Micronuclei and Genome Chaos: Changing the System Inheritance
Abstract
1. Introduction
2. Brief History of Micronuclei Research
3. Examples of Linking Micronuclei to Biological Functions and Cancer
- -
- Link between micronuclei and cancer risk: Micronuclei are much more easily quantifiable than other types of chromosomal aberrations and the protocol and scoring of the CBMN assay is much easier than that of assessing metaphase chromosomes. A strong link has developed between chromosomal instability and cancer, with micronucleus formation being indicative of CIN. The current consensus is that micronuclei are linked to cancer risk, although there is a fair amount of variability in predictive power among cancer types [49]. The HUMN collaborative proved a useful tool to develop a clinical test based on micronuclei using their previously published criteria for scoring micronuclei in peripheral lymphocytes. Through analysis of data on micronucleus frequency studies in cancer patients and controls, there was found to be a strong link between urogenital and colon cancer development and patients who had micronuclei frequencies defined as medium to high [49]. Additionally, increased micronucleus frequency in lymphocytes has had variable correlations with lung and pancreatic cancer risks that may become validated with further studies [50,51]. Micronuclei are also being evaluated in cervical cells as a possible result of HPV infection and link to cervical cancer development [52]. Epithelial cells, due to the ease of access, have long been an attractive target for such a clinical test, but due to the common nuclear anomalies seen in these cell types that could have been mistaken for micronuclei, new methods had to be developed [53]. This work has laid the basis for further investigation of epithelial cell micronuclei. The HUMN collaborative has worked on a validation and coordination process for using buccal epithelial cells in the CBMN-Cyt protocol to develop an even easier method to access tissue as a clinical test for cancer risk [54]. A comparison of the developing technique between three laboratories showed a 20-fold increase in MN frequency among cancer patients compared to controls [55].
- -
- Tumor radiosensitivity: Tumor cells have been exposed to differing doses of radiation in vitro; the more micronuclei per binucleate cell after cytokinesis blocking, the less the surviving fraction of cells [56]. These results imply that micronucleus formation is correlated with cell death, a point corroborated by other studies that linked micronuclei formation during mitosis to eventual apoptosis [57]. There have been issues drawing conclusions about the micronuclei link to radiosensitivity in vivo.
- -
- Post-therapy assessment of tumors: Increased chromosomal instability has generally been associated with progression and poor outcome [58] but the link between increased micronuclei formation and apoptosis has opened up some therapeutic possibilities, resulting in a paradox: CIN has been viewed as beneficial in some studies and harmful in others. Is there an explanation? Some studies show that an increase in already rather unstable tumor cells may “tip the balance” toward apoptosis, resulting in tumor reduction [59,60]. This highlights the importance of being able to measure CIN.
- -
- Elimination of double minutes: The amplified genes represented by double minutes have been shown to be able to be removed from the cell by micronucleus formation [61,62]. As many elements contributing to invasiveness and poor outcome are carried in double minutes, this represents a possible therapeutic goal. Radiation therapy at low levels has been shown to expedite the removal of double minutes via micronucleus formation [63].
- -
- Relationship with genome chaos: Micronucleus formation may be a driver of CIN and chromosomal damage, not just a consequence of it. Micronuclei frequencies have been shown to increase in cells that have undergone checkpoint adaptation, entering mitosis with damaged DNA, a process normally prevented via the p53 pathway [64]. When the p53 pathway has been inactivated, numerous structural aneuploidies including micronuclei are able to form [65]. The resulting micronuclei may contain DNA that becomes damaged further via nuclear envelope collapse [66]. This damaged DNA may then be reintegrated into the genome, resulting in a localized chromosome rearrangement known as chromothripsis, a common feature of many cancers [9,67,68]. Some early studies have in fact shown that not all micronuclei can simply be eliminated, but can perform biological functions such as DNA synthesis and rejoining into other nuclei and further contributing to abnormal karyotypes [69,70]. However, like most NCCAs, they were regarded by the research community as insignificant background noise and have largely been ignored [7,71] Recently, time lapse microscopy has confirmed the fact that many micronuclei are actively involving the CIN process. However, without the theoretical framework to explain the meaning of altered karyotypes and how MN essentially contribute to new karyotypes, such observations would have failed to illustrate the importance of the micronuclei in cancer. The discovery of the association between punctuated microcellular evolution and karyotype chaos or genome chaos highlighted the importance of studying various unclassified types of chromosomal and nuclear abnormalities, including micronuclei [11,12]. The later realization that increased genome alterations are essential for cellular adaptation also put forth a positive perspective to this idea [12,13,14,29]. It was also realized that the altered karyotype represents a new genomic information package, as the order of genes along chromosomes represent genomic coding [23,72]. Thus, like many other types of NCCAs, the micronuclei are not just the products of biological errors or “bad” outcomes, but are also important components of the dynamics of the systems essential for cellular adaptation [13,14,21,22]. Recently, micronuclei have been linked to various genome chaos-related chromosomal and nuclear abnormalities, including “budding/bursting” and horizontal transfer [73,74,75,76]. Despite their different mechanisms of induction, they share the same evolutionary mechanism of creating new genomic information (more see later discussions). In fact, it has long been known that micronuclei have been detected in cells from patients with chromosomal instability/cancer susceptibility syndromes (e.g., Ataxia telangiectasia and Bloom syndrome) [77,78]. It is also known that elevated CIN can be detected from these patients. The linkage between CIN and micronuclei is obvious.
- -
- Link between immuno-systems: Recent studies have established that micronuclei activate the innate immune response [2,3,4,28]. Specifically, exposing fragmented DNA to the cytoplasm triggers the activation of IFN-I and IFN-stimulated inflammatory genes, which suggest the presence of micronuclei can be sensed by the cell defense machinery. Similarly, it was shown that double-stranded DNA breaks lead to the formation of micronuclei, which precede the activation of inflammatory signaling and are a repository for the pattern-recognition receptor cyclic GMP–AMP synthase. Such links between DNA damage, micronuclei, and immunity offer a new avenue to studying cancer evolution and drug treatment response.
- -
- Some key features of micronuclei differ from primary nuclei: Interestingly, not just the size difference, but both the degree of chromatin condensation and nuclear envelope composition differ between micronuclei and primary nuclei. Micronuclei also lack active proteasomes [26]. Together, the relationship between micronuclei and primary nuclei provides an excellent example of the emergent relationship between parts vs. entire system, as such emergence is not just about the quantitative difference, but different systems.
