Back to the Salt Mines: Genome and Transcriptome Comparisons of the Halophilic Fungus Aspergillus salisburgensis and Its Halotolerant Relative Aspergillus sclerotialis
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Cultivation
2.2. Whole Genome Sequencing
2.3. Total RNA Extraction
2.4. RNA Library Preparation
2.5. Genome Annotation and Assembly
2.6. Comparative Genomics
2.7. Differential Expression
3. Results and Discussion
3.1. Highly Complete Genome and Gene Set
3.2. Gene Conservation
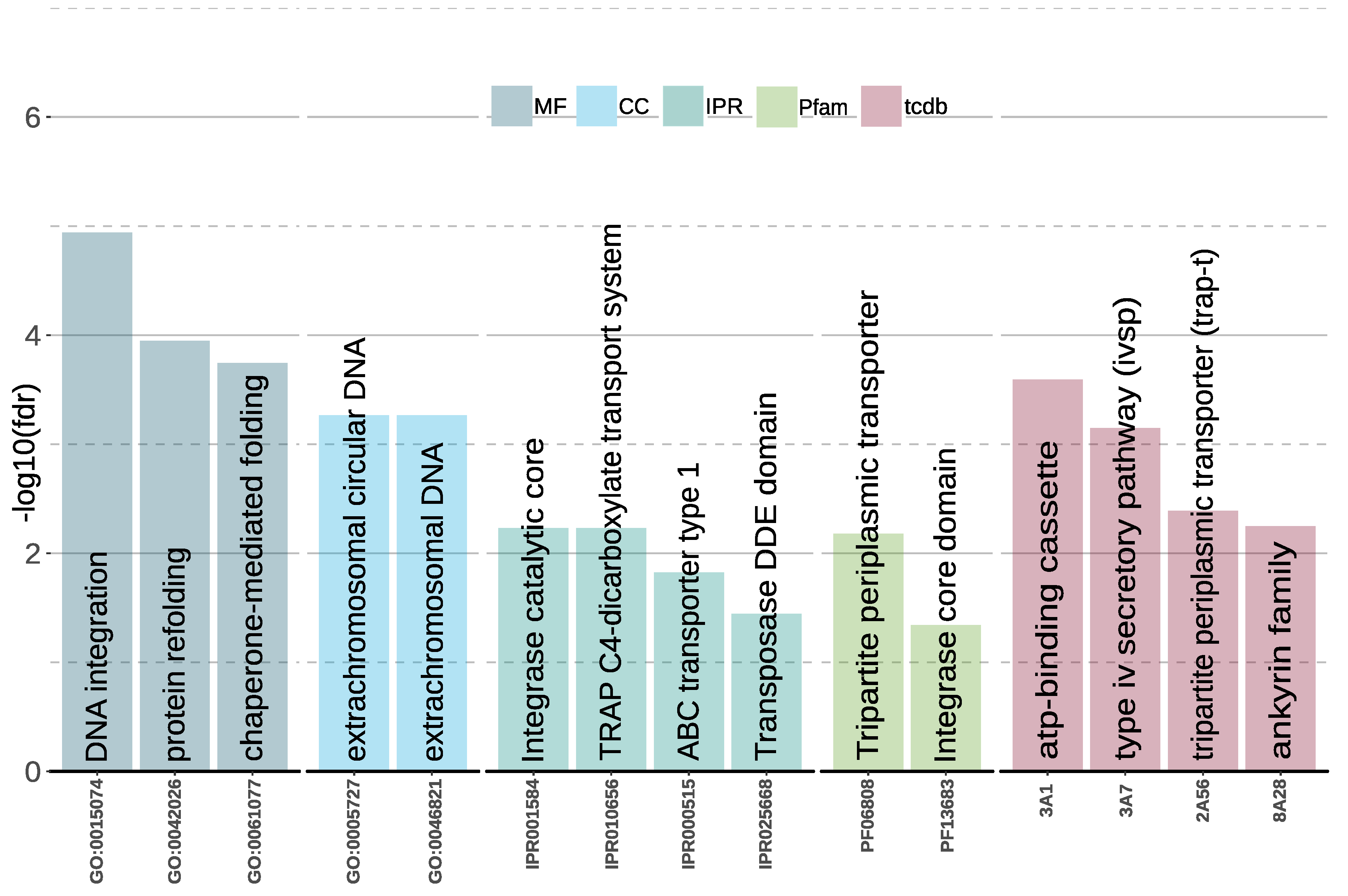
3.3. Gene Family Evolution
Gene Family Enrichment
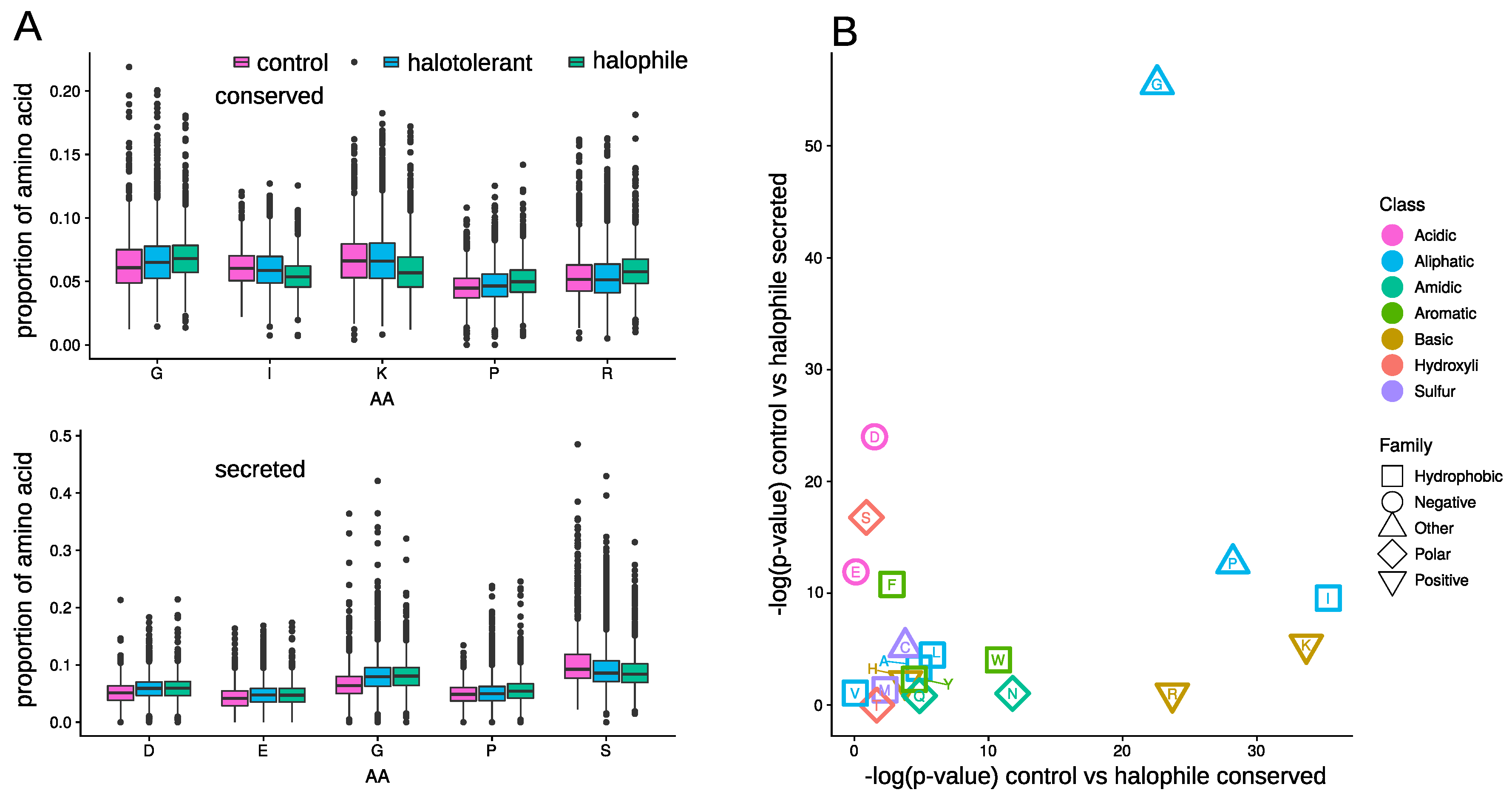
3.4. Amino Acid Composition
3.5. Differential Expression between 5% and 20% NaCl
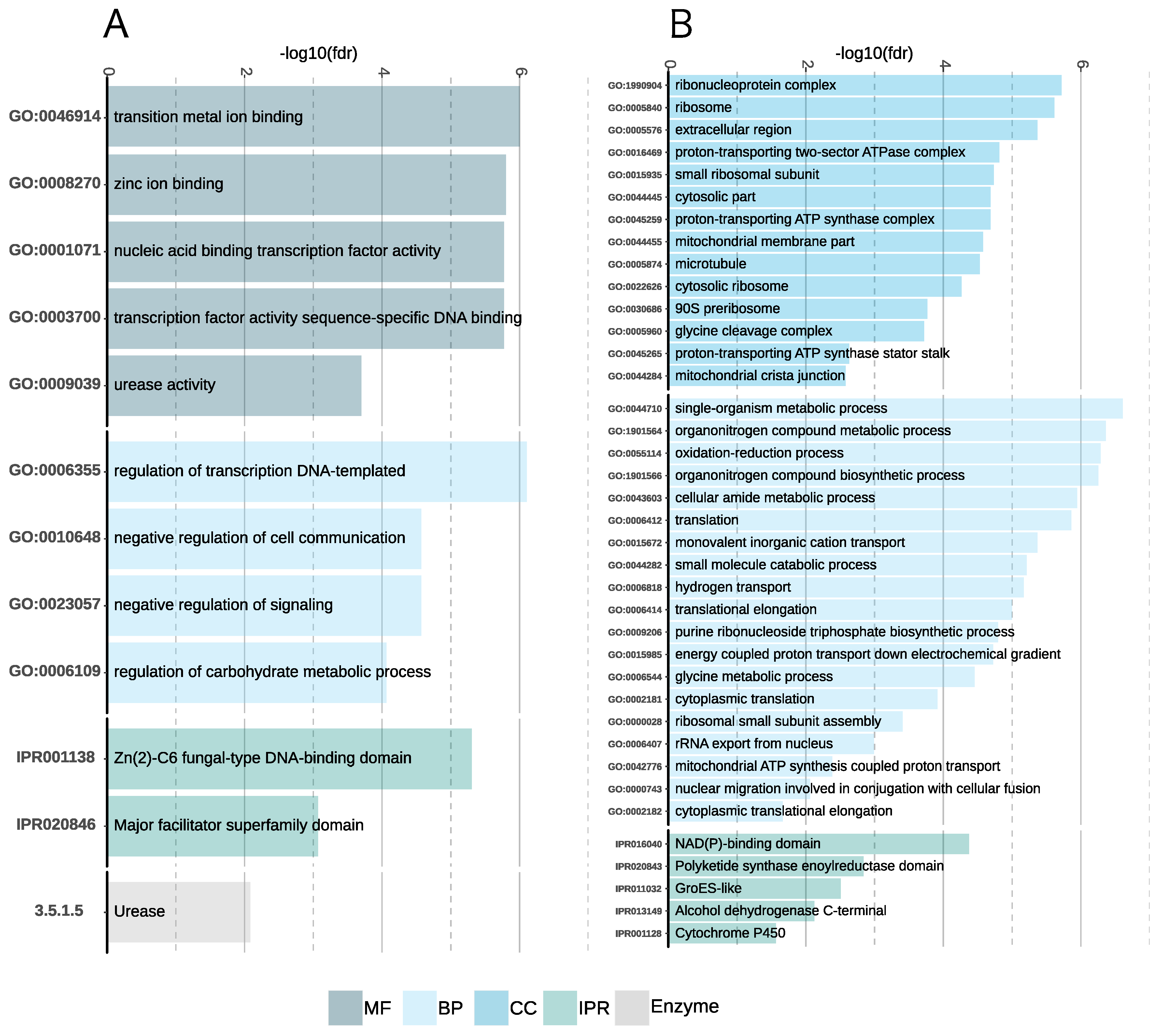
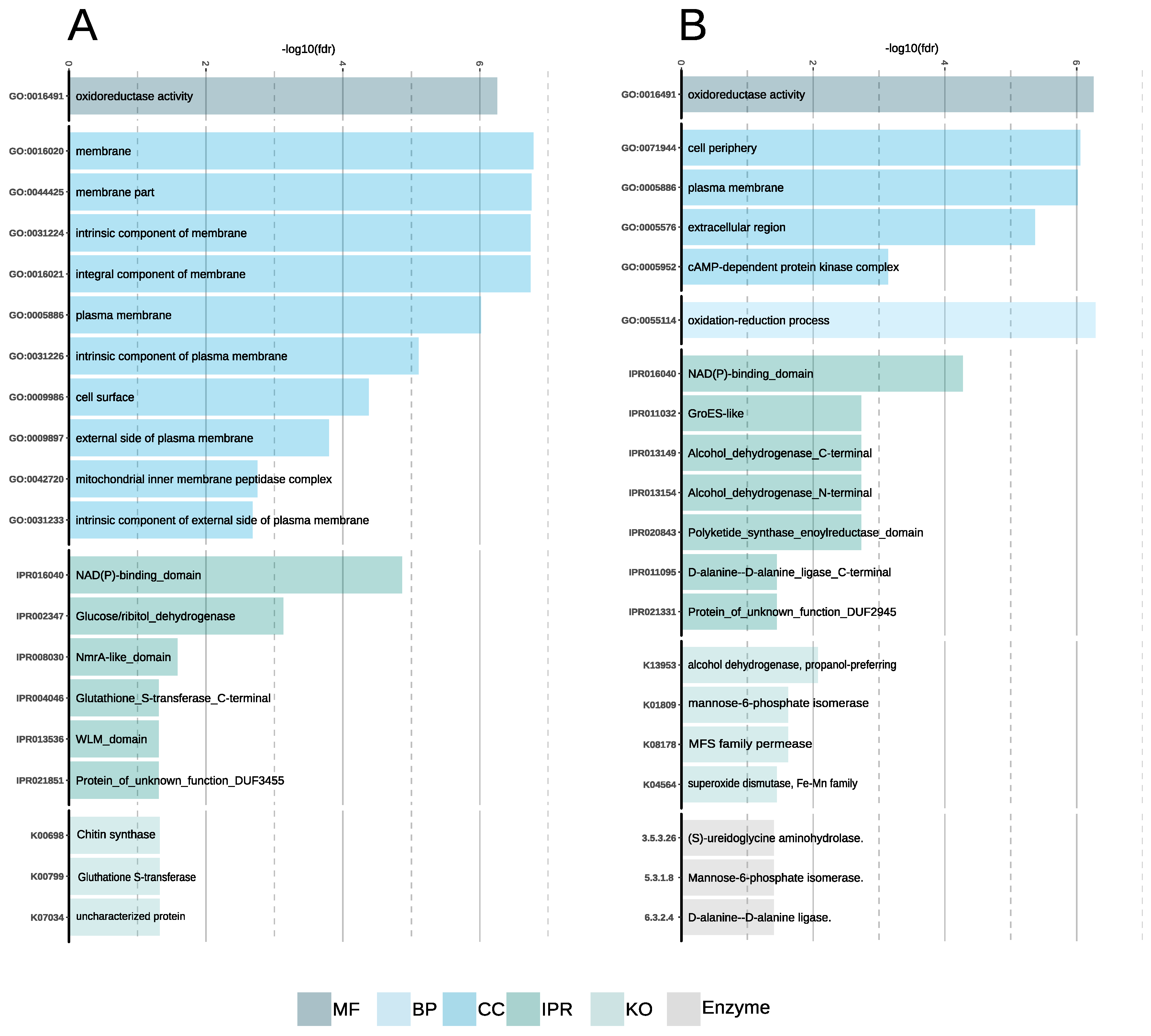
3.5.1. Aspergillus Sclerotialis
3.5.2. A. salisburgensis
3.5.3. Comparative Transcriptomics
3.6. Comparison to Fungal Osmoadaptation and Osmoregulation
3.6.1. Osmosensing
3.6.2. Ion Homeostasis
3.6.3. Cellular Respiration
3.6.4. Stress Response
3.6.5. Cell Interface
3.6.6. Compatible Solute Management
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gunde-Cimerman, N.; Zalar, P. Extremely halotolerant and halophilic fungi inhabit brine in solar salterns around the globe. Food Technol. Biotechnol. 2014, 52, 170–179. [Google Scholar]
- Gunde-Cimerman, N.; Zalar, P.; De Hoog, S.; Plemenitaš, A. Hypersaline waters in salterns—Natural ecological niches for halophilic black yeasts. FEMS Microbiol. Ecol. 2000, 32, 235–240. [Google Scholar] [CrossRef]
