1. Introduction
The dysfunction and death of photoreceptors are the main cause of vision loss in inherited retinal diseases (IRDs), a group of mendelian rare disorders with high genetic and clinical heterogeneity. After more than 30 years of intense clinical and genetic studies to identify IRD genes, around 300 causative genes have been identified that cause the dysfunction of photoreceptors or alter the function of the adjacent retinal pigment epithelium (RPE), thus leading to the progressive attrition of photoreceptors [
1,
2].
From the clinical side and most relevant to patients, the main challenge is to devise effective treatments to halt the progression of the disease or regain visual capacity. Depending on the gene and type of mutation, different molecular approaches for therapy can be devised; but, at least for autosomal recessive retinal diseases, the most straightforward strategy is the restoration of a fully functional version of the protein via DNA-based gene therapy, even though gene delivery to fully differentiated and mature cells is not an easy task [
3,
4]. On the other hand, the eye is an excellent target for gene therapy since it is accessible, amenable to non-invasive examination, possesses a well-defined anatomy and is relatively immune privileged [
3,
4,
5]. Ocular gene therapy for retinal disorders is starting to be developed and the first commercial gene therapy to treat a severe infantile genetic blindness (Leber congenital amaurosis caused by bi-allelic mutations in the
RPE65 gene) was approved last year. These and other ongoing therapies in different clinical trial phases are based on adeno-associated virus (AAV) vectors [
6]. However, viral gene delivery also raises several concerns, such as small size capacity (maximum packaging size for 5 kb), high production costs, probability of immunogenicity and inflammatory responses and their invasive route of administration to the retina (subretinal microinjection), which have fostered basic research on non-viral vectors such as nanoparticles (NPs) for gene delivery.
NPs are highly customizable, and can be designed and optimized for cellular uptake, bypassing the degradative machinery of the cells and improving gene expression in the nucleus [
7]. Although a priori NPs might yield lower transgene expression levels compared to viral vectors, they overcome most safety concerns of viral vectors, are cost-effective and easily customizable and, most relevant, have a large DNA capacity, critical for gene delivery of large ocular disease genes [
4,
8]. All these desirable characteristics make the study of NPs for gene delivery to ocular tissues (cornea, retina and retinal pigment epithelium (RPE)) a research priority in order to explore viable alternatives to viral vectors. Nanoparticles made from different materials that display distinct properties, such as inorganic NPs, liposomes, solid lipids and polymeric NPs, have potential use in retinal cells [
3,
4,
9,
10]. Some successful attempts using compact DNA polycationic nanoparticles for gene delivery into the retina of pre-clinical animal models have also been reported [
11,
12].
However, more basic research is still needed to provide a full toolbox of NPs for different uses in the retina, including gene therapy. For instance, liposomal-based NPs, particularly those with positive charges, show cytotoxicity at the large quantities required for efficient gene delivery [
13]. The use of polyethilenimine (PEI)-coated NPs increases cellular uptake but PEI is also cytotoxic [
3,
9]. Gold NPs are mostly non-toxic, but cellular uptake strongly depends on their size and diameter. Several reports indicate that gold NPs ranging between 20 and 50 nm are non-toxic [
10]. Indeed, NPs display different physicochemical properties depending on the size, shape, surface charge and hydrophilicity, and these parameters directly impact cellular uptake by different types of vesicular and non-vesicular entry pathways, thereby determining the endocytic route and the final destination of NPs. In gene delivery, the internalization of NPs through the endosomes and eventual fusion of late endosomes with lysosomes should be avoided since it involves the destruction of nucleic acids by lysosomal hydrolases before the genetic material reaches the nucleus [
7,
14]. Moreover, it has been shown that the lysosomal accumulation of inorganic NPs, elicited by the acidic conditions of the lysosomal cellular compartment, enhance surface instability and ion release. Intracellular ion release is responsible for the cascading events associated with nanoparticle-induced intracellular toxicity [
15] and therefore, accumulation of NPs in lysosomes should be avoided.
In this work, we explored the feasibility of gene delivery in cultured differentiated RPE cells using DNA-wrapped gold NPs (DNA-gold NPs). In particular, we focused on cellular uptake and intracellular endosomal trafficking routes of pristine gold NPs and DNA-gold NPs, showing that a higher proportion of DNA-gold NPs (compared to pristine gold NPs) are internalized through clathrin-independent routes that do not end up in late endosomes, thereby avoiding lysosomal degradation. These uptake routes most probably allow faster gene delivery into the nucleus as measured by an early expression of the green fluorescent protein (GFP) reporter gene.