4. New Challenges and Opportunities for Micronuclei Research
4.1. Re-Organizing the Genome: A New Genomic and Evolutionary Framework for Understanding the Function of Micronuclei
4.1.1. The Recently Realized Genomic Information Context: System Inheritance and Fuzzy Inheritance
4.1.2. Evolutionary Context: Stress Induced Genomic Variants and Its Evolutionary Significance Coupled with Trade-Off
4.1.3. Limitation of Specific Pathways Used to Explain a Highly Complex System Behavior: Lower Level Agents vs. Higher Level Emergence
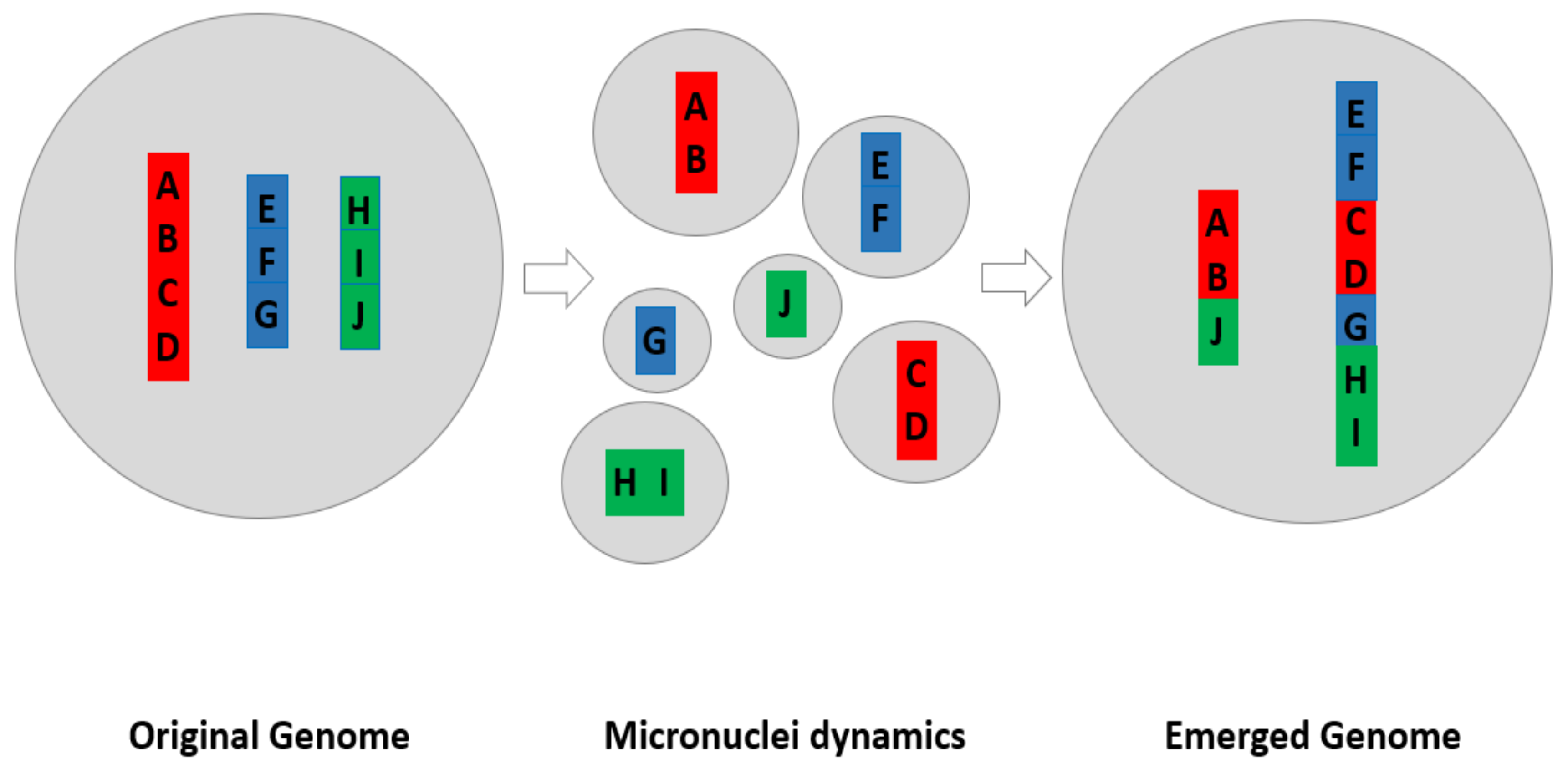
4.1.4. The Model of How Micronuclei Contribute to New Genome Systems by Creating New Genomic Coding
4.2. The Need for Further Research to Clarify Important Issues
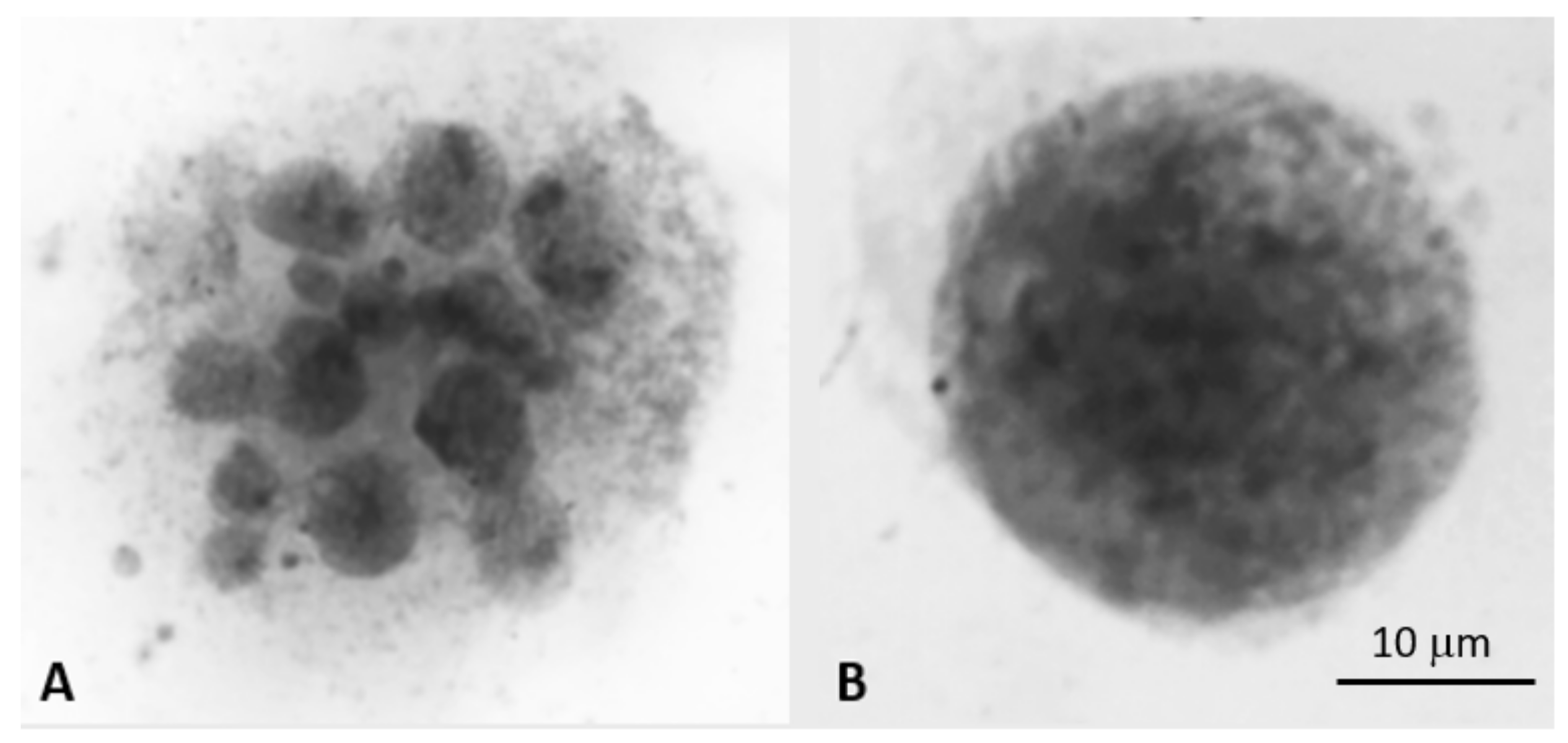
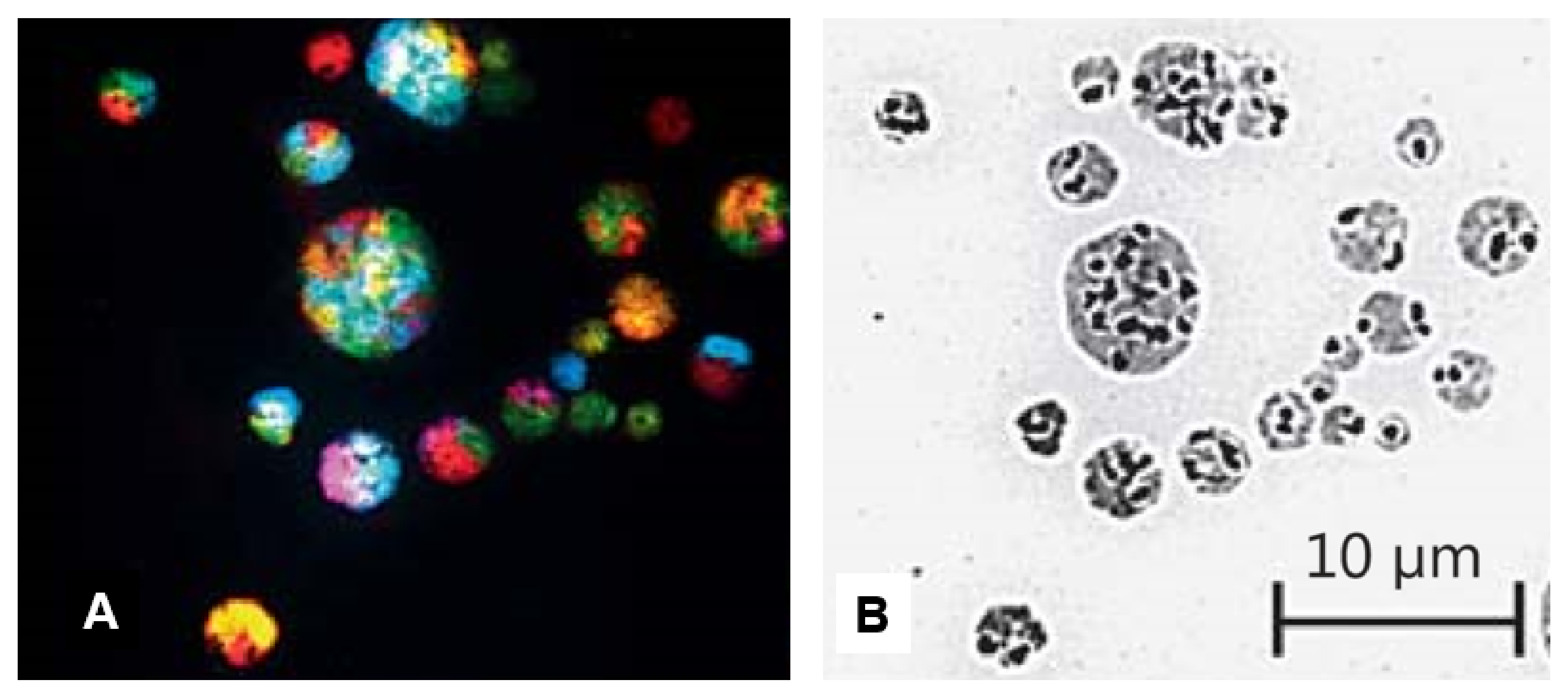
4.2.1. Differentiating Micronuclei Generated from Relatively Stable Genomes and Micronuclei Clusters from Unstable Genomes
4.2.2. Awareness to Confusing Issues
- a.
- Micronuclei and cell death:
- b.
- Fragmentations of DNA, chromosomes and nuclei:
- c.