- Northolt, M.D.; Van Egmond, H.P.; Paulsch, W.E. Patulin Production by Some Fungal Species in Relation to Water Activity and Temperature. J. Food Prot. 1978, 41, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, K.A.; Hocking, A.D. Interactions among xerophilic fungi associated with dried salted fish. J. Appl. Bacteriol. 1993, 74, 164–169. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage; Springer: Boston, MA, USA, 2009. [Google Scholar]
- Gunde-Cimerman, N.; Ramos, J.; Plemenitaš, A. Halotolerant and halophilic fungi. In Biodiversity of Fungi: Their Role in Human Life; Deshmukh, S.K., Rai, M., Eds.; Science Publishers: Enfield, NH, USA, 2005; pp. 69–127. [Google Scholar]
- Sklenář, F.; Jurjević, Ž.; Zalar, P.; Frisvad, J.; Visagie, C.; Kolařík, M.; Houbraken, J.; Chen, A.; Yilmaz, N.; Seifert, K.; et al. Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Stud. Mycol. 2017, 88, 161–236. [Google Scholar] [CrossRef] [PubMed]
- Sterflinger, K.; Voitl, C.; Lopandic, K.; Piñar, G.; Tafer, H. Big Sound and Extreme Fungi—Xerophilic, Halotolerant Aspergilli and Penicillia with Low Optimal Temperature as Invaders of Historic Pipe Organs. Life 2018, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 2002, 28, 56–63. [Google Scholar] [CrossRef]
- Kis-Papo, T.; Weig, A.R.; Riley, R.; Peršoh, D.; Salamov, A.; Sun, H.; Lipzen, A.; Wasser, S.P.; Rambold, G.; Grigoriev, I.V.; et al. Genomic adaptations of the halophilic Dead Sea filamentous fungus Eurotium rubrum. Nat. Commun. 2014, 5, 3745. [Google Scholar] [CrossRef]
- Martinelli, L.; Zalar, P.; Gunde-Cimerman, N.; Azua-Bustos, A.; Sterflinger, K.; Piñar, G. Aspergillus atacamensis and A. salisburgensis: Two new halophilic species from hypersaline/arid habitats with a phialosimplex-like morphology. Extremophiles 2017, 21, 755–773. [Google Scholar] [CrossRef]