2. Materials and Methods
2.1. ARPE-19 Cell Culture and Differentiation Conditions
Human ARPE-19 cells, acquired from ATCC (CRL_2302), were cultured in 1:1 Dulbecco’s Modified Eagle’s Medium (DMEM) (ATCC, Manassas, VA, USA) and Ham’s F-12 Nutrient Mix (Life Technologies, Carlsbad, CA, USA) supplemented with 10% foetal bovine serum (FBS) (Life Technologies, Carlsbad, CA, USA) and 1% penicillin-streptomycin (Life Technologies, Carlsbad, CA, USA) in a 5% CO2 humidified chamber at 37 °C. For immunocytochemistry, human ARPE-19 cells (1.5 × 105cells/well) were seeded in growth medium without antibiotics onto poly-L-lysine-coated coverslips in 24-well plates. To induce differentiation, ARPE-19 cells were deprived of FBS for 48 h and maintained in a 1:1 media of DMEM and Ham’s F-12 Nutrient Mix. Subsequently, 48 h post-differentiation, cells were transfected with either nanoparticles or liposomes. For dynasore treatment, the media was changed at 48 h of differentiation, cells were washed once in PBS 1×, and dynasore was added at 80 μM (final concentration) for 30 min. Cell medium was then changed and cells were immediately nanotransfected.
2.2. Lipofection
After 48 h of differentiation, cells were co-transfected with the pEGFP reporter vector (500 ng/well), using Lipotransfectine (Niborlab, Guillena, Spain) (DNA: Lipotransfectine ratio of 1:3) in differentiation medium without antibiotics. This medium was replaced 5 h post-transfection with differentiation medium with antibiotics. After 16 h or 48 h of transfection, cells were fixed with 4% PFA for 20 min at RT and used for immunocytochemistry.
2.3. Gold NP Production and Nanotransfection
Gold nanoparticles (40 nm) at OD 1, stabilized in sodium citrate, were purchased from Sigma-Aldrich (St. Louis, MO, USA) (S-741981). One hundred microliters of gold nanoparticles at OD1 were spun down and resuspended in 10 μL of Milli-Q water and mixed with 1 μL (0.250 μg/μL) of pEGFP plasmid DNA per well. Plasmid DNA-gold hybrid structures were produced by allowing conditions in which double-stranded DNA (dsDNA) loops wrap over the surface of the nanoparticle. The dsDNA-nanoparticle complexes made by this method have an excellent dispersion in different solutions from water to cell culture medium [
16]. DNA-wrapped gold nanoparticles were introduced in each well with seeded cells. Photothermal plasmon resonance of the DNA-gold nanoparticles was activated by white light irradiation [
17]. Cells were allowed to grow for either 2 h (for immunocytochemistry), or 16 h/48 h (for EGFP expression). For transfection efficiency studies, cells were allowed to grow for 4 h before medium was changed. The details of the wrapping reaction and cell-transfection are currently subject to a patent and published under standard free patent procedures [
18].
2.4. Immunocytochemistry
After 2 h, 16 h or 48 h post-transfection with either pristine gold NPs, DNA-wrapped gold NPs or liposomes, cells were fixed with 4% PFA for 20 min at RT, washed in PBS (3 × 5 min), permeabilized in 0.2% Triton X-100 (St. Louis, MO, USA) in PBS (20 min at RT), and blocked for 1 h in 4% sheep serum in PBS. Primary antibodies were incubated overnight at 4 °C in a 1:200 dilution in blocking solution (rabbit anti-GFP, rabbit anti-EEA1, mouse anti-RAB7, all from Abcam, Cambridge, UK). After incubation, coverslips with cells were rinsed in PBS 1× (3 × 5 min), incubated with the corresponding secondary antibodies conjugated to either Alexa Fluor 488, 568 or 647 (Life Technologies, Grand Island, NY, USA) (1:400) at RT (1 h) in blocking solution, nuclei were stained with DAPI (Roche Diagnostics, Indianapolis, IN, USA) (1:1000), washed again in PBS 1× (3 × 5 min), mounted in Mowiol 4–88 (Merck, Darmstadt, Germany) and analysed by confocal microscopy and optical transmission microscopy (Zeiss LSM 880, Thornwood, NY, USA). To quantify GFP-positive cells, coverslips were visualized in a ZOE™ Fluorescent Cell Imager (Bio-Rad, Hercules, CA, USA).
