC-ESI-MS/MS Analysis
2.6. Library Generation and SWATH Data File Process
3. Results and Discussion
3.1. Physiological Indicators under Pb Stress
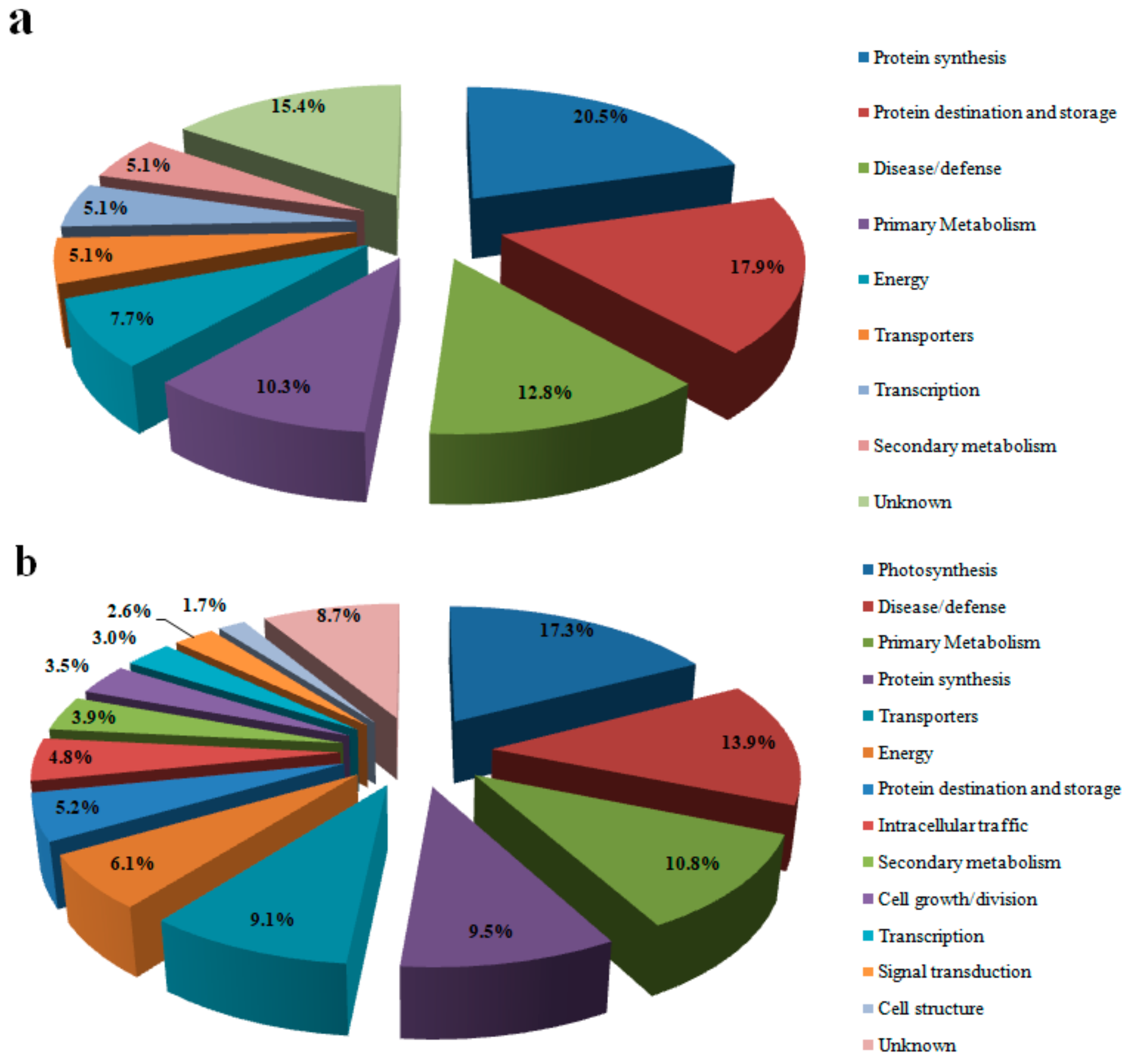
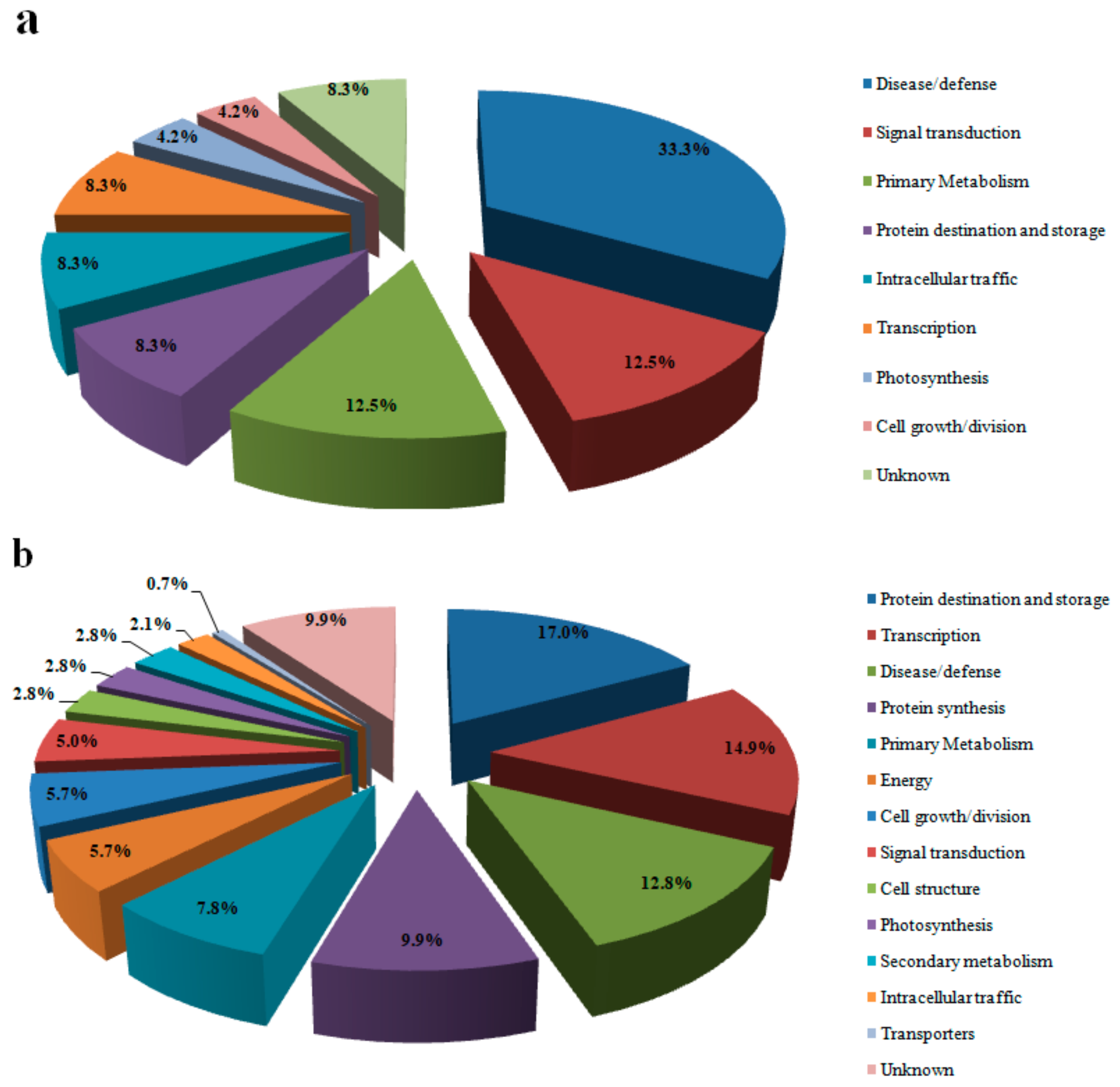
3.2. Protein Identification and Analysis
3.3. Proteins Related to Energy Metabolism
3.4. Proteins Related to Photosynthesis
3.5. Primary and Secondary Metabolism-Related Proteins
3.6. Protein Related to Disease/Defense
3.7. Proteins Related to Protein Synthesis and Transcription
3.8. Proteins Related to Transport and Signal Transduction
3.9. Proteins Related to Protein Destination and Storage
3.10. Proteins Related to Cell Growth/Division, Cell Structure and Intracellular Traffic
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kumar, P.B.; Dushenkov, V.; Motto, H.; Raskin, I. Phytoextraction: The use of plants to remove heavy metals from soils. Environ. Sci. Technol. 1995, 29, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.P. Lead pollution—A risk factor for cardiovascular disease in Asian developing countries. Pak. J. Pharm. Sci. 2012, 25, 289–294. [Google Scholar]
- Michalak, E.; Wierzbicka, M. Differences in lead tolerance between Allium cepa plants developing from seeds and bulbs. Plant Soil 1998, 199, 251–260. [Google Scholar] [CrossRef]
- Cappa, J.J.; Pilon-Smits, E.A.H. Evolutionary aspects of elemental hyperaccumulation. Planta 2014, 239, 267–275. [Google Scholar] [CrossRef]
- Gallego, S.M.; Pena, L.B.; Barcia, R.A.; Azpilicueta, C.E.; Lannone, M.F.; Rosales, E.P.; Zawoznik, M.S.; Groppa, M.D.; Benavides, M.P. Unravelling cadmium toxicity and tolerance in plants: Insight into regulatory mechanisms. Environ. Exp. Bot. 2012, 83, 33–46. [Google Scholar] [CrossRef]
- Milner, M.J.; Kochian, L.V. Investigating heavy-metal hyperaccumulation using Thlaspi caerulescens as a model system. Ann. Bot.-Lond. 2008, 102, 3–13. [Google Scholar] [CrossRef]
- Van der Ent, A.; Baker, A.J.M.; Reeves, R.D.; Pollard, A.J.; Schat, H. Hyperaccumulators of metal and metalloid trace elements: Facts and fiction. Plant Soil 2013, 362, 319–334. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals-Concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Arazi, T.; Sunkar, R.; Kaplan, B.; Fromm, H. A tobacco plasma membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. Plant J. 1999, 20, 171–182. [Google Scholar] [CrossRef]
- Song, W.Y.; Sohn, E.J.; Martinoia, E.; Lee, Y.J.; Yang, Y.Y.; Jasinski, M.; Forestier, C.; Hwang, I.; Lee, Y. Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat. Biotechnol. 2003, 21, 914–919. [Google Scholar] [CrossRef]