- Piñar, G.; Dalnodar, D.; Voitl, C.; Reschreiter, H.; Sterflinger, K. Biodeterioration Risk Threatens the 3100 Year Old Staircase of Hallstatt (Austria): Possible Involvement of Halophilic Microorganisms. PLoS ONE 2016, 11, e0148279. [Google Scholar] [CrossRef]
- Zajc, J.; Liu, Y.; Dai, W.; Yang, Z.; Hu, J.; Gostin Ar, C.; Gunde-Cimerman, N.; Gostinčar, C.; Gunde-Cimerman, N. Genome and transcriptome sequencing of the halophilic fungus Wallemia ichthyophaga: Haloadaptations present and absent. BMC Genom. 2013, 14, 617. [Google Scholar] [CrossRef]
- Attaby, H.S.H. Influence of salinity stress on the growth, biochemical changes, and response to gamma irradiation of Penicillium chrysogenum. Pak. J. Biol. Sci. 2001, 4, 703–706. [Google Scholar]
- Plemenitaš, A.; Lenassi, M.; Konte, T.; Kejžar, A.; Zajc, J.; Gostinčar, C.; Gunde-Cimerman, N. Adaptation to high salt concentrations in halotolerant/halophilic fungi: A molecular perspective. Front. Microbiol. 2014, 5, 199. [Google Scholar] [CrossRef]
- Krauke, Y.; Sychrova, H. Four pathogenic Candida species differ in salt tolerance. Curr. Microbiol. 2010, 61, 335–339. [Google Scholar] [CrossRef]
- Michán, C.; Martínez, J.L.; Alvarez, M.C.; Turk, M.; Sychrova, H.; Ramos, J. Salt and oxidative stress tolerance in Debaryomyces hansenii and Debaryomyces fabryi. FEMS Yeast Res. 2013, 13, 180–188. [Google Scholar] [CrossRef]
- Salmerón-Santiago, K.G.; Pardo, J.P.; Flores-Herrera, O.; Mendoza-Hernández, G.; Miranda-Arango, M.; Guerra-Sánchez, G. Response to osmotic stress and temperature of the fungus Ustilago maydis. Arch. Microbiol. 2011, 193, 701–709. [Google Scholar] [CrossRef]
- Sert, H.B.; Sterflinger, K. A new Coniosporium species from historical marble monuments. Mycol. Prog. 2010, 9, 353–359. [Google Scholar] [CrossRef]
