Genome-Wide Association Studies Based on Equine Joint Angle Measurements Reveal New QTL Affecting the Conformation of Horses
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Phenotyping
2.2.1. Photograph Selection
2.2.2. Horse Shape Data
2.2.3. Phenotype Concordance
2.2.4. Quality Control of the Phenotype
2.3. Genetic Analyses
2.3.1. Franches-Montagnes Horses
2.3.2. Lipizzan Horses
2.3.3. Merging the Genetic Data Sets (Franches-Montagnes and Lipizzan Horses)
2.4. Genome-Wide Association Studies
2.5. Functional Annotation
3. Results
3.1. Phenotypic Analyses
3.2. Population Structure
3.3. Genome-Wide Association Studies for Joint Angle Measurements
3.3.1. Poll Angle Associations
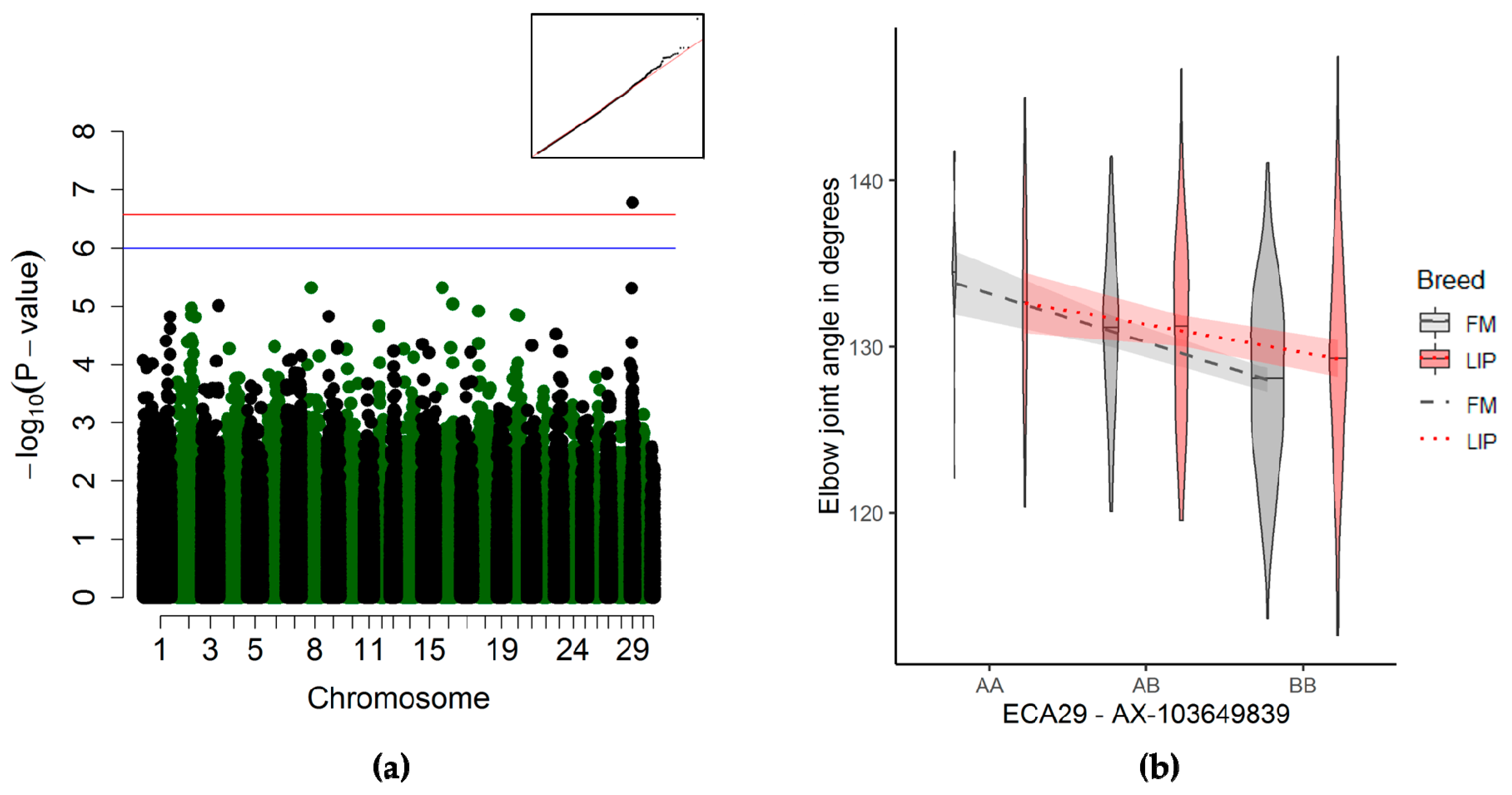
3.3.2. Elbow Joint Angle Association
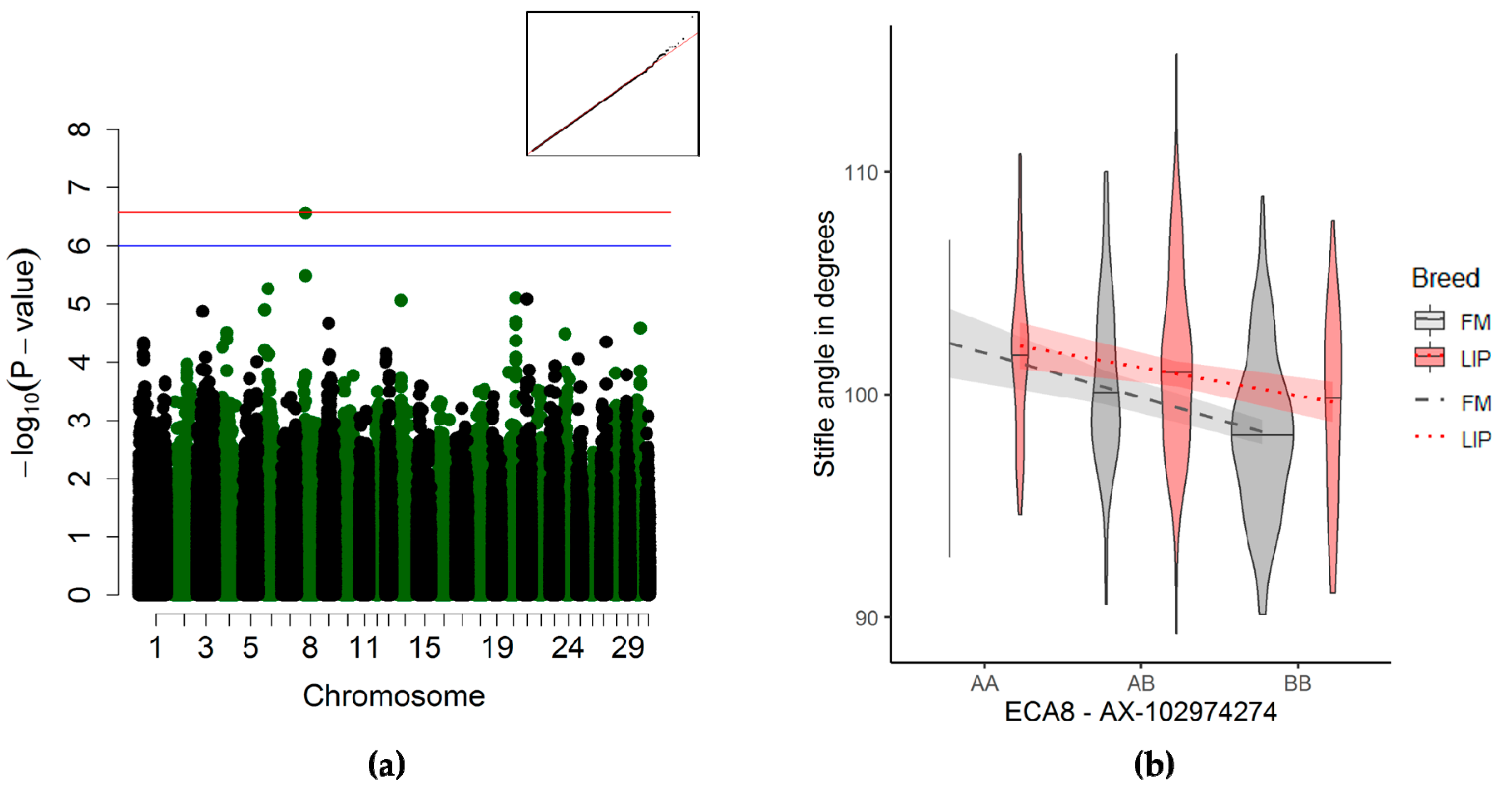
3.3.3. Stifle Joint Angle Association
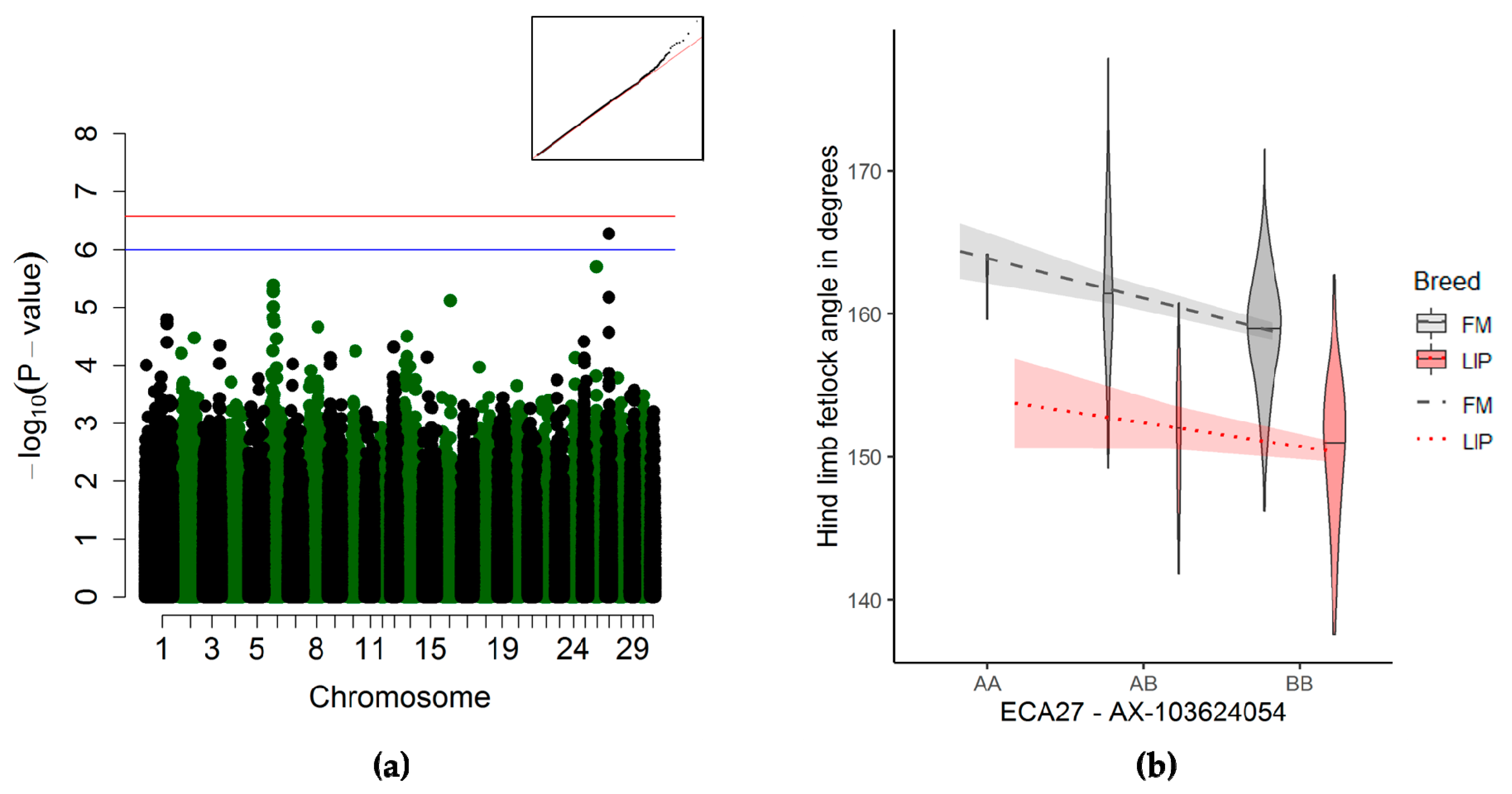
3.3.4. Fetlock Joint Angle of the Hind Limb Association
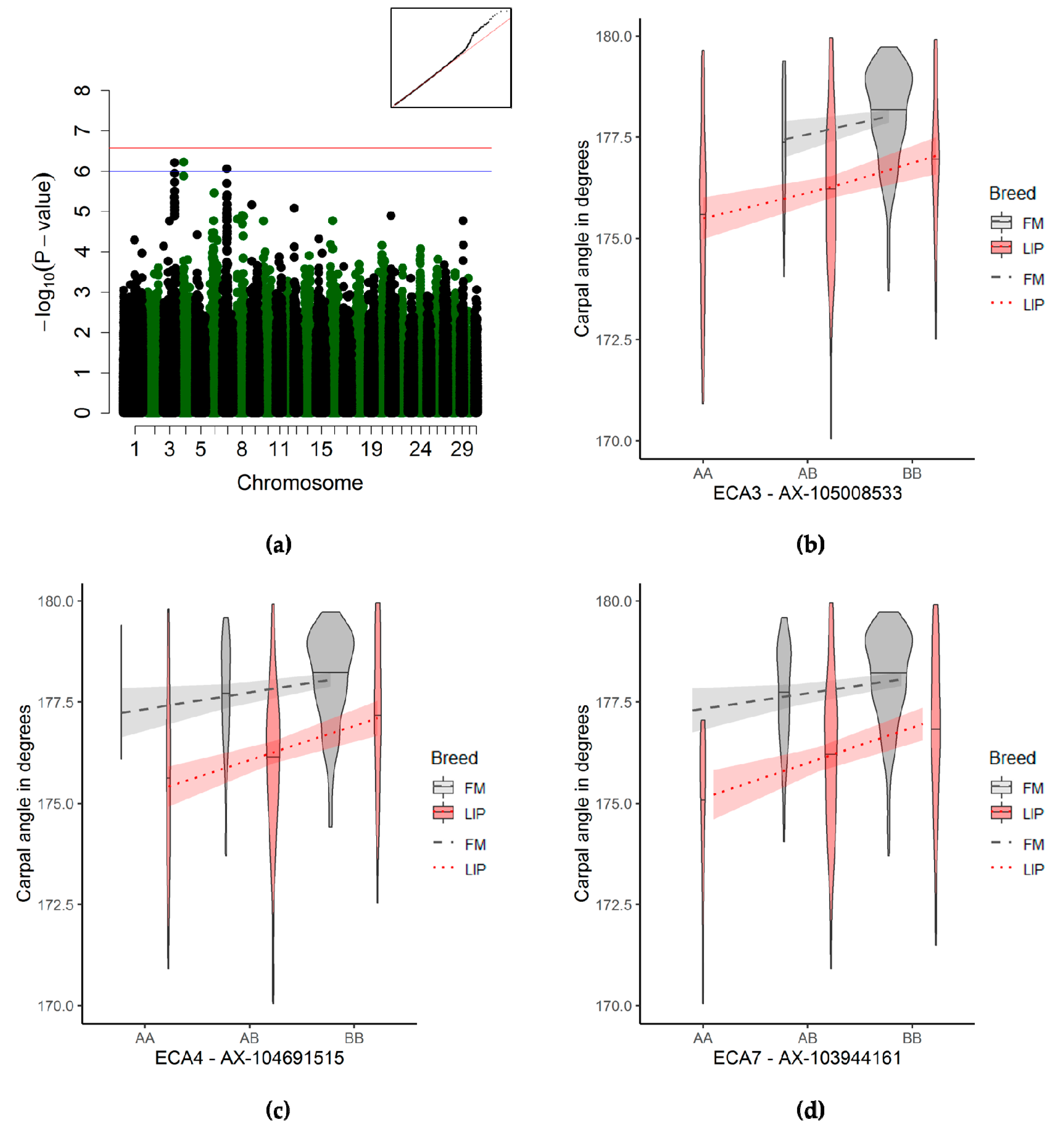
3.3.5. Carpal Joint Angle Associations
4. Discussion
4.1. Phenotypes
4.2. Genome-Wide Association Studies
4.2.1. Identification and Interpretation of Relevant Quantitative Trait Loci
4.2.2. Poll Angle
4.2.3. Elbow Joint Angle
4.2.4. Stifle Joint Angle
4.2.5. Fetlock Joint Angle of the Hind Limb
4.2.6. Carpal Joint Angle
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Saastamoinen, M.T.; Barrey, E. Genetics of conformation, locomotion and physiological traits. In The Genetics of the Horse; Bowling, A., Ruvinsky, A., Eds.; CABI: Wallingford, UK, 2000; pp. 439–472. [Google Scholar]
- Holmström, M.; Back, W. The effects of conformation. In Equine Locomotion, 2nd ed.; Back, W., Clayton, H., Eds.; WB Saunders: London, UK, 2013; pp. 229–243. [Google Scholar]
- Thomas, H.S. The Horse Conformation Handbook; Storey Publishing: North Adams, MA, USA, 2005. [Google Scholar]
- Reginald Harrison, S.; Goody, P.C. Horse Structure and Movement; J.A. Allen: London, UK, 1993; pp. 152–202. [Google Scholar]
- Smythe, R.H. What Makes a Good Horse: Its Structure and Performance, 1st ed.; Country Life LTD: London, UK, 1957. [Google Scholar]
- Druml, T.; Dobretsberger, M.; Brem, G. Ratings of equine conformation–new insights provided by shape analysis using the example of Lipizzan stallions. Arch. Anim. Breed. 2016, 59, 309–317. [Google Scholar] [CrossRef]