- Micronuclei can be linked to genome chaos (including chromothripsis), as well as many other stress related factors.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fenech, M.; Knasmueller, S.; Bolognesi, C.; Bonassi, S.; Holland, N.; Migliore, L.; Palitti, F.; Natarajan, A.T.; Kirsch-Volders, M. Molecular mechanisms by which in vivo exposure to exogenous chemical genotoxic agents can lead to micronucleus formation in lymphocytes in vivo and ex vivo in humans. Mutat. Res. 2016, 770, 12–25. [Google Scholar] [CrossRef]
- Mackenzie, K.J.; Carroll, P.; Martin, C.A.; Murina, O.; Fluteau, A.; Simpson, D.J.; Olova, N.; Sutcliffe, H.; Rainger, J.K.; Leitch, A.; et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017, 548, 461–465. [Google Scholar] [CrossRef]
- Harding, S.M.; Benci, J.L.; Irianto, J.; Discher, D.E.; Minn, A.J.; Greenberg, R.A. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017, 548, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, K.; Knittler, K.; Borowski, C.; Rudnik, S.; Damme, M.; Aden, K.; Spehlmann, M.E.; Frey, N.; Saftig, P.; Chalaris, A.; et al. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum. Mol. Genet. 2017, 26, 3960–3972. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, N. Molecular mechanisms of the origin of micronuclei from extrachromosomal elements. Mutagenesis 2011, 26, 119–123. [Google Scholar] [CrossRef]
- Erenpreisa, J.E.; Ivanov, A.; Dekena, G.; Vitina, A.; Krampe, R.; Freivalds, T.; Selivanova, G.; Roach, H.I. Arrest in metaphase and anatomy of mitotic catastrophe: mild heat shock in two human osteosarcoma cell lines. Cell. Biol. Int. 2000, 24, 61–70. [Google Scholar] [CrossRef]
- Ambros, I.M.; Rumpler, S.; Luegmayr, A.; Hattinger, C.M.; Strehl, S.; Kovar, H.; Gadner, H.; Ambros, P.F. Neuroblastoma cells can actively eliminate supernumerary MYCN gene copies by micronucleus formation—Sign of tumour cell revertance? Eur. J. Cancer 1997, 33, 2043–2049. [Google Scholar] [CrossRef]
- Heng, H.H.; Liu, G.; Bremer, S.; Ye, K.J.; Stevens, J.; Ye, C.J. Clonal and non-clonal chromosome aberrations and genome variation and aberration. Genome 2006, 49, 195–204. [Google Scholar] [CrossRef]
- Stevens, J.B.; Liu, G.; Bremer, S.W.; Ye, K.J.; Xu, W.; Xu, J.; Sun, Y.; Wu, G.S.; Savasan, S.; Krawetz, S.A.; et al. Mitotic cell death by chromosome fragmentation. Cancer Res. 2007, 67, 7686–7694. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mercado-Uribe, I.; Xing, Z.; Sun, B.; Kuang, J.; Liu, J. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene 2014, 33, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Stevens, J.B.; Horne, S.D.; Abdallah, B.Y.; Ye, K.J.; Bremer, S.W.; Ye, C.J.; Chen, D.J.; Heng, H.H. Genome chaos: Survival strategy during crisis. Cell Cycle 2014, 13, 528–537. [Google Scholar] [CrossRef]
- Heng, H.H.; Liu, G.; Stevens, J.B.; Abdallah, B.Y.; Horne, S.D.; Ye, K.J.; Bremer, S.W.; Chowdhury, S.K.; Ye, C.J. Karyotype heterogeneity and unclassified chromosomal abnormalities. Cytogenet. Genome Res. 2013, 139, 144–157. [Google Scholar] [CrossRef]
- Heng, H.H. Debating Cancer: The Paradox in Cancer Research; World Scientific Publishing Co.: Singapore, 2015; ISBN 978-981-4520-84-3. [Google Scholar]
- Heng, H.H. Genome Chaos: Rethinking Genetics, Evolution, and Molecular Medicine; Academic Press Elsevier: Cambridge, MA, USA, 2019; ISBN 978-012-8136-35-5. [Google Scholar]
- Ye, C.J.; Liu, G.; Heng, H.H. Experimental induction of genome chaos. Methods Mol. Biol. 2018, 1769, 337–352. [Google Scholar] [CrossRef]
- Stevens, J.B.; Abdallah, B.Y.; Liu, G.; Horne, S.D.; Bremer, S.W.; Ye, K.J.; Huang, J.Y.; Kurkinen, M.; Ye, C.J.; Heng, H.H. Heterogeneity of cell death. Cytogenet. Genome Res. 2013, 139, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.B.; Abdallah, B.Y.; Liu, G.; Ye, C.J.; Horne, S.D.; Wang, G.; Savasan, S.; Shekhar, M.; Krawetz, S.A.; Hüttemann, M.; et al. Diverse system stresses: Common mechanisms of chromosome fragmentation. Cell Death Dis. 2011, 2, e178. [Google Scholar] [CrossRef]
- Heng, H.H.; Bremer, S.W.; Stevens, J.; Ye, K.J.; Miller, F.; Liu, G.; Ye, C.J. Cancer progression by non-clonal chromosome aberrations. J. Cell Biochem. 2006, 98, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.; Stevens, J.B.; Liu, G.; Bremer, S.W.; Ye, K.J.; Reddy, P.V.; Wu, G.S.; Wang, Y.A.; Tainsky, M.A.; Ye, C.J. Stochastic cancer progression driven by non-clonal chromosome aberrations. J. Cell Physiol. 2006, 208, 461–472. [Google Scholar] [CrossRef]
- Horne, S.D.; Pollick, S.A.; Heng, H.H. Evolutionary mechanism unifies the hallmarks of cancer. Int. J. Cancer 2015, 136, 2012–2021. [Google Scholar] [CrossRef]
- Heng, H.H.; Regan, S.M.; Liu, G.; Ye, C.J. Why it is crucial to analyze non clonal chromosome aberrations or NCCAs? Mol. Cytogenet. 2016, 9, 15. [Google Scholar] [CrossRef]
- Ye, C.J.; Regan, S.; Liu, G.; Alemara, S.; Heng, H.H. Understanding aneuploidy in cancer through the lens of system inheritance, fuzzy inheritance and emergence of new genome systems. Mol. Cytogenet. 2018, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H. The genome-centric concept: Resynthesis of evolutionary theory. Bioessays 2009, 31, 512–525. [Google Scholar] [CrossRef]