- Poyntner, C.; Blasi, B.; Arcalis, E.; Mirastschijski, U.; Sterflinger, K.; Tafer, H. The Transcriptome of Exophiala dermatitidis during Ex-vivo Skin Model Infection. Front. Cell. Infect. Microbiol. 2016, 6, 136. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006, 34, 435–439. [Google Scholar] [CrossRef]
- Hoff, K.J.; Lange, S.; Lomsadze, A.; Borodovsky, M.; Stanke, M. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 2016, 32, 767–769. [Google Scholar] [CrossRef]
- Lomsadze, A.; Burns, P.D.; Borodovsky, M. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 2014, 42, e119. [Google Scholar] [CrossRef]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.Y.; Dosztányi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef]
- Saier, M.H.; Tran, C.V.; Barabote, R.D. TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006, 34, 181–186. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Waller, M.; Barrett, A.J.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014, 42, 503–509. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Findeiß, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (co-) orthologs in large-scale analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef]
- Tamura, K.; Tao, Q.; Kumar, S. Theoretical Foundation of the RelTime Method for Estimating Divergence Times from Variable Evolutionary Rates. Mol. Biol. Evol. 2018, 35, 1770–1782. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- R Core Team. R: A Language And Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 888. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W. ggplot2: An Implementation of the Grammar of Graphics. R Package Version 0.7. 2008. Available online: http://CRAN.R-project. org/package=ggplot2 (accessed on 20 November 2018).