- Holmström, M.; Magnusson, L.E.; Philipsson, J. Variation in conformation of Swedish Warmblood horses and conformational characteristics of elite sport horses. Equine Vet. J. 1990, 22, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Holmström, M.; Fredricson, I.; Drevemo, S. Variation in angular pattern adaptation from trot in hand to passage and piaffe in the grand prix dressage horse. Equine Vet. J. 1995, 27, 132–137. [Google Scholar] [CrossRef]
- Holmström, M.; Philipsson, J. Relationships between conformation, performance and health in 4-year-old Swedish warmblood riding horses. Livest. Prod. Sci. 1993, 33, 293–312. [Google Scholar] [CrossRef]
- Wallin, L.; Strandberg, E.; Philipsson, J. Phenotypic relationship between test results of Swedish Warmblood horses as 4-year-olds and longevity. Livest. Prod. Sci. 2001, 68, 97–105. [Google Scholar] [CrossRef]
- Love, S.; Wyse, C.; Stirk, A.; Stear, M.; Calver, P.; Voute, L.; Mellor, D. Prevalence, heritability and significance of musculoskeletal conformational traits in Thoroughbred yearlings. Equine Vet. J. 2006, 38, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, L.; Egenvall, A.; Roepstorff, L.; Näsholm, A.; Dalin, G.; Philipsson, J. Associations of health status and conformation with longevity and lifetime competition performance in young Swedish warmblood riding horses: 8,238 cases (1983–2005). J. Am. Vet. Med. Assoc. 2014, 244, 1449–1461. [Google Scholar] [CrossRef]
- Anderson, T.; McIlwraith, C.; Douay, P. The role of conformation in musculoskeletal problems in the racing Thoroughbred. Equine Vet. J. 2004, 36, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Axelsson, M.; Björnsdottir, S.; Eksell, P.; Häggström, J.; Sigurdsson, H.; Carlsten, J. Risk factors associated with hindlimb lameness and degenerative joint disease in the distal tarsus of Icelandic horses. Equine Vet. J. 2001, 33, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Rustin, M.; Janssens, S.; Buys, N.; Gengler, N. Multi-trait animal model estimation of genetic parameters for linear type and gait traits in the Belgian warmblood horse. J. Anim. Breed. Genet. 2009, 126, 378–386. [Google Scholar] [CrossRef]
- Viklund, A.; Eriksson, S. Genetic analyses of linear profiling data on 3-year-old Swedish Warmblood horses. J. Anim. Breed. Genet. 2018, 135, 62–72. [Google Scholar] [CrossRef]
- Samoré, A.; Pagnacco, G.; Miglior, F. Genetic parameters and breeding values for linear type traits in the Haflinger horse. Livest. Prod. Sci. 1997, 52, 105–111. [Google Scholar] [CrossRef]