- Gisbert, C.; Ros, R.; De Haro, A.; Walker, D.J.; Bernal, M.P.; Serrano, R.; Navarro-Avino, J. A plant genetically modified that accumulates Pb is especially promising for phytoremediation. Biochem. Biophys. Res. Commun. 2003, 303, 440–445. [Google Scholar] [CrossRef]
- Liu, F.H.; Hu, H.R.; Du, G.H.; Deng, G.; Yang, Y. Ethnobotanical research on origin, cultivation, distribution and utilization of hemp (Cannabis sativa L.) in China. Indian J. Tradit. Knowl. 2017, 16, 235–242. [Google Scholar]
- Xu, Y.P.; Chen, X.; Guo, M.B.; Zhang, Q.Y.; He, X.Y.; Guo, H.Y.; Yang, M. The effects of 4 heavy metals stress on seed germination of industrial hemp. J. West China For. Sci. 2014, 4, 78–82. [Google Scholar]
- Angelova, V.; Ivanova, R.; Delibaltova, V.; Ivanov, K. Bio-accumulation and distribution of heavy metals in fibre crops (flax, cotton and hemp). Ind. Crop. Prod. 2004, 19, 197–205. [Google Scholar] [CrossRef]
- Jin, N.N.; Su, W.H.; Zhang, G.F.; Zhou, R. Cannabis sativa cultivation experiments on heavy metal zinc contaminated land. J. Anhui Agric. Sci. 2013, 5, 1994–1996. [Google Scholar]
- Linger, P.; Müssig, J.; Fischer, H.; Kobert, J. Industrial hemp (Cannabis sativa L.) growing on heavy metal contaminated soil: Fibre quality and phytoremediation potential. Ind. Crop. Prod. 2002, 16, 33–42. [Google Scholar] [CrossRef]
- Wang, X.K. Experimental Principle and Technology of Plant Physiology and Biochemistry, 3rd ed.; Higher Education Press: Beijing, China, 2015. [Google Scholar]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Bevan, M.; Bancroft, I.; Bent, E.; Love, K.; Goodman, H.; Dean, C.; Bergkamp, R.; Dirkse, W.; van Staveren, M.; Stiekema, W.; et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998, 391, 485–488. [Google Scholar]
- Romanowska, E.; Pokorska, B.; Siedlecka, M. The effects of oligomycin on content of adenylates in mesophyll protoplasts, chloroplasts and mitochondria from Pb2+ treated pea and barley leaves. Acta Physiol. Plant. 2005, 27, 29–36. [Google Scholar] [CrossRef]
- Romanowska, E.; Wroblewska, B.; Drozak, A.; Siedlecka, A. High light intensity protects photosynthetic apparatus of pea plants against exposure to lead. Plant Physiol. Biochem. 2006, 44, 387–394. [Google Scholar] [CrossRef]
- Dzeja, P.; Terzic, A. Adenylate Kinase and AMP Signaling Networks: Metabolic Monitoring, Signal Communication and Body Energy Sensing. Int. J. Mol. Sci. 2009, 10, 1729–1772. [Google Scholar] [CrossRef] [PubMed]
- Brandt, U. Energy converting NADH: Quinone oxidoreductase (Complex I). Annu. Rev. Biochem. 2006, 75, 69–92. [Google Scholar] [CrossRef]