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Kuncic, M.K.; Kralj Kuncic, M.; Kogej, T.; Drobne, D.; Gunde-Cimerman, N. Morphological Response of the Halophilic Fungal Genus Wallemia to High Salinity. Appl. Environ. Microbiol. 2010, 76, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Lages, F.; Silva-Graça, M.; Lucas, C. Active glycerol uptake is a mechanism underlying halotolerance in yeasts: A study of 42 species. Microbiology 1999, 145, 2577–2585. [Google Scholar] [CrossRef]
- Klement, T.; Milker, S.; Jäger, G.; Grande, P.M.; Domínguez de María, P.; Büchs, J. Biomass pretreatment affects Ustilago maydis in producing itaconic acid. Microb. Cell Fact. 2012, 11, 43. [Google Scholar] [CrossRef]
- Grammann, K.; Volke, A.; Kunte, H.J. New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581(T). J. Bacteriol. 2002, 184, 3078–3085. [Google Scholar] [CrossRef]
- Verghese, J.; Abrams, J.; Wang, Y.; Morano, K.A. Biology of the heat shock response and protein chaperones: Budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 2012, 76, 115–158. [Google Scholar] [CrossRef]
- Brennan, R.G.; Matthews, B.W. The helix-turn-helix DNA binding motif. J. Biol. Chem. 1989, 264, 1903–1906. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, TimeTrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Cui, X.; Lin, S.; Gan, S.; Xing, H.; Dou, D. GmCYP82A3, a Soybean Cytochrome P450 Family Gene Involved in the Jasmonic Acid and Ethylene Signaling Pathway, Enhances Plant Resistance to Biotic and Abiotic Stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef] [PubMed]
- Gahlot, S.; Joshi, A.; Singh, P.; Tuteja, R.; Dua, M.; Jogawat, A.; Kumar, M.; Raj, S.; Dayaman, V.; Johri, A.K.; et al. Isolation of genes conferring salt tolerance from Piriformospora indica by random overexpression in Escherichia coli. World J. Microbiol. Biotechnol. 2015, 31, 1195–1209. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Ehrlich, K.C. Genome-wide analysis of the Zn(II)2Cys6 zinc cluster-encoding gene family in Aspergillus flavus. Appl. Microbiol. Biotechnol. 2013, 97, 4289–4300. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Jack, D.L.; Saier, M.H. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 2000, 146, 1797–1814. [Google Scholar] [CrossRef]
- Saier, M.H., Jr. Active transport in communication, protection and nutrition. J. Mol. Microbiol. Biotechnol. 2007, 12, 161–164. [Google Scholar] [CrossRef]
- Gilkes, N.R.; Henrissat, B.; Kilburn, D.G.; Miller, R.C., Jr.; Warren, R.A. Domains in microbial beta-1, 4-glycanases: Sequence conservation, function, and enzyme families. Microbiol. Rev. 1991, 55, 303–315. [Google Scholar]
- Sweigard, J.A.; Chumley, F.G.; Valent, B. Cloning and analysis of CUT1, a cutinase gene from Magnaporthe grisea. Mol. Gen. Genet. 1992, 232, 174–182. [Google Scholar]
- Fu, H.L.; Meng, Y.; Ordóñez, E.; Villadangos, A.F.; Bhattacharjee, H.; Gil, J.A.; Mateos, L.M.; Rosen, B.P. Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalliredigens and Corynebacterium glutamicum. J. Biol. Chem. 2009, 284, 19887–19895. [Google Scholar] [CrossRef]
- Paul, S.; Bag, S.K.; Das, S.; Harvill, E.T.; Dutta, C. Molecular signature of hypersaline adaptation: Insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008, 9, R70. [Google Scholar] [CrossRef]
- Tadeo, X.; López-Méndez, B.; Trigueros, T.; Laín, A.; Castaño, D.; Millet, O. Structural basis for the aminoacid composition of proteins from halophilic archea. PLoS Biol. 2009, 7, e1000257. [Google Scholar] [CrossRef] [PubMed]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Fukuchi, S.; Yoshimune, K.; Wakayama, M.; Moriguchi, M.; Nishikawa, K. Unique amino acid composition of proteins in halophilic bacteria. J. Mol. Biol. 2003, 327, 347–357. [Google Scholar] [CrossRef]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar]
- Temple, B.; Horgen, P.A.; Bernier, L.; Hintz, W.E. Cerato-ulmin, a hydrophobin secreted by the causal agents of Dutch elm disease, is a parasitic fitness factor. Fungal Genet. Biol. 1997, 22, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Sigler, L.; Sutton, D.A.; Gibas, C.F.C.; Summerbell, R.C.; Noel, R.K.; Iwen, P.C. Phialosimplex, a new anamorphic genus associated with infections in dogs and having phylogenetic affinity to the Trichocomaceae. Med. Mycol. 2010, 48, 335–345. [Google Scholar] [CrossRef]
- Liu, X.; Baird, W.M. Differential expression of genes regulated in response to drought or salinity stress in sunflower. Crop Sci. 2003, 43, 678–687. [Google Scholar]