- Hellsten, E.T.; Viklund, Å.; Koenen, E.; Ricard, A.; Bruns, E.; Philipsson, J. Review of genetic parameters estimated at stallion and young horse performance tests and their correlations with later results in dressage and show-jumping competition. Livest. Sci. 2006, 103, 1–12. [Google Scholar] [CrossRef]
- Duensing, J.; Stock, K.F.; Krieter, J. Implementation and prospects of linear profiling in the warmblood horse. J. Equine Vet. Sci. 2014, 34, 360–368. [Google Scholar] [CrossRef]
- Signer-Hasler, H.; Flury, C.; Haase, B.; Burger, D.; Simianer, H.; Leeb, T.; Rieder, S. A genome-wide association study reveals loci influencing height and other conformation traits in horses. PLoS ONE 2012, 7, e37282. [Google Scholar] [CrossRef]
- Frischknecht, M.; Signer-Hasler, H.; Leeb, T.; Rieder, S.; Neuditschko, M. Genome-wide association studies based on sequence-derived genotypes reveal new QTL associated with conformation and performance traits in the Franches–Montagnes horse breed. Anim. Genet. 2016, 47, 227–229. [Google Scholar] [CrossRef]
- Stock, K.F.; Distl, O. Genetic correlations between conformation traits and radiographic findings in the limbs of German warmblood riding horses. Gen. Sel. Evol. 2006, 38, 657. [Google Scholar]
- Schroderus, E.; Ojala, M. Estimates of genetic parameters for conformation measures and scores in finnhorse and standardbred foals. J. Anim. Breed. Gen. 2010, 127, 395–403. [Google Scholar] [CrossRef] [PubMed]
- van Bergen, H.M.; van Arendonk, J.A. Genetic parameters for linear type traits in Shetland Ponies. Livest. Prod. Sci. 1993, 36, 273–284. [Google Scholar] [CrossRef]
- Meira, C.T.; Farah, M.M.; Fortes, M.R.; Moore, S.S.; Pereira, G.L.; Silva, J.A.I.V.; da Mota, M.D.S.; Curi, R.A. A genome-wide association study for morphometric traits in quarter horse. J. Equine Vet. Sci. 2014, 34, 1028–1031. [Google Scholar] [CrossRef]
- Staiger, E.A.; Al Abri, M.; Pflug, K.M.; Kalla, S.; Ainsworth, D.; Miller, D.; Raudsepp, T.; Sutter, N.B.; Brooks, S.A. Skeletal variation in Tennessee Walking Horses maps to the LCORL/NCAPG gene region. Physiol. Genomics 2016, 48, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Pfammatter, M. Le Système de Description Linéaire au Sein de la Race des Franches-Montagnes. Bachelor’s Thesis, Haute École des Sciences Agronomiques, Forestières et Alimentaires, Zollikofen, Switzerland, 2017. [Google Scholar]