- Heng, H.H.; Bremer, S.W.; Stevens, J.B.; Horne, S.D.; Liu, G.; Abdallah, B.Y.; Ye, K.J.; Ye, C.J. Chromosomal instability (CIN): What it is and why it is crucial to cancer evolution. Cancer Metastasis Rev. 2013, 32, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.; Stevens, J.B.; Bremer, S.W.; Ye, K.J.; Liu, G.; Ye, C.J. The evolutionary mechanism of cancer. J. Cell Biochem. 2010, 109, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Maass, K.K.; Rosing, F.; Ronchi, P.; Willmund, K.V.; Devens, F.; Hergt, M.; Herrmann, H.; Lichter, P.; Ernst, A. Altered nuclear envelope structure and proteasome function of micronuclei. Exp. Cell Res. 2018, 371, 353–363. [Google Scholar] [CrossRef]
- Liu, S.; Kwon, M.; Mannino, M.; Yang, N.; Renda, F.; Khodjakov, A.; Pellman, D. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 2018, 561, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Gekara, N.O. DNA damage-induced immune response: Micronuclei provide key platform. J. Cell Biol. 2017, 216, 2999–3001. [Google Scholar] [CrossRef]
- Horne, S.D.; Chowdhury, S.K.; Heng, H.H. Stress, genomic adaptation, and the evolutionary trade-off. Front. Genet. 2014, 5, 92. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.J.; Stevens, J.B.; Liu, G.; Bremer, S.W.; Jaiswal, A.S.; Ye, K.J.; Lin, M.F.; Lawrenson, L.; Lancaster, W.D.; Kurkinen, M.; et al. Genome based cell population heterogeneity promotes tumorigenicity: The evolutionary mechanism of cancer. J. Cell Physiol. 2009, 219, 288–300. [Google Scholar] [CrossRef]
- Heng, H.H.; Stevens, J.B.; Bremer, S.W.; Liu, G.; Abdallah, B.Y.; Ye, C.J. Evolutionary mechanisms and diversity in cancer. Adv. Cancer Res. 2011, 112, 217–253. [Google Scholar] [CrossRef]
- Heng, H.H.; Horne, S.D.; Stevens, J.B.; Abdallah, B.Y.; Liu, G.; Chowdhury, S.K.; Bremer, S.W.; Zhang, K.; Ye, C.J. Heterogeneity mediated system complexity: The ultimate challenge for studying common and complex diseases. In The Value of Systems and Complexity Sciences for Healthcare; Sturmberg, J.P., Ed.; Springer International Publishing: New York, NY, USA, 2016; pp. 107–120. ISBN 978-3-319-26221-5. [Google Scholar]
- Heng, H.H. The conflict between complex systems and reductionism. JAMA 2008, 300, 1580–1581. [Google Scholar] [CrossRef] [PubMed]
- Sears, D.A.; Udden, M.M. Howell-Jolly bodies: A brief historical review. Am. J. Med. Sci. 2012, 343, 407–409. [Google Scholar] [CrossRef]
- Evans, H.J.; Neary, G.J.; Williamson, F.S. The relative biological efficiency of single doses of fast neutrons and gamma-rays on Vicia faba roots and the effect of oxygen. Part II. Chromosome damage: The production of micronuclei. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1959, 1, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Schmid, W. The micronucleus test. Mutat. Res. 1975, 31, 9–15. [Google Scholar] [CrossRef]
- Countryman, P.I.; Heddle, J.A. The production of micronuclei from chromosome aberrations in irradiated cultures of human lymphocytes. Mutat. Res. 1976, 41, 321–332. [Google Scholar] [CrossRef]
- Countryman, P.I.; Heddle, J.A. A true microculture technique for human lymphocytes. Hum. Genet. 1977, 35, 197–200. [Google Scholar] [CrossRef]
- Fenech, M.; Morley, A. Solutions to the kinetic problem in the micronucleus assay. Cytobios 1985, 43, 233–246. [Google Scholar]
- Fenech, M. The in vitro micronucleus technique. Mutat. Res. 2000, 455, 81–95. [Google Scholar] [CrossRef]
- Fenech, M. The advantages and disadvantages of the cytokinesis-block micronucleus method. Mutat. Res. 1997, 392, 11–18. [Google Scholar] [CrossRef]
- Kirsch-Volders, M.; Fenech, M. Inclusion of micronuclei in non-divided mononuclear lymphocytes and necrosis/apoptosis may provide a more comprehensive cytokinesis block micronucleus assay for biomonitoring purposes. Mutagenesis 2001, 16, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Chang, W.P.; Kirsch-Volders, M.; Holland, N.; Bonassi, S.; Zeiger, E.; project, H.M. HUMN project: Detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat. Res. 2003, 534, 65–75. [Google Scholar] [CrossRef]
- Bonassi, S.; Fenech, M.; Lando, C.; Lin, Y.P.; Ceppi, M.; Chang, W.P.; Holland, N.; Kirsch-Volders, M.; Zeiger, E.; Ban, S.; et al. HUman MicroNucleus project: International database comparison for results with the cytokinesis-block micronucleus assay in human lymphocytes: I. Effect of laboratory protocol, scoring criteria, and host factors on the frequency of micronuclei. Environ. Mol. Mutagen 2001, 37, 31–45. [Google Scholar] [CrossRef]
- Fenech, M. Cytokinesis-block micronucleus assay evolves into a “cytome” assay of chromosomal instability, mitotic dysfunction and cell death. Mutat. Res. 2006, 600, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Bull, C.F.; Beetstra-Hill, S.; Benassi-Evans, B.J.; Crott, J.W.; Kimura, M.; Teo, T.; Wu, J.; Fenech, M.F. Application and adaptation of the in vitro micronucleus assay for the assessment of nutritional requirements of cells for DNA damage prevention. Mutagenesis 2011, 26, 193–197. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lyulko, O.V.; Garty, G.; Randers-Pehrson, G.; Turner, H.C.; Szolc, B.; Brenner, D.J. Fast image analysis for the micronucleus assay in a fully automated high-throughput biodosimetry system. Radiat. Res. 2014, 181, 146–161. [Google Scholar] [CrossRef]
- Thompson, S.L.; Bakhoum, S.F.; Compton, D.A. Mechanisms of chromosomal instability. Curr. Biol. 2010, 20, R285–R295. [Google Scholar] [CrossRef]
- Bonassi, S.; Znaor, A.; Ceppi, M.; Lando, C.; Chang, W.P.; Holland, N.; Kirsch-Volders, M.; Zeiger, E.; Ban, S.; Barale, R.; et al. An increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of cancer in humans. Carcinogenesis 2007, 28, 625–631. [Google Scholar] [CrossRef] [PubMed]