- Shelest, E. Transcription Factors in Fungi: TFome Dynamics, Three Major Families, and Dual-Specificity TFs. Front. Genet. 2017, 8, 53. [Google Scholar] [CrossRef]
- Yashiroda, H.; Tanaka, K. But1 and But2 proteins bind to Uba3, a catalytic subunit of E1 for neddylation, in fission yeast. Biochem. Biophys. Res. Commun. 2003, 311, 691–695. [Google Scholar] [CrossRef]
- Lopez, F.; Leube, M.; Gil-Mascarell, R.; Navarro-Aviñó, J.P.; Serrano, R. The yeast inositol monophosphatase is a lithium- and sodium-sensitive enzyme encoded by a non-essential gene pair. Mol. Microbiol. 1999, 31, 1255–1264. [Google Scholar] [CrossRef]
- Hermand, D. F-box proteins: More than baits for the SCF? Cell Div. 2006, 1, 30. [Google Scholar] [CrossRef]
- Sadeghi, M.; Dehghan, S.; Fischer, R.; Wenzel, U.; Vilcinskas, A.; Kavousi, H.R.; Rahnamaeian, M. Isolation and characterization of isochorismate synthase and cinnamate 4-hydroxylase during salinity stress, wounding, and salicylic acid treatment in Carthamus tinctorius. Plant Signal. Behav. 2013, 8, e27335. [Google Scholar] [CrossRef]
- Bu, Y.; Kou, J.; Sun, B.; Takano, T.; Liu, S. Adverse effect of urease on salt stress during seed germination in Arabidopsis thaliana. FEBS Lett. 2015, 589, 1308–1313. [Google Scholar] [CrossRef]
- Kovács, Z.; Simon-Sarkadi, L.; Vashegyi, I.; Kocsy, G. Different accumulation of free amino acids during short- and long-term osmotic stress in wheat. Sci. World J. 2012, 2012, 216521. [Google Scholar] [CrossRef]
- Vale, R.D. The molecular motor toolbox for intracellular transport. Cell 2003, 112, 467–480. [Google Scholar] [CrossRef]
- Leger, R.J.S.; Joshi, L.; Roberts, D.W. Adaptation of proteases and carbohydrases of saprophytic, phytopathogenic and entomopathogenic fungi to the requirements of their ecological niches. Microbiology 1997, 143, 1983–1992. [Google Scholar] [CrossRef]
- Muszewska, A.; Stepniewska-Dziubinska, M.M.; Steczkiewicz, K.; Pawlowska, J.; Dziedzic, A.; Ginalski, K. Fungal lifestyle reflected in serine protease repertoire. Sci. Rep. 2017, 7, 9147. [Google Scholar] [CrossRef]
- Liu, J.X.; Srivastava, R.; Che, P.; Howell, S.H. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. Cell Mol. Biol. 2007, 51, 897–909. [Google Scholar] [CrossRef]
- Figueiredo, J.; Sousa Silva, M.; Figueiredo, A. Subtilisin-like proteases in plant defence: The past, the present and beyond. Mol. Plant Pathol. 2018, 19, 1017–1028. [Google Scholar] [CrossRef]
- Chen, J.H.; Jiang, H.W.; Hsieh, E.J.; Chen, H.Y.; Chien, C.T.; Hsieh, H.L.; Lin, T.P. Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 2012, 158, 340–351. [Google Scholar] [CrossRef]
- Szopinska, A.; Degand, H.; Hochstenbach, J.F.; Nader, J.; Morsomme, P. Rapid response of the yeast plasma membrane proteome to salt stress. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef]
- Ene, I.V.; Walker, L.A.; Schiavone, M.; Lee, K.K.; Martin-Yken, H.; Dague, E.; Gow, N.A.R.; Munro, C.A.; Brown, A.J.P. Cell Wall Remodeling Enzymes Modulate Fungal Cell Wall Elasticity and Osmotic Stress Resistance. mBio 2015, 6, e00986. [Google Scholar] [CrossRef]
- Gostinčar, C.; Gunde-Cimerman, N. Overview of Oxidative Stress Response Genes in Selected Halophilic Fungi. Genes 2018, 9, 143. [Google Scholar] [CrossRef]
- Ortiz-Bermúdez, P.; Srebotnik, E.; Hammel, K.E. Chlorination and cleavage of lignin structures by fungal chloroperoxidases. Appl. Environ. Microbiol. 2003, 69, 5015–5018. [Google Scholar] [CrossRef]
- Dence, C.W. Halogenation and nitration. In Lignins. Occurrence, Formation, Structure and Reactions; Wiley-Interscience: New York, NY, USA, 1971; pp. 373–432. [Google Scholar]
- García-Martínez, J.; Castrillo, M.; Avalos, J. The gene cutA of Fusarium fujikuroi, encoding a protein of the haloacid dehalogenase family, is involved in osmotic stress and glycerol metabolism. Microbiology 2014, 160, 26–36. [Google Scholar] [CrossRef]
- Hara, M.; Furukawa, J.; Sato, A.; Mizoguchi, T.; Miura, K. Abiotic stress and role of salicylic acid in plants. In Abiotic Stress Responses in Plants; Ahmad, P., Prasad, M.N.V., Eds.; Springer: New York, NY, USA, 2012; pp. 235–251. [Google Scholar]
- Miura, K.; Tada, Y. Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 2014, 5, 4. [Google Scholar] [CrossRef]
- Hohmann, S. Control of high osmolarity signaling in the yeast Saccharomyces cerevisiae. FEBS Lett. 2009, 583, 4025–4029. [Google Scholar] [CrossRef]