- Gmel, A.I.; Druml, T.; Portele, K.; von Niederhäusern, R.; Neuditschko, M. Repeatability, reproducibility and consistency of horse shape data and its association with linearly described conformation traits in Franches-Montagnes stallions. PLoS ONE 2018, 13, e0202931. [Google Scholar] [CrossRef] [PubMed]
- Druml, T.; Dobretsberger, M.; Brem, G. The use of novel phenotyping methods for validation of equine conformation scoring results. Animal 2015, 1–10. [Google Scholar] [CrossRef]
- Rohlf, F.J. The Tps Utility Program; 1.68 v.; Departement of Ecology & Evolution, State University of New York SUNY at Stony Brook: New York, NY, USA, 2016. [Google Scholar]
- Rohlf, F.J. The Tps Dig2; 2.26 v.; Departement of Ecology & Evolution, State University of New York SUNY at Stony Brook: New York, NY, USA, 2016. [Google Scholar]
- Wolak, M.E.; Fairbairn, D.J.; Paulsen, Y.R. Guidelines for estimating repeatability. Methods Ecol. Evol. 2012, 3, 129–137. [Google Scholar] [CrossRef]
- Cicchetti, D.V. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol. Assess. 1994, 6, 284. [Google Scholar] [CrossRef]
- Frischknecht, M.; Neuditschko, M.; Jagannathan, V.; Drögemüller, C.; Tetens, J.; Thaller, G.; Leeb, T.; Rieder, S. Imputation of sequence level genotypes in the Franches-Montagnes horse breed. Gen. Sel. Evol. 2014, 46, 63. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jagannathan, V.; Gerber, V.; Rieder, S.; Tetens, J.; Thaller, G.; Drögemüller, C.; Leeb, T. Comprehensive characterization of horse genome variation by whole-genome sequencing of 88 horses. Anim. Genet. 2018. [Google Scholar] [CrossRef]
- Purcell, S. PLINK; 1.07, v.; Center for Human Genetic Research (CHGR), Massachusetts General Hospital (MGH), the Broad Institute of Harvard & MIT: Boston, MA, USA, 2017. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; Van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [PubMed]
- Rönnegård, L.; Shen, X.; Alam, M. hglm: A package for fitting hierarchical generalized linear models. R J. 2010, 2, 20–28. [Google Scholar] [CrossRef]
- Lee, Y.; Nelder, J.A. Hierarchical generalized linear models. J. R. Stat. Soc. Ser. B 1996, 58, 619–678. [Google Scholar] [CrossRef]