- El-Zein, R.A.; Schabath, M.B.; Etzel, C.J.; Lopez, M.S.; Franklin, J.D.; Spitz, M.R. Cytokinesis-blocked micronucleus assay as a novel biomarker for lung cancer risk. Cancer Res. 2006, 66, 6449–6456. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Li, Y.; Li, D. Micronuclei levels in peripheral blood lymphocytes as a potential biomarker for pancreatic cancer risk. Carcinogenesis 2011, 32, 210–215. [Google Scholar] [CrossRef]
- Adam, M.L.; Pini, C.; Túlio, S.; Cantalice, J.C.; Torres, R.A.; Dos Santos Correia, M.T. Assessment of the association between micronuclei and the degree of uterine lesions and viral load in women with human papillomavirus. Cancer Genom. Proteomics 2015, 12, 67–71. [Google Scholar]
- Tolbert, P.E.; Shy, C.M.; Allen, J.W. Micronuclei and other nuclear anomalies in buccal smears: Methods development. Mutat. Res. 1992, 271, 69–77. [Google Scholar] [CrossRef]
- Holland, N.; Bolognesi, C.; Kirsch-Volders, M.; Bonassi, S.; Zeiger, E.; Knasmueller, S.; Fenech, M. The micronucleus assay in human buccal cells as a tool for biomonitoring DNA damage: The HUMN project perspective on current status and knowledge gaps. Mutat. Res. 2008, 659, 93–108. [Google Scholar] [CrossRef]
- Bolognesi, C.; Bonassi, S.; Knasmueller, S.; Fenech, M.; Bruzzone, M.; Lando, C.; Ceppi, M. Clinical application of micronucleus test in exfoliated buccal cells: A systematic review and metanalysis. Mutat. Res. 2015, 766, 20–31. [Google Scholar] [CrossRef]
- Shibamoto, Y.; Streffer, C.; Fuhrmann, C.; Budach, V. Tumor radiosensitivity prediction by the cytokinesis-block micronucleus assay. Radiat. Res. 1991, 128, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Utani, K.; Kohno, Y.; Okamoto, A.; Shimizu, N. Emergence of micronuclei and their effects on the fate of cells under replication stress. PLoS ONE 2010, 5, e10089. [Google Scholar] [CrossRef] [PubMed]
- McClelland, S.E. Role of chromosomal instability in cancer progression. Endocr. Relat. Cancer 2017, 24, T23–T31. [Google Scholar] [CrossRef] [PubMed]
- Birkbak, N.J.; Eklund, A.C.; Li, Q.; McClelland, S.E.; Endesfelder, D.; Tan, P.; Tan, I.B.; Richardson, A.L.; Szallasi, Z.; Swanton, C. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011, 71, 3447–3452. [Google Scholar] [CrossRef]
- Vargas-Rondón, N.; Villegas, V.E.; Rondón-Lagos, M. The role of chromosomal instability in cancer and therapeutic responses. Cancers 2017, 10, 4. [Google Scholar] [CrossRef]
- Valent, A.; Bénard, J.; Clausse, B.; Barrois, M.; Valteau-Couanet, D.; Terrier-Lacombe, M.J.; Spengler, B.; Bernheim, A. In vivo elimination of acentric double minutes containing amplified MYCN from neuroblastoma tumor cells through the formation of micronuclei. Am. J. Pathol. 2001, 158, 1579–1584. [Google Scholar] [CrossRef]
- Ji, W.; Bian, Z.; Yu, Y.; Yuan, C.; Liu, Y.; Yu, L.; Li, C.; Zhu, J.; Jia, X.; Guan, R.; et al. Expulsion of micronuclei containing amplified genes contributes to a decrease in double minute chromosomes from malignant tumor cells. Int. J. Cancer 2014, 134, 1279–1288. [Google Scholar] [CrossRef]
- Schoenlein, P.V.; Barrett, J.T.; Kulharya, A.; Dohn, M.R.; Sanchez, A.; Hou, D.Y.; McCoy, J. Radiation therapy depletes extrachromosomally amplified drug resistance genes and oncogenes from tumor cells via micronuclear capture of episomes and double minute chromosomes. Int. J. Radiat. Oncol. Biol. Phys. 2003, 55, 1051–1065. [Google Scholar] [CrossRef]
- Lewis, C.W.; Golsteyn, R.M. Cancer cells that survive checkpoint adaptation contain micronuclei that harbor damaged DNA. Cell Cycle 2016, 15, 3131–3145. [Google Scholar] [CrossRef]
- Soto, M.; Raaijmakers, J.A.; Bakker, B.; Spierings, D.C.J.; Lansdorp, P.M.; Foijer, F.; Medema, R.H. p53 Prohibits propagation of chromosome segregation errors that produce structural aneuploidies. Cell Rep. 2017, 19, 2423–2431. [Google Scholar] [CrossRef]
- Hatch, E.M.; Fischer, A.H.; Deerinck, T.J.; Hetzer, M.W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 2013, 154, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Hatch, E.M.; Hetzer, M.W. Linking micronuclei to chromosome fragmentation. Cell 2015, 161, 1502–1504. [Google Scholar] [CrossRef]
- Kramer, J.; Schaich-Walch, G.; Nüsse, M. DNA synthesis in radiation-induced micronuclei studied by bromodeoxyuridine (BrdUrd) labelling and anti-BrdUrd antibodies. Mutagenesis 1990, 5, 491–495. [Google Scholar] [CrossRef]
- Gustavino, B.; Degrassi, F.; Filipponi, R.; Modesti, D.; Tanzarella, C.; Rizzoni, M. Mitotic indirect non-disjunction in phytohemagglutinin stimulated human lymphocytes. Mutagenesis 1994, 9, 17–21. [Google Scholar] [CrossRef]
- Abdallah, B.Y.; Horne, S.D.; Stevens, J.B.; Liu, G.; Ying, A.Y.; Vanderhyden, B.; Krawetz, S.A.; Gorelick, R.; Heng, H.H. Single cell heterogeneity: Why unstable genomes are incompatible with average profiles. Cell Cycle 2013, 12, 3640–3649. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.; Liu, G.; Stevens, J.B.; Bremer, S.W.; Ye, K.J.; Abdallah, B.Y.; Horne, S.D.; Ye, C.J. Decoding the genome beyond sequencing: The new phase of genomic research. Genomics 2011, 98, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. Coming full circle-from endless complexity to simplicity and back again. Cell 2014, 157, 267–271. [Google Scholar] [CrossRef]
- Mirzayans, R.; Andrais, B.; Murray, D. Roles of polyploid/multinucleated giant cancer cells in metastasis and disease relapse following anticancer treatment. Cancers 2018, 10, E118. [Google Scholar] [CrossRef]
- Díaz-Carballo, D.; Saka, S.; Klein, J.; Rennkamp, T.; Acikelli, A.H.; Malak, S.; Jastrow, H.; Wennemuth, G.; Tempfer, C.; Schmitz, I.; et al. A distinct oncogenerative multinucleated cancer cell serves as a source of stemness and tumor heterogeneity. Cancer Res. 2018, 78, 2318–2331. [Google Scholar] [CrossRef]