- Wu, J.; Steinebrunner, I.; Sun, Y.; Butterfield, T.; Torres, J.; Arnold, D.; Gonzalez, A.; Jacob, F.; Reichler, S.; Roux, S.J. Apyrases (nucleoside triphosphate-diphosphohydrolases) play a key role in growth control in Arabidopsis. Plant Physiol. 2007, 144, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Clark, G.; Fraley, D.; Steinebrunner, I.; Cervantes, A.; Onyirimba, J.; Liu, A.; Torres, J.; Tang, W.Q.; Kim, J.; Roux, S.J. Extracellular Nucleotides and Apyrases Regulate Stomatal Aperture in Arabidopsis. Plant Physiol. 2011, 156, 1740–1753. [Google Scholar] [CrossRef]
- Granot, D.; Kelly, G.; Stein, O.; David-Schwartz, R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 2014, 65, 809–819. [Google Scholar] [CrossRef]
- Zorb, C.; Schmitt, S.; Muhling, K.H. Proteomic changes in maize roots after short-term adjustment to saline growth conditions. Proteomics 2010, 10, 4441–4449. [Google Scholar] [CrossRef]
- Fulda, S.; Mikkat, S.; Stegmann, H.; Horn, R. Physiology and proteomics of drought stress acclimation in sunflower (Helianthus annuus L.). Plant Biol. 2011, 13, 632–642. [Google Scholar] [CrossRef]
- Guglielminetti, L.; Morita, A.; Yamaguchi, J.; Loreti, E.; Perata, P.; Alpi, A. Differential expression of two fructokinases in Oryza sativa seedlings grown under aerobic and anaerobic conditions. J. Plant Res. 2006, 119, 351–356. [Google Scholar] [CrossRef]
- Bailey, J.M.; Fishman, P.H.; Pentchev, P.G. Mutarotase in higher plants: Distribution and properties. Science 1966, 152, 1270–1272. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Flexas, J.; Bota, J.; Loreto, F.; Cornic, G.; Sharkey, T.D. Diffusive and metabolic limitations to photosynthesis under drought and salinity in C(3) plants. Plant Biol. 2004, 6, 269–279. [Google Scholar] [CrossRef]
- Sengar, R.S.; Gautam, M.; Sengar, R.S.; Garg, S.K.; Sengar, K.; Chaudhary, R. Lead Stress Effects on Physiobiochemical Activities of Higher Plants. Rev. Environ. Contam. Toxicol. 2008, 196, 73–93. [Google Scholar]
- Xiong, Z.T.; Zhao, F.; Li, M.J. Lead toxicity in Brassica pekinensis rupr.: Effect on nitrate assimilation and growth. Environ. Toxicol. 2006, 21, 147–153. [Google Scholar] [CrossRef]
- Fromme, P.; Mathis, P. Unraveling the Photosystem I reaction center: A history, or the sum of many efforts. Photosynth. Res. 2004, 80, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Loll, B.; Kern, J.; Saenger, W.; Zouni, A.; Biesiadka, J. Towards complete cofactor arrangement in the 3.0 angstrom resolution structure of photosystem II. Nature 2005, 438, 1040–1044. [Google Scholar] [CrossRef]