- Chen, W.-M.; Abecasis, G.R. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 2007, 81, 913–926. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Tanaka, T.; Okada, Y. Empirical estimation of genome-wide significance thresholds based on the 1000 Genomes Project data set. J. Hum. Genet. 2016, 61, 861. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Genome Data Viewer. Available online: https://www.ncbi.nlm.nih.gov/genome/gdv/ (accessed on 6 May 2019).
- Holland, P.W.; Booth, H.A.F.; Bruford, E.A. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007, 5, 47. [Google Scholar] [CrossRef]
- Uz, E.; Alanay, Y.; Aktas, D.; Vargel, I.; Gucer, S.; Tuncbilek, G.; von Eggeling, F.; Yilmaz, E.; Deren, O.; Posorski, N. Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: Expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia. Am. J. Hum. Genet. 2010, 86, 789–796. [Google Scholar] [CrossRef]
- Funato, N.; Nakamura, M. Identification of shared and unique gene families associated with oral clefts. Int. J. Oral Sci. 2017, 9, 104. [Google Scholar] [CrossRef]
- Bordbari, M.; Penedo, M.; Aleman, M.; Valberg, S.; Mickelson, J.; Finno, C.J. Deletion of 2.7 kb near HOXD 3 in an Arabian horse with occipitoatlantoaxial malformation. Anim. Genet. 2017, 48, 287–294. [Google Scholar] [CrossRef]
- Grilz-Seger, G.; Druml, T.; Neuditschko, M.; Dobretsberger, M.; Horna, M.; Brem, G. High-resolution population structure and runs of homozygosity reveal the genetic architecture of complex traits in the Lipizzan horse. BMC Genomics 2019, 20, 174. [Google Scholar] [CrossRef]
- Grilz-Seger, G.; Mesarič, M.; Cotman, M.; Neuditschko, M.; Druml, T.; Brem, G. Runs of homozygosity and population history of three horse breeds with small population size. J. Equine Vet. Sci. 2018, 71, 27–34. [Google Scholar] [CrossRef]
- Metzger, J.; Karwath, M.; Tonda, R.; Beltran, S.; Águeda, L.; Gut, M.; Gut, I.G.; Distl, O. Runs of homozygosity reveal signatures of positive selection for reproduction traits in breed and non-breed horses. BMC Genomics 2015, 16, 764. [Google Scholar] [CrossRef] [PubMed]
- Meijlink, F.; Beverdam, A.; Brouwer, A.; Oosterveen, T.C.; Berge, D. Vertebrate aristaless-related genes. Int. J. Dev. Biol. 2004, 43, 651–663. [Google Scholar]
- Wilkins, A.S.; Wrangham, R.W.; Fitch, W.T. The “domestication syndrome” in mammals: A unified explanation based on neural crest cell behavior and genetics. Genetics 2014, 197, 795–808. [Google Scholar] [CrossRef]
- Librado, P.; Gamba, C.; Gaunitz, C.; Der Sarkissian, C.; Pruvost, M.; Albrechtsen, A.; Fages, A.; Khan, N.; Schubert, M.; Jagannathan, V. Ancient genomic changes associated with domestication of the horse. Science 2017, 356, 442–445. [Google Scholar] [CrossRef]