- Mirzayans, R.; Andrais, B.; Kumar, P.; Murray, D. The growing complexity of cancer cell response to DNA-damaging agents: Caspase 3 mediates cell death or survival? Int. J. Mol. Sci. 2016, 17, 708. [Google Scholar] [CrossRef]
- Tomanin, R.; Sarto, F.; Mazzotti, D.; Giacomelli, L.; Raimondi, F.; Trevisan, C. Louis-Bar syndrome: Spontaneous and induced chromosomal aberrations in lymphocytes and micronuclei in lymphocytes, oral mucosa and hair root cells. Hum. Genet. 1990, 85, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Rosin, M.P.; German, J. Evidence for chromosome instability in vivo in Bloom syndrome: Increased numbers of micronuclei in exfoliated cells. Hum. Genet. 1985, 71, 187–191. [Google Scholar] [CrossRef]
- Chen, J.; Niu, N.; Zhang, J.; Qi., L.; Shen, W.; Donkena, K.V.; Feng, Z.; Liu, J. Polyploid giant cancer cells (PGCCs): The evil roots of cancer. Curr. Cancer Drug Targets 2018, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, A.A.; Heng, H.H. Transient and stable vector transfection: Pitfalls, off-target effects, artifacts. Mutat. Res. 2017, 773, 91–103. [Google Scholar] [CrossRef]
- Högstedt, B.; Karlsson, A. The size of micronuclei in human lymphocytes varies according to inducing agent used. Mutat. Res. 1985, 156, 229–232. [Google Scholar] [CrossRef]
- Hashimoto, K.; Nakajima, Y.; Matsumura, S.; Chatani, F. An in vitro micronucleus assay with size-classified micronucleus counting to discriminate aneugens from clastogens. Toxicol. In Vitro 2010, 24, 208–216. [Google Scholar] [CrossRef]
- Yasui, M.; Koyama, N.; Koizumi, T.; Senda-Murata, K.; Takashima, Y.; Hayashi, M.; Sugimoto, K.; Honma, M. Live cell imaging of micronucleus formation and development. Mutat. Res. 2010, 692, 12–18. [Google Scholar] [CrossRef]
- Heng, H.H.; Stevens, J.B.; Liu, G.; Bremer, S.W.; Ye, C.J. Imaging genome abnormalities in cancer research. Cell Chromosome 2004, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.L.; Yuen, K.L.; Tang, H.M.; Fung, M.C. Reversibility of apoptosis in cancer cells. Br. J. Cancer 2009, 100, 118–122. [Google Scholar] [CrossRef]
- Tang, H.M.; Talbot, C.C.; Fung, M.C.; Tang, H.L. Molecular signature of anastasis for reversal of apoptosis. F1000 Res. 2017, 6, 43. [Google Scholar] [CrossRef]
- Gong, Y.N.; Guy, C.; Olauson, H.; Becker, J.U.; Yang, M.; Fitzgerald, P.; Linkermann, A.; Green, D.R. ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell 2017, 169, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Hamann, J.C.; Surcel, A.; Chen, R.; Teragawa, C.; Albeck, J.G.; Robinson, D.N.; Overholtzer, M. Entosis is induced by glucose starvation. Cell. Rep. 2017, 20, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.B.; Liu, G.; Abdallah, B.Y.; Horne, S.D.; Ye, K.J.; Bremer, S.W.; Ye, C.J.; Krawetz, S.A.; Heng, H.H. Unstable genomes elevate transcriptome dynamics. Int. J. Cancer 2014, 134, 2074–2087. [Google Scholar] [CrossRef]
- Stevens, J.B.; Abdallah, B.Y.; Regan, S.M.; Liu, G.; Bremer, S.W.; Ye, C.J.; Heng, H.H. Comparison of mitotic cell death by chromosome fragmentation to premature chromosome condensation. Mol. Cytogenet. 2010, 3, 20. [Google Scholar] [CrossRef]
- Zhang, C.Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Niederwieser, C.; Nicolet, D.; Carroll, A.J.; Kolitz, J.E.; Powell, B.L.; Kohlschmidt, J.; Stone, R.M.; Byrd, J.C.; Mrózek, K.; Bloomfield, C.D. Chromosome abnormalities at onset of complete remission are associated with worse outcome in patients with acute myeloid leukemia and an abnormal karyotype at diagnosis: CALGB 8461 (Alliance). Haematologica 2016, 101, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Rangel, N.; Forero-Castro, M.; Rondón-Lagos, M. New insights in the cytogenetic practice: Karyotypic chaos, non-clonal chromosomal alterations and chromosomal instability in human cancer and therapy response. Genes 2017, 8, 155. [Google Scholar] [CrossRef]
- Frias, S.; Ramos, S.; Salas, C.; Molina, B.; Sánchez, S.; Rivera-Luna, R. Nonclonal chromosome aberrations and genome chaos in somatic and germ cells from patients and survivors of hodgkin lymphoma. Genes (Basel) 2019, 10, 37. [Google Scholar] [CrossRef]
- Iourov, I.Y.; Vorsanova, S.G.; Yurov, Y.B. Chromosomal mosaicism goes global. Mol. Cytogenet. 2008, 1, 26. [Google Scholar] [CrossRef] [PubMed]
- Vorsanova, S.G.; Yurov, Y.B.; Soloviev, I.V.; Iourov, I.Y. Molecular cytogenetic diagnosis and somatic genome variations. Curr. Genom. 2010, 11, 440–446. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heng, H.H.; Liu, G.; Alemara, S.; Regan, S.; Armstrong, Z.; Ye, CJ. The mechanisms of how genomic heterogeneity impacts bio-emergent properties: The challenges for precision medicine. In Embracing Complexity in Health; Springer: Basel, Switzerland, 2019. [Google Scholar]
| Genome Chaos |
|
| The relationship between genome chaos and other types of chromosomal or nuclear abnormalities |
|
| The relationship between stress and genome alteration-mediated evolution |
|