- Huang, H.; Yang, M.; Su, Y.E.; Qu, L.J.; Deng, X.W. Arabidopsis Atypical Kinases ABC1K1 and ABC1K3 Act Oppositely to Cope with Photodamage Under Red Light. Mol. Plant 2015, 8, 1122–1124. [Google Scholar] [CrossRef][Green Version]
- Yang, S.G.; Zeng, X.Q.; Li, T.; Liu, M.; Zhang, S.C.; Gao, S.J.; Wang, Y.Q.; Peng, C.L.; Li, L.; Yang, C.W. AtACDO1, an ABC1-like kinase gene, is involved in chlorophyll degradation and the response to photooxidative stress in Arabidopsis. J. Exp. Bot. 2012, 63, 3959–3973. [Google Scholar] [CrossRef] [PubMed]
- Coschigano, K.T.; Melo-Oliveira, R.; Lim, J.; Coruzzi, G.M. Arabidopsis gls mutants and distinct Fd-GOGAT genes. Implications for photorespiration and primary nitrogen assimilation. Plant Cell 1998, 10, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Assche, F.; Clijsters, H. Effects of metals on enzyme activity in plants. Plant Cell Environ. 1990, 13, 195–206. [Google Scholar] [CrossRef]
- Jaimes, R.; Kuzmiak-Glancy, S.; Brooks, D.M.; Swift, L.M.; Posnack, N.G.; Kay, M.W. Functional response of the isolated, perfused normoxic heart to pyruvate dehydrogenase activation by dichloroacetate and pyruvate. Pflug. Arch. Eur. J. Physiol. 2016, 468, 131–142. [Google Scholar] [CrossRef]
- Lam, H.M.; Wong, P.; Chan, H.K.; Yam, K.M.; Chen, L.; Chow, C.M.; Coruzzi, G.M. Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol. 2003, 132, 926–935. [Google Scholar] [CrossRef]
- Sampedro, J.; Gianzo, C.; Iglesias, N.; Guitian, E.; Revilla, G.; Zarra, I. AtBGAL10 Is the Main Xyloglucan β-Galactosidase in Arabidopsis, and Its Absence Results in Unusual Xyloglucan Subunits and Growth Defects. Plant Physiol. 2012, 158, 1146–1157. [Google Scholar] [CrossRef]
- Leasure, C.D.; Tong, H.Y.; Hou, X.W.; Shelton, A.; Minton, M.; Esquerra, R.; Roje, S.; Hellmann, H.; He, Z.H. root uv-b sensitive Mutants Are Suppressed by Specific Mutations in ASPARTATE AMINOTRANSFERASE2 and by Exogenous Vitamin B6. Mol. Plant 2011, 4, 759–770. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tan, Y.F.; O’Toole, N.; Taylor, N.L.; Millar, A.H. Divalent Metal Ions in Plant Mitochondria and Their Role in Interactions with Proteins and Oxidative Stress-Induced Damage to Respiratory Function. Plant Physiol. 2010, 152, 747–761. [Google Scholar] [CrossRef]
- Marschner, H.; Marschner, P. Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Elsevier: London, UK; Academic Press: Waltham, MA, USA, 2012. [Google Scholar]