2.5. Imaging and Statistical Analyses
Image analyses were performed using ImageJ (FIJI) software 1.52i. Stack images from five consecutive planes (0.46 μm of separation each) centred on the subcellular location with maximum vesicle intensity were retrieved, segmented in the different channels (clathrin, EEA1, RAB7 or NP), measured and analysed. In the case that the total concentration of the protein of interest had to be determined, the fluorescence threshold value was manually selected and converted into a binary mask. Then, the binary mask of each protein was converted to area values. In the case that the area of the nanoparticles had to be determined, the threshold value was restricted to the dark spots produced by the plasmon resonance of the nanoparticles. The binary mask of NPs was superimposed over the image of interest—for example, the mask of the NP field was superimposed over the image of the clathrin-positive vesicles’ signal. Then, the intersection area between the two selected channels generated a new image, of which the total intensity was determined. To calculate the percentage of NPs in each type of intracellular vesicles, the area of intersection image (e.g., NPs on clathrin-positive cells) was measured and compared to the total area covered by NPs. For statistical analysis, three independent replicates were performed per condition, and three representative different regions of interest (ROIs) per condition and replicate were quantified. Values, ratios and statistical significance were analysed using GraphPad Prism 7.03 (San Diego, CA, USA).
4. Discussion
Gene augmentation therapy in recessive mendelian disorders caused by loss of function mutations is, in principle, a plausible option for treatment. Addition of a wild-type copy of the gene should be effective, provided that the gene is delivered and correctly expressed at the target tissue. In IRDs, retinal degeneration is caused by mutations in either genes expressed in photoreceptors or genes expressed in the adjacent RPE, as it is the case with mutations in RPE65, MERTK or LRAT. AAV-based vectors are being used for gene delivery to photoreceptors, but non-viral vectors based on NPs should be concurrently explored for cases in which viral approaches may not be desirable [
3,
4,
8]. Several reports have shown that RPE is particularly amenable for transfection with NPs. This apparent feasibility may be due to the phagocytic nature of RPE cells, which easily engulf and internalize photoreceptor outer segments as well as other exogenous particles [
4,
9]. Phagocytosis involves the formation of large vesicles, but most small particles enter the cell via pinocytosis, which involve internalization routes mediated by different protein and lipid interactions, for example, clathrin- and caveolae-mediated vesicles as well as clathrin- and caveolae-independent endocytosis [
7,
14]. The endocytic entry route is key to the final outcome of the internalized particles, since cargo internalized via clathrin vesicles mainly end up being degraded by the lysosome, whereas cargo internalized by caveolin-mediated vesicles or clathrin-independent routes bypass lysosomal degradation. In this context, we have explored the potential use of gold NPs to transfect RPE cells in culture, focusing on the uptake routes and the intracellular endosomal trafficking pathways.
Several authors have reported that in vitro biocompatibility of gold nanoparticles largely depended on their shape and size, and determined that sizes between 5 and 30 nm were more cytotoxic, even though the rate of internalization was higher than that of the less toxic NPs of 50–100 nm (reviewed in [
10]). We have used NPs of 40 nm as a compromise between cytotoxicity and internalization. In addition, surface chemical modification of NPs is a critical step that decreases toxicity, increases stability, reduces aggregation and modulates cellular uptake. Most NPs use positive charges at the surface to facilitate the entry at the negatively charged cell membranes, but positively charged NPs induce cell death, whereas negatively charged surfaces induce internalization by clathrin/caveolae-independent endocytosis [
7,
14]. In this context, we have used plasmid DNA wrapping over gold NPs for two different but relevant reasons: as a means to stabilize gold NPs [
8] and favour clathrin-independent cellular uptake, as well as to provide the DNA molecule to be delivered.