| Fuzzy inheritance: Understanding the diverse morphological variations of chromosomes and nuclei | It has been very confusing why there are many stochastic chromosomal variants. Traditionally, it was thought that these non-clonal abnormal structures were insignificant “noise”. They also were considered as the results of bio-errors. Following mapping of the non-clonal chromosomal aberrations into the dynamic phase of cancer evolution, it was realized that all these non-clonal variants represent evolutionary potential. Furthermore, the search for biological meaning of these highly diverse genome level variants has finally led to the concept of fuzzy inheritance [13,14,21,22]. Based on a new understanding of the genomic basis of bio-heterogeneity, it is now realized that the inheritance itself needs to be heterogenous as well. In contrast to the gene theory, which states that a gene codes for a specific, fixed phenotype, the genome theory suggests that most genes code for a range of potential phenotypes depending on context provided by other genes and the environment. From this “fuzzy” range of phenotypes, the respective environment can then allow the best-suited status to be “chosen” [22]. Such inheritance that codes for a range of phenotypes, not just a fixed phenotype, is named as fuzzy inheritance. Fuzzy inheritance can be observed at the gene, epigenetic and genome level. Furthermore, genome instability can increase the fuzziness of the inheritance.Equally important, the fuzzy inheritance explains why there are so many diverse nuclear variations including all types of micronuclei, especially under high stress or within the phase of macro-evolution. It is now clear that despite different morphological features and different mechanisms to produce them (either directly from near 2n cell or giant cells with >4n), these fundamentally represent fuzzy inheritance at the somatic cell level, with the evolutionary function remaining the same, changing the genome encoded genomic information. |
| Genome Theory | Departing from gene theory where genes determine the individual’s characteristics, and represent the independent informational unit, genome theory considers that the genomic topology serves as the context of the gene interactions, and genomic inheritance is about the network structure determined by chromosomal coding (gene order along and among chromosomes). There are 12 principles for genome theory. Among them, that the genome organizes the interactive relationship among genes, the genome is the main platform for evolutionary selection, and the genome functions as a main constraint for a given species are some of the important ones [13,14,23]. |
| Characteristic | HUMN Standardization Criteria: Peripheral Blood Lymphocytes [43] | Tolbert 1992 Exfoliated Cells [53] | Heddle 1976 Peripheral Blood Lymphocytes [37] | HUMN Laboratory Survey [44] | Comments |
|---|---|---|---|---|---|
| Size | 1/16 to 1/3 of diameter of main nucleus | Less than 1/3 of diameter of main nucleus; no lower limit if shape and color are discernable | Less than 1/3 of main nucleus | 1/16 to 1/3 of diameter of main nucleus (91% of labs surveyed) | Multinucleated cell division and fusion as seen in single cancer cells and genome chaos may alter these criteria, as several nuclei much larger than the average micronucleus may result. |
| Shape | Round or oval with own membrane | Round and smooth, suggests a membrane | N/A | Round or oval (99%) Morphologically identical to main nucleus (62%) | Micronuclei may have abnormal shapes depending on mechanism and integrity of the nuclear envelope. A disordered/disrupted nuclear envelope in a micronucleus-type structure likely leads to DNA content loss but is still a reflection of chromosomal instability. |
| Stain | Same staining intensity as main nuclei, but can be slightly darker | Mostly same as main nuclei | Mostly same as main nuclei or lighter | Same intensity as main nucleus (83%) Same color as main nucleus (85%) | Micronuclei may be on a separate “condensation schedule” than the main nucleus which may contribute to DNA damage to their genetic content |
| Overlap/Contact | May not overlap with main nucleus but can touch itMust have a distinguishable boundary with no bridge, not linked or connected | No overlap or bridge with main nuclei | No overlap or contact with main nuclei | Overlapping with or touching main nuclei is allowed as long as it is distinguishable (49%) No bridge to main nuclei (73%) | HUMN regards micronucleus-like structures continuous with the main nuclear membrane as nuclear buds (NBUDS); these are often formed in interphase from double minutes. Inclusion of NBUDs in scoring systems often treats them as a separate phenomenon. |
| Other | Non-refractile | Same focal plane as main nuclei Feulgen-positive | Non-refractile Within 3 to 4 nuclear diameters of main nuclei No more than 2 micronuclei associated with same nucleus | Non-refractile (95%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, C.J.; Sharpe, Z.; Alemara, S.; Mackenzie, S.; Liu, G.; Abdallah, B.; Horne, S.; Regan, S.; Heng, H.H. Micronuclei and Genome Chaos: Changing the System Inheritance. Genes 2019, 10, 366. https://doi.org/10.3390/genes10050366
Ye CJ, Sharpe Z, Alemara S, Mackenzie S, Liu G, Abdallah B, Horne S, Regan S, Heng HH. Micronuclei and Genome Chaos: Changing the System Inheritance. Genes. 2019; 10(5):366. https://doi.org/10.3390/genes10050366
Chicago/Turabian StyleYe, Christine J., Zachary Sharpe, Sarah Alemara, Stephanie Mackenzie, Guo Liu, Batoul Abdallah, Steve Horne, Sarah Regan, and Henry H. Heng. 2019. "Micronuclei and Genome Chaos: Changing the System Inheritance" Genes 10, no. 5: 366. https://doi.org/10.3390/genes10050366
APA StyleYe, C. J., Sharpe, Z., Alemara, S., Mackenzie, S., Liu, G., Abdallah, B., Horne, S., Regan, S., & Heng, H. H. (2019). Micronuclei and Genome Chaos: Changing the System Inheritance. Genes, 10(5), 366. https://doi.org/10.3390/genes10050366