- Ma, D.; Li, R. Current understanding of HOG-MAPK pathway in Aspergillus fumigatus. Mycopathologia 2013, 175, 13–23. [Google Scholar] [CrossRef]
- Lenassi, M.; Plemenitaš, A. Novel group VII histidine kinase HwHhk7B from the halophilic fungi Hortaea werneckii has a putative role in osmosensing. Curr. Genet. 2007, 51, 393–405. [Google Scholar] [CrossRef]
- Mortimer, R.K.; Schild, D.; Contopoulou, C.R.; Kans, J.A. Genetic map of Saccharomyces cerevisiae, edition 10. Yeast 1989, 5, 321–403. [Google Scholar] [CrossRef]
- Ono, T.; Suzuki, T.; Anraku, Y.; Iida, H. The MID2 gene encodes a putative integral membrane protein with a Ca(2+)-binding domain and shows mating pheromone-stimulated expression in Saccharomyces cerevisiae. Gene 1994, 151, 203–208. [Google Scholar] [CrossRef]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef]
- Stratford, M.; Steels, H.; Novodvorska, M.; Archer, D.B.; Avery, S.V. Extreme Osmotolerance and Halotolerance in Food-Relevant Yeasts and the Role of Glycerol-Dependent Cell Individuality. Front. Microbiol. 2019, 9, 3238. [Google Scholar] [CrossRef] [PubMed]
- Wurgler-Murphy, S.M.; Maeda, T.; Witten, E.A.; Saito, H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol. Cell. Biol. 1997, 17, 1289–1297. [Google Scholar] [CrossRef]
- Smith, A.; Ward, M.P.; Garrett, S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998, 17, 3556–3564. [Google Scholar] [CrossRef] [PubMed]
- Ferrigno, P.; Posas, F.; Koepp, D.; Saito, H.; Silver, P.A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998, 17, 5606–5614. [Google Scholar] [CrossRef]
- Kruppa, M.; Calderone, R. Two-component signal transduction in human fungal pathogens. FEMS Yeast Res. 2006, 6, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Prista, C.; Loureiro-Dias, M.C.; Montiel, V.; García, R.; Ramos, J. Mechanisms underlying the halotolerant way of Debaryomyces hansenii. FEMS Yeast Res. 2005, 5, 693–701. [Google Scholar] [CrossRef]
- Haro, R.; Garciadeblas, B.; Rodríguez-Navarro, A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991, 291, 189–191. [Google Scholar] [CrossRef]
- Pannala, V.R.; Dash, R.K. Mechanistic characterization of the thioredoxin system in the removal of hydrogen peroxide. Free Radic. Biol. Med. 2015, 78, 42–55. [Google Scholar] [CrossRef]
- Madeo, F.; Herker, E.; Maldener, C.; Wissing, S.; Lächelt, S.; Herlan, M.; Fehr, M.; Lauber, K.; Sigrist, S.J.; Wesselborg, S.; et al. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 2002, 9, 911–917. [Google Scholar] [CrossRef]
- Lee, R.E.C.; Brunette, S.; Puente, L.G.; Megeney, L.A. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. USA 2010, 107, 13348–13353. [Google Scholar] [CrossRef] [PubMed]
- Gille, G.; Sigler, K. Oxidative stress and living cells. Folia Microbiol. 1995, 40, 131–152. [Google Scholar] [CrossRef]
- Petrovic, U. Role of oxidative stress in the extremely salt-tolerant yeast Hortaea werneckii. FEMS Yeast Res. 2006, 6, 816–822. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guo, F.; Stanevich, V.; Wlodarchak, N.; Sengupta, R.; Jiang, L.; Satyshur, K.A.; Xing, Y. Structural basis of PP2A activation by PTPA, an ATP-dependent activation chaperone. Cell Res. 2014, 24, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Levine, R.L. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 2003, 25, 207–218. [Google Scholar] [CrossRef]
- Saito, H.; Posas, F. Response to hyperosmotic stress. Genetics 2012, 192, 289–318. [Google Scholar] [CrossRef]
- Kuznetsov, E.; Kučerová, H.; Váchová, L.; Palková, Z. SUN family proteins Sun4p, Uth1p and Sim1p are secreted from Saccharomyces cerevisiae and produced dependently on oxygen level. PLoS ONE 2013, 8, e73882. [Google Scholar] [CrossRef] [PubMed]
- Arispe, N.; De Maio, A. ATP and ADP modulate a cation channel formed by Hsc70 in acidic phospholipid membranes. J. Biol. Chem. 2000, 275, 30839–30843. [Google Scholar] [CrossRef] [PubMed]
- Katz, A.; Waridel, P.; Shevchenko, A.; Pick, U. Salt-induced changes in the plasma membrane proteome of the halotolerant alga Dunaliella salina as revealed by blue native gel electrophoresis and nano-LC-MS/MS analysis. Mol. Cell. Proteom. 2007, 6, 1459–1472. [Google Scholar] [CrossRef] [PubMed]