- Niu, T.; Xu, X. Candidate genes for osteoporosis. Am. J. Pharm. 2001, 1, 11–19. [Google Scholar] [CrossRef]
- Garvican, E.; Clegg, P. Clinical aspects of the equine shoulder and elbow joints. UK Vet. Companion Anim. 2008, 13, 5–12. [Google Scholar] [CrossRef]
- Stover, S.M. Stress fracture diagnosis in racehorses. In Robinson’s Current Therapy in Equine Medicine, 7th ed.; Sprayberry, K.A., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Baccarin, R.Y. Accessing the equine elbow joint: New insights on an old approach. Vet. Rec. 2016, 179, 170–172. [Google Scholar] [CrossRef]
- Guérin, M.; Thariat, J.; Ouali, M.; Bouvier, C.; Decouvelaere, A.-V.; Cassagnau, E.; Aubert, S.; Lepreux, S.; Coindre, J.-M.; Valmary-Degano, S. A new subtype of high-grade mandibular osteosarcoma with RASAL1/MDM2 amplification. Hum. Pathol. 2016, 50, 70–78. [Google Scholar] [CrossRef]
- Tabareau-Delalande, F.; Collin, C.; Gomez-Brouchet, A.; Bouvier, C.; Decouvelaere, A.-V.; De Muret, A.; Pagès, J.-C.; De Pinieux, G. Chromosome 12 long arm rearrangement covering MDM2 and RASAL1 is associated with aggressive craniofacial juvenile ossifying fibroma and extracranial psammomatoid fibro-osseous lesions. Mod. Pathol. 2015, 28, 48. [Google Scholar] [CrossRef]
- Peng, G.; Westerfield, M. Lhx5 promotes forebrain development and activates transcription of secreted Wnt antagonists. Development 2006, 133, 3191–3200. [Google Scholar] [CrossRef]
- Hunter, C.S.; Rhodes, S.J. LIM-homeodomain genes in mammalian development and human disease. Mol. Biol. Rep. 2005, 32, 67–77. [Google Scholar] [CrossRef]
- Hanel, M.L.; Wuebbles, R.D.; Jones, P.L. Muscular dystrophy candidate gene FRG1 is critical for muscle development. Dev. Dyn. 2009, 238, 1502–1512. [Google Scholar] [CrossRef]
- Dolvik, N.I.; Klemetsdal, G. Arthritis in the carpal joints of Norwegian trotter—heritability, effects of inbreeding and conformation. Livest. Prod. Sci. 1994, 39, 283–290. [Google Scholar] [CrossRef]
- Makvandi-Nejad, S.; Hoffman, G.E.; Allen, J.J.; Chu, E.; Gu, E.; Chandler, A.M.; Loredo, A.I.; Bellone, R.R.; Mezey, J.G.; Brooks, S.A. Four loci explain 83% of size variation in the horse. PLoS ONE 2012, 7, e39929. [Google Scholar] [CrossRef]