- Cheyns, K.; Peeters, S.; Delcourt, D.; Smolders, E. Lead phytotoxicity in soils and nutrient solutions is related to lead induced phosphorus deficiency. Environ. Pollut. 2012, 164, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.C.; Song, H.; Liu, H.W.; Lu, P.H. Current development in isoprenoid precursor biosynthesis and regulation. Curr. Opin. Chem. Biol. 2013, 17, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.K.; Nicoloso, F.T.; Schetinger, M.R.C.; Rossato, L.V.; Pereira, L.B.; Castro, G.Y.; Srivastava, S.; Tripathi, R.D. Antioxidant defense mechanism in hydroponically grown Zea mays seedlings under moderate lead stress. J. Hazard. Mater. 2009, 172, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K. Heavy metals toxicity in plants: An overview on the role of glutathione and phytochelatins in heavy metal stress tolerance of plants. S. Afr. J. Bot. 2010, 76, 167–179. [Google Scholar] [CrossRef]
- Vellosillo, T.; Martinez, M.; Lopez, M.A.; Vicente, J.; Cascon, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 2007, 19, 831–846. [Google Scholar] [CrossRef]
- Bannenberg, G.; Martinez, M.; Hamberg, M.; Castresana, C. Diversity of the Enzymatic Activity in the Lipoxygenase Gene Family of Arabidopsis thaliana. Lipids 2009, 44, 85–95. [Google Scholar] [CrossRef]
- Deponte, M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. BBA-Gen. Subj. 2013, 1830, 3217–3266. [Google Scholar] [CrossRef] [PubMed]
- Marshall, S.D.G.; Putterill, J.J.; Plummer, K.M.; Newcomb, R.D. The carboxylesterase gene family from Arabidopsis thaliana. J. Mol. Evol. 2003, 57, 487–500. [Google Scholar]
- Smith-Harding, T.J.; Beardall, J.; Mitchell, J.G. The role of external carbonic anhydrase in photosynthesis during growth of the marine diatom Chaetoceros muelleri. J. Phycol. 2017, 53, 1159–1170. [Google Scholar] [CrossRef]
- Riemenschneider, A.; Wegele, R.; Schmidt, A.; Papenbrock, J. Isolation and characterization of a D-cysteine desulfhydrase protein from Arabidopsis thaliana. FEBS J. 2005, 272, 1291–1304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lin, F.; Mao, T.L.; Nie, J.N.; Yan, M.; Yuan, M.; Zhang, W.H. Phosphatidic Acid Regulates Microtubule Organization by Interacting with MAP65-1 in Response to Salt Stress in Arabidopsis. Plant Cell 2012, 24, 4555–4576. [Google Scholar] [CrossRef]
- Song, W.Y.; Martinoia, E.; Lee, J.; Kim, D.; Kim, D.Y.; Vogt, E.; Shim, D.; Choi, K.S.; Hwang, I.; Lee, Y. A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 2004, 135, 1027–1039. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Choi, K.S.; Kim, D.Y.; Geisler, M.; Park, J.; Vincenzetti, V.; Schellenberg, M.; Kim, S.H.; Lim, Y.P.; Noh, E.W.; et al. Arabidopsis PCR2 Is a Zinc Exporter Involved in Both Zinc Extrusion and Long-Distance Zinc Transport. Plant Cell 2010, 22, 2237–2252. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.Y.; Ni, Z.F.; Du, J.K.; Wang, X.L.; Wu, H.Y.; Sun, Q.X. Isolation and characterization of 15 genes encoding ribosomal proteins in wheat (Triticum aestivum L.). Plant Sci. 2006, 170, 579–586. [Google Scholar] [CrossRef]