- Douglas, L.M.; Wang, H.X.; Keppler-Ross, S.; Dean, N.; Konopka, J.B. Sur7 promotes plasma membrane organization and is needed for resistance to stressful conditions and to the invasive growth and virulence of Candida albicans. mBio 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Vögler, O.; Barceló, J.M.; Ribas, C.; Escribá, P.V. Membrane interactions of G proteins and other related proteins. Biochim. Biophys. Acta 2008, 1778, 1640–1652. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Brickner, J.H.; Aguilar, P.S.; Bernales, S.; Pantoja, C.; Walter, P. Eisosomes mark static sites of endocytosis. Nature 2006, 439, 998–1003. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.; van Voorst, F.; Martins, A.; Neves, L.; Oliveira, R.; Kielland-Brandt, M.C.; Lucas, C.; Brandt, A. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol. Biol. Cell 2005, 16, 2068–2076. [Google Scholar] [CrossRef] [PubMed]
- Luyten, K.; Albertyn, J.; Skibbe, W.F.; Prior, B.A.; Ramos, J.; Thevelein, J.M.; Hohmann, S. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 1995, 14, 1360–1371. [Google Scholar] [CrossRef] [PubMed]
- Lambou, K.; Pennati, A.; Valsecchi, I.; Tada, R.; Sherman, S.; Sato, H.; Beau, R.; Gadda, G.; Latgé, J.P. Pathway of glycine betaine biosynthesis in Aspergillus fumigatus. Eukaryot. Cell 2013, 12, 853–863. [Google Scholar] [CrossRef]
- Brown, A.D. Microbial Water Stress Physiology. Principles and Perspectives; John Wiley & Sons: Chichester, UK, 1990. [Google Scholar]
- Sterflinger, K. Temperature and NaCl-tolerance of rock-inhabiting meristematic fungi. Antonie Van Leeuwenhoek 1998, 74, 271–281. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Krell, A. Salt Stress Tolerance in the Psychrophilic Diatom Fragilariopsis Cylindrus. Ph.D. Thesis, Bremen University, Bremen, Germany, 2006. [Google Scholar]
- Pade, N.; Michalik, D.; Ruth, W.; Belkin, N.; Hess, W.R.; Berman-Frank, I.; Hagemann, M. Trimethylated homoserine functions as the major compatible solute in the globally significant oceanic cyanobacterium Trichodesmium. Proc. Natl. Acad. Sci. USA 2016, 113, 13191–13196. [Google Scholar] [CrossRef] [PubMed]
- Saum, S.H.; Sydow, J.F.; Palm, P.; Pfeiffer, F.; Oesterhelt, D.; Müller, V. Biochemical and molecular characterization of the biosynthesis of glutamine and glutamate, two major compatible solutes in the moderately halophilic bacterium Halobacillus halophilus. J. Bacteriol. 2006, 188, 6808–6815. [Google Scholar] [CrossRef] [PubMed]
- Stepkowski, T.; Legocki, A.B. Reduction of bacterial genome size and expansion resulting from obligate intracellular lifestyle and adaptation to soil habitat. Acta Biochim. Pol. 2001, 48, 367–381. [Google Scholar] [PubMed]


| Species | Salt Concentration | Type | Reference |
|---|---|---|---|
| A. salisburgensis | 5–30%,optimal at 20% | HH | This publication |
| Aspergillus ruber | >10%, optimal at 18% | HH | [10] |
| Wallemia ichthyophaga | >8%, optimal 18% | HH | [13] |
| A. sclerotialis | 0–20%, optimal at 10% | H | This publication |
| Hortaea werneckii | 0–32%, optimal 3–9% | H | [15] |
| Penicillium chrysogenum | 0–18%, optimal at 10% | H | [14] |
| Candida parapsilosis | 0–12%, optimal 0 % | H | [16] |
| Debaryomyces fabryi | 0–16%, optimal 0 % | H | [17] |
| Debaryomyces hansenii | 0–24%, optimal 0 % | H | [17] |
| Wallemia mellicola | 0–27%, optimal 0 % | H | [47] |
| Saccharomyces cerevisiae | 0–8%, optimal 0 % | C | [48] |
| Schizosaccharomyces pombe | 0–5%, optimal 0 % | C | [48] |
| Ustilago maydis | 0–7%, optimal 0 % | C | [18,49] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tafer, H.; Poyntner, C.; Lopandic, K.; Sterflinger, K.; Piñar, G. Back to the Salt Mines: Genome and Transcriptome Comparisons of the Halophilic Fungus Aspergillus salisburgensis and Its Halotolerant Relative Aspergillus sclerotialis. Genes 2019, 10, 381. https://doi.org/10.3390/genes10050381
Tafer H, Poyntner C, Lopandic K, Sterflinger K, Piñar G. Back to the Salt Mines: Genome and Transcriptome Comparisons of the Halophilic Fungus Aspergillus salisburgensis and Its Halotolerant Relative Aspergillus sclerotialis. Genes. 2019; 10(5):381. https://doi.org/10.3390/genes10050381
Chicago/Turabian StyleTafer, Hakim, Caroline Poyntner, Ksenija Lopandic, Katja Sterflinger, and Guadalupe Piñar. 2019. "Back to the Salt Mines: Genome and Transcriptome Comparisons of the Halophilic Fungus Aspergillus salisburgensis and Its Halotolerant Relative Aspergillus sclerotialis" Genes 10, no. 5: 381. https://doi.org/10.3390/genes10050381
APA StyleTafer, H., Poyntner, C., Lopandic, K., Sterflinger, K., & Piñar, G. (2019). Back to the Salt Mines: Genome and Transcriptome Comparisons of the Halophilic Fungus Aspergillus salisburgensis and Its Halotolerant Relative Aspergillus sclerotialis. Genes, 10(5), 381. https://doi.org/10.3390/genes10050381