- Boyko, A.R.; Brooks, S.A.; Behan-Braman, A.; Castelhano, M.; Corey, E.; Oliveira, K.C.; Swinburne, J.E.; Todhunter, R.J.; Zhang, Z.; Ainsworth, D.M. Genomic analysis establishes correlation between growth and laryngeal neuropathy in Thoroughbreds. BMC Genomics 2014, 15, 259. [Google Scholar] [CrossRef] [PubMed]
- Tetens, J.; Widmann, P.; Kühn, C.; Thaller, G. A genome-wide association study indicates LCORL/NCAPG as a candidate locus for withers height in German Warmblood horses. Anim. Genet. 2013, 44, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.-A.; Murray, J.E.; Carroll, P.; Leitch, A.; Mackenzie, K.J.; Halachev, M.; Fetit, A.E.; Keith, C.; Bicknell, L.S.; Fluteau, A. Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 2016, 30, 2158–2172. [Google Scholar] [CrossRef]
- Pollitt, C.; Anderson, L.V.; Pogue, R.; Davison, K.; Pyle, A.; Bushby, K. The phenotype of calpainopathy: Diagnosis based on a multidisciplinary approach. Neuromuscul. Disord. 2001, 11, 287–296. [Google Scholar] [CrossRef]
- Wilson, L.O.; Fahrer, A.M. Condensins, chromatin remodeling and gene transcription. In Chromatin Remodelling; Radzioch, D., Ed.; IntechOpen: London, UK, 2013. [Google Scholar]


| Joint Angle | Mean | SD | Min | Max | h2 | SE | ICC (20 LIP) |
|---|---|---|---|---|---|---|---|
| Poll | 102.55 | 4.70 | 85.35 | 117.29 | 0.38 | 0.098 | 0.92 |
| Neck–shoulder blade | 81.86 | 6.56 | 62.77 | 99.47 | 0.42 | 0.086 | 0.16 |
| Shoulder joint | 94.00 | 6.61 | 77.09 | 114.94 | 0.42 | 0.090 | 0.13 |
| Elbow joint | 129.50 | 5.79 | 112.60 | 147.50 | 0.22 | 0.076 | 0.52 |
| Carpal joint | 177.30 | 1.81 | 170.00 | 179.90 | 0.35 | 0.088 | 0.49 |
| Fetlock joint of the forelimb | 149.10 | 4.08 | 136.80 | 162.00 | 0.32 | 0.089 | 0.79 |
| Hip joint | 78.22 | 3.01 | 68.80 | 91.17 | 0.24 | 0.092 | 0.58 |
| Stifle joint | 99.76 | 4.08 | 89.28 | 115.26 | 0.40 | 0.089 | 0.90 |
| Hock | 152.90 | 2.30 | 145.00 | 159.50 | 0.25 | 0.095 | 0.96 |
| Fetlock joint of the hind limb | 155.80 | 6.43 | 137.60 | 177.90 | 0.58 | 0.086 | 0.81 |
| Joint Angle | Stud Farm | Birth Year Category | Age Category | Sex | Head in Relation to Camera | Position of Front Limb | Position of Hind Limb | Body Alignment to the Camera | Tail Position |
|---|---|---|---|---|---|---|---|---|---|
| Poll | ** | * | ** | * | ** | ||||
| Elbow joint | *** | ** | |||||||
| Carpal joint | *** | ||||||||
| Fetlock joint of the forelimb | ** | * | |||||||
| Hip joint | ** | * | *** | *** | |||||
| Stifle joint | *** | * | *** | *** | |||||
| Hock | *** | ||||||||