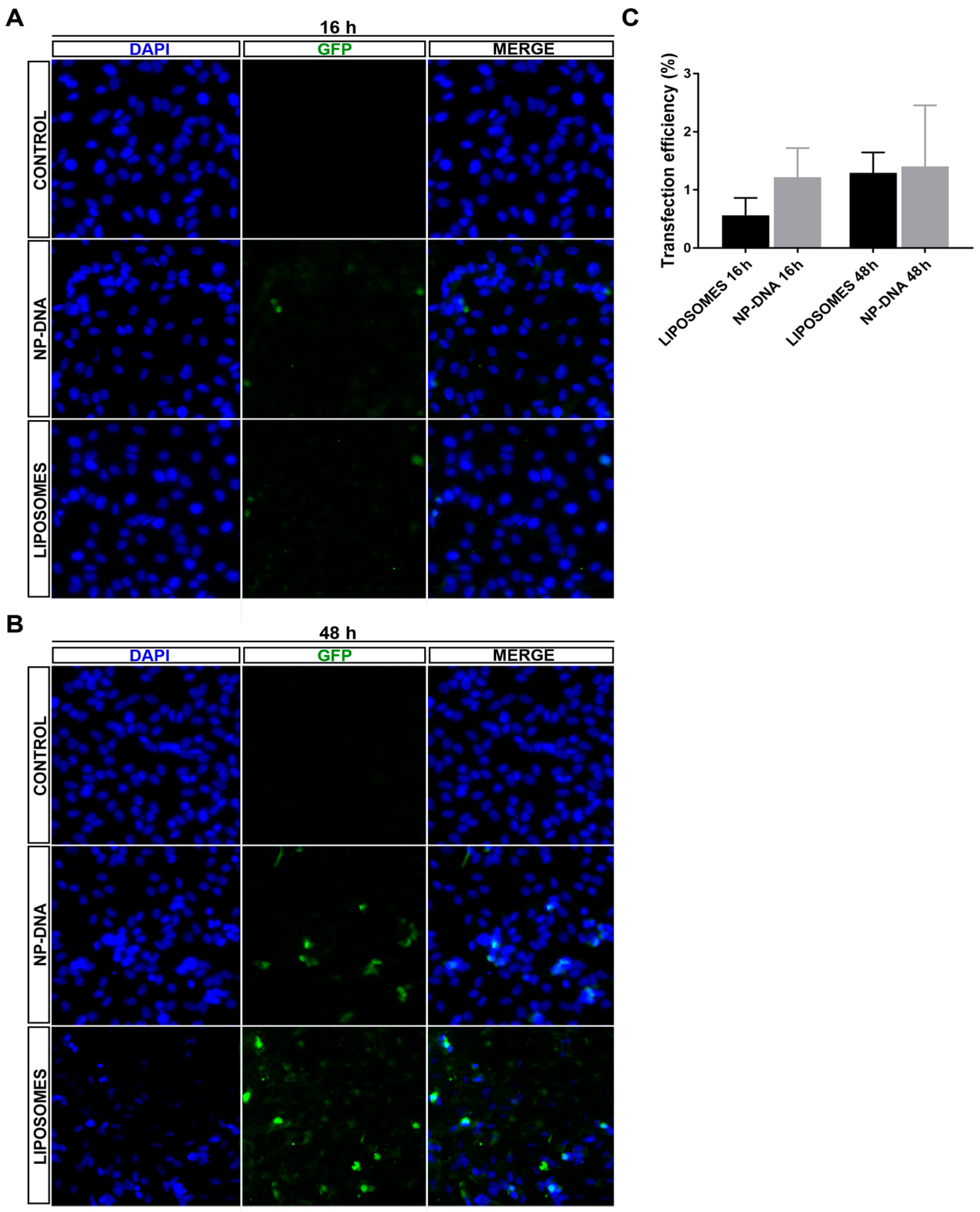
In differentiated ARPE-19 cells, at least up to 48 h post-transfection, DNA-wrapped gold NPs did not show any toxicity. DAPI staining did not detect any pyknotic cells, and the transfection efficiency was equivalent for both systems (
Figure 1 and
Figure S1), even though NPs required half the amount of DNA for the same rate of transfection, thus indicating that this point could be further optimized in the future. Interestingly, in cells transfected with DNA-gold NPs, the same number of cells expressed the reporter GFP gene at 16 h as at 48 h, which suggested that DNA-gold NPs might use a faster uptake/internalization route than cationic liposome-DNA complexes, which are mostly internalized via clathrin vesicles [
14,
19,
20,
21].