- Cheng, X.; Deng, G.; Su, Y.; Liu, J.J.; Yang, Y.; Du, G.H.; Chen, Z.Y.; Liu, F.H. Protein mechanisms in response to NaCl-stress of salt-tolerant and salt-sensitive industrial hemp based on iTRAQ technology. Ind. Crop. Prod. 2016, 83, 444–452. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Networks in Response to Abiotic Stresses in Arabidopsis and Grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef]
- Eisenhut, M.; Planchais, S.; Cabassa, C.; Guivarc’h, A.; Justin, A.M.; Taconnat, L.; Renou, J.P.; Linka, M.; Gagneul, D.; Timm, S.; et al. Arabidopsis A BOUT DE SOUFFLE is a putative mitochondrial transporter involved in photorespiratory metabolism and is required for meristem growth at ambient CO2 levels. Plant J. 2013, 73, 836–849. [Google Scholar] [CrossRef]
- Kaldenhoff, R.; Fischer, M. Functional aquaporin diversity in plants. BBA-Biomembr. 2006, 1758, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Peterman, T.K.; Ohol, Y.M.; McReynolds, L.J.; Luna, E.J. Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 2004, 136, 3080–3094, discussion 3001—3082. [Google Scholar] [CrossRef]
- Picault, N.; Palmieri, L.; Pisano, I.; Hodges, M.; Palmieri, F. Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 2002, 277, 24204–24211. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Miao, Q.; Sun, D.; Yang, G.; Wu, C.; Huang, J.; Zheng, C. The mitochondrial phosphate transporters modulate plant responses to salt stress via affecting ATP and gibberellin metabolism in Arabidopsis thaliana. PLoS ONE 2012, 7, e43530. [Google Scholar] [CrossRef] [PubMed]
- Peyro, M.; Soheilypour, M.; Lee, B.L.; Mofrad, M.R.K. Evolutionarily Conserved Sequence Features Regulate the Formation of the FG Network at the Center of the Nuclear Pore Complex. Sci. Rep. 2015, 5, 15795. [Google Scholar] [CrossRef]
- Peters, R. Introduction to nucleocytoplasmic transport: Molecules and mechanisms. Methods Mol. Biol. 2006, 322, 235–258. [Google Scholar]
- Li, J.; Zhao-Hui, C.; Batoux, M.; Nekrasov, V.; Roux, M.; Chinchilla, D.; Zipfel, C.; Jones, J.D.G. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. USA 2009, 106, 15973–15978. [Google Scholar] [CrossRef]
- Saijo, Y.; Tintor, N.; Lu, X.L.; Rauf, P.; Pajerowska-Mukhtar, K.; Haweker, H.; Dong, X.N.; Robatzek, S.; Schulze-Lefert, P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009, 28, 3439–3449. [Google Scholar] [CrossRef]