| Fetlock joint of the hind limb | * | *** | ** |
| Joint Angle | ECA | Position on EquCab2.0 | Position on EquCab 3.0 | #SNP | SNP with the Lowest p-Value | p-Value | Number of Genotyped Horses | Allele Frequency | ||
|---|---|---|---|---|---|---|---|---|---|---|
| AAFM,LIP | ABFM,LIP | BBFM,LIP | ||||||||
| Poll | 28 | 12,101,898–12,106,363 | 13,129,017–13,133,483 | 3 | AX-103978374 | 1.36 × 10−07 | 495 | 4911,38 | 19797,100 | 249176,73 |
| 1 | 124,405,158 | 125,551,151 | 1 | AX-103779310 | 6.83 × 10−07 | 495 | 138,5 | 13370,63 | 349206,143 | |
| Elbow joint | 29 | 18,799,958 | 19,878,299 | 1 | AX-103649839 | 1.69 × 10−07 | 490 | 3712,25 | 15469,85 | 299198,101 |
| Stifle joint | 8 | 19,266,146 | 21,704,931 | 1 | AX-102974274 | 2.77 × 10−07 | 473 | 454,41 | 18680,106 | 242178,64 |
| Fetlock joint of the hind limb | 27 | 22,021,462 | 22,068,848 | 1 | AX-103624054 | 5.42 × 10−07 | 495 | 53,2 | 10270,32 | 388211,177 |
| Carpal joint | 4 | 37,412,203 | 37,460,405 | 1 | AX-104691515 | 6.07 × 10−07 | 479 | 463,43 | 16054,106 | 273211,62 |
| 3 | 106,128,177 | 107,955,102 | 1 | AX-105008533 | 6.24 × 10−07 | 493 | 390,39 | 14328,115 | 311254,57 | |
| 7 | 42,659,817 | 43,694,770 | 1 | AX-103944161 | 8.83 × 10−07 | 495 | 222,20 | 18675,111 | 287207,80 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gmel, A.I.; Druml, T.; von Niederhäusern, R.; Leeb, T.; Neuditschko, M. Genome-Wide Association Studies Based on Equine Joint Angle Measurements Reveal New QTL Affecting the Conformation of Horses. Genes 2019, 10, 370. https://doi.org/10.3390/genes10050370
Gmel AI, Druml T, von Niederhäusern R, Leeb T, Neuditschko M. Genome-Wide Association Studies Based on Equine Joint Angle Measurements Reveal New QTL Affecting the Conformation of Horses. Genes. 2019; 10(5):370. https://doi.org/10.3390/genes10050370
Chicago/Turabian StyleGmel, Annik Imogen, Thomas Druml, Rudolf von Niederhäusern, Tosso Leeb, and Markus Neuditschko. 2019. "Genome-Wide Association Studies Based on Equine Joint Angle Measurements Reveal New QTL Affecting the Conformation of Horses" Genes 10, no. 5: 370. https://doi.org/10.3390/genes10050370
APA StyleGmel, A. I., Druml, T., von Niederhäusern, R., Leeb, T., & Neuditschko, M. (2019). Genome-Wide Association Studies Based on Equine Joint Angle Measurements Reveal New QTL Affecting the Conformation of Horses. Genes, 10(5), 370. https://doi.org/10.3390/genes10050370