- Wang, W.H.; Yi, X.Q.; Han, A.D.; Liu, T.W.; Chen, J.; Wu, F.H.; Dong, X.J.; He, J.X.; Pei, Z.M.; Zheng, H.L. Calcium-sensing receptor regulates stomatal closure through hydrogen peroxide and nitric oxide in response to extracellular calcium in Arabidopsis. J. Exp. Bot. 2012, 63, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.H.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef]
- Guo, H.Q.; Li, L.; Ye, H.X.; Yu, X.F.; Algreen, A.; Yin, Y.H. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 7648–7653. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.E.; Hemmingsen, S.M. Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones 2001, 6, 190–200. [Google Scholar] [CrossRef]
- Haussuhl, K.; Andersson, B.; Adamska, I. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 2001, 20, 713–722. [Google Scholar]
- Wetzel, C.M.; Harmacek, L.D.; Yuan, L.H.; Wopereis, J.L.; Chubb, R.; Turini, P. Loss of chloroplast protease SPPA function alters high light acclimation processes in Arabidopsis thaliana L. (Heynh.). J. Exp. Bot. 2009, 60, 1715–1727. [Google Scholar] [CrossRef]
- Rautengarten, C.; Steinhauser, D.; Bussis, D.; Stintzi, A.; Schaller, A.; Kopka, J.; Altmann, T. Inferring hypotheses on functional relationships of genes: Analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput. Biol. 2005, 1, e40. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef]
- Michaels, S.D.; He, Y.H.; Scortecci, K.C.; Amasino, R.M. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 10102–10107. [Google Scholar] [CrossRef]
- Johnson, K.L.; Jones, B.J.; Bacic, A.; Schultz, C.J. The fasciclin-like arabinogalactan proteins of arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 2003, 133, 1911–1925. [Google Scholar] [CrossRef]
- Baumberger, N.; Steiner, M.; Ryser, U.; Keller, B.; Ringli, C. Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 2003, 35, 71–81. [Google Scholar] [CrossRef]
- Liu, D.; Li, T.Q.; Jin, X.F.; Yang, X.E.; Islam, E.; Mahmood, Q. Lead induced changes in the growth and antioxidant metabolism of the lead accumulating and non-accumulating ecotypes of Sedum alfredii. J. Integr. Plant Biol. 2008, 50, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Alford, E.R.; Pilon-Smits, E.A.H.; Paschke, M.W. Metallophytes-a view from the rhizosphere. Plant Soil 2010, 337, 33–50. [Google Scholar] [CrossRef]
- Friso, G.; Giacomelli, L.; Ytterberg, A.J.; Peltier, J.B.; Rudella, A.; Sun, Q.; van Wijk, K.J. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 2004, 16, 478–499. [Google Scholar] [CrossRef] [PubMed]
- Verhey, K.J.; Gaertig, J. The tubulin code. Cell Cycle 2007, 6, 2152–2160. [Google Scholar] [CrossRef]
- Watt, G.; Leoff, C.; Harper, A.D.; Bar-Peled, M. A bifunctional 3,5-epimerase/4-keto reductase for nucleotide-rhamnose synthesis in Arabidopsis. Plant Physiol. 2004, 134, 1337–1346. [Google Scholar] [CrossRef]
- Singh, T.; Hayashi, M.; Mano, S.; Arai, Y.; Goto, S.; Nishimura, M. Molecular components required for the targeting of PEX7 to peroxisomes in Arabidopsis thaliana. Plant J. 2009, 60, 488–498. [Google Scholar] [CrossRef]
- Lister, R.; Carrie, C.; Duncan, O.; Ho, L.H.M.; Howell, K.A.; Murcha, M.W.; Whelan, J. Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 2007, 19, 3739–3759. [Google Scholar] [CrossRef]

| Treatment | Photo (μmolm−2·s−1) | Cond (mmol·m−2·s−1) | Ci (μl·L−1) | Tromol (g·m−2·h−1) | Chlorophyll (mg/g) | Soluble Sugar (mg/g) | SOD (U/g) | MDA (nmol/g) | |
|---|---|---|---|---|---|---|---|---|---|
| BM | Pb | 14.3 ± 0.46 | 0.51 ± 0.02 | 342.00 ± 6.00 * | 9.54 ± 0.29 | 68.31 ± 0.06 | 295.14 ± 0.65 | 193.12 ± 5.63 | 0.98 ± 0.38 |
| CK | 15.0 ± 0.50 | 0.43 ± 0.09 | 310.33 ± 9.87 | 8.99 ± 0.32 | 68.75 ± 0.03 * | 304.13 ± 0.42 * | 195.00 ± 1.88 | 0.94 ± 0.17 | |
| IR | -4.7% | 17.6% | 10.2% | 6.1% | -0.6% | -3.0% | -1.0% | 4.0% | |
| Y1 | Pb | 14.03 ± 1.00 | 0.48 ± 0.02 | 322.67 ± 4.16 * | 8.24 ± 0.53 | 68.51 ± 0.11 | 288.47 ± 1.64 | 200.62 ± 3.75 | 0.86 ± 0.19 * |
| CK | 17.33 ± 1.14 * | 0.55 ± 0.05 | 306.00 ± 2.65 | 9.74 ± 0.26 * | 68.92 ± 0.10 * | 292.94 ± 4.25 | 203.12 ± 9.44 | 0.53 ± 0.08 | |
| IR | −19.0% | −12.8% | 5.4% | −14.4% | −0.6% | −1.5% | −1.2% | 62.3% | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, C.; Hong, L.; Yang, Y.; Yanping, X.; Xing, H.; Gang, D. Protein Changes in Response to Lead Stress of Lead-Tolerant and Lead-Sensitive Industrial Hemp Using SWATH Technology. Genes 2019, 10, 396. https://doi.org/10.3390/genes10050396
Xia C, Hong L, Yang Y, Yanping X, Xing H, Gang D. Protein Changes in Response to Lead Stress of Lead-Tolerant and Lead-Sensitive Industrial Hemp Using SWATH Technology. Genes. 2019; 10(5):396. https://doi.org/10.3390/genes10050396
Chicago/Turabian StyleXia, Cheng, Li Hong, Yang Yang, Xu Yanping, Huang Xing, and Deng Gang. 2019. "Protein Changes in Response to Lead Stress of Lead-Tolerant and Lead-Sensitive Industrial Hemp Using SWATH Technology" Genes 10, no. 5: 396. https://doi.org/10.3390/genes10050396
APA StyleXia, C., Hong, L., Yang, Y., Yanping, X., Xing, H., & Gang, D. (2019). Protein Changes in Response to Lead Stress of Lead-Tolerant and Lead-Sensitive Industrial Hemp Using SWATH Technology. Genes, 10(5), 396. https://doi.org/10.3390/genes10050396