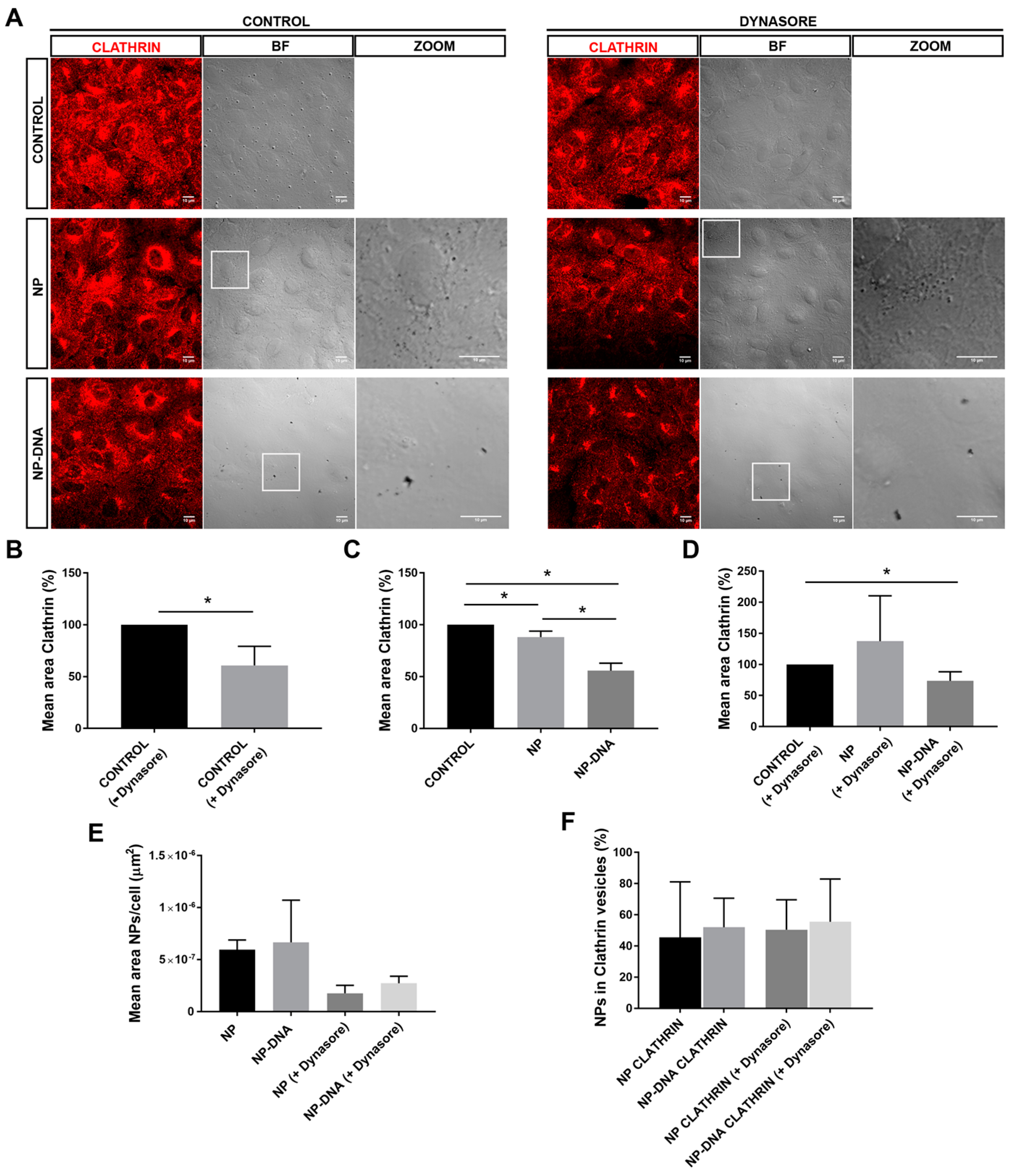
Our assays also determined that, in vitro, RPE cells responded differently to pristine gold NPs and DNA-gold NPs, since the area of clathrin vesicles was reduced in the presence of DNA-gold NPs. These differences might be due to changes in the membrane–NP interface. On the one hand, at least 50% of the pool of pristine gold NPs and DNA-gold NPs were internalized via clathrin-coated vesicles, which most probably ended up in the endo-lysosomal degradation pathway (
Figure 2). It is likely that this 50% of 40 nm NPs internalized by clathrin-dependent endocytosis corresponded to soluble NPs. On the other hand, the other half of the pool of pristine gold NPs and DNA-gold NPs uses different alternative endocytic routes for internalization, as shown by different localization/trafficking to endosomal compartments. At least 30% of pristine gold NPs are internalized by non-vesicular routes and localized neither in early nor in late endosomes. Since after dynasore treatment this alternative entry route was inhibited and all pristine NPs were detected in the endosomal pathway, the internalization of a significant pool of gold NPs was through lipid rafts or diffusion by destabilization of the cell membrane [
22,
23]. In contrast, the pool of DNA-gold NPs that was not detected in the endosomal pathway was much lower. A preference for vesicular entry could be explained by the larger size of the DNA-gold NP complexes. Interestingly, DNA-gold NPs are equivalently distributed in early and late endosomes, irrespective of dynasore treatment, thus indicating that a pool of DNA-gold NPs are found in early endosomes that will not mature into late endosome vesicles. These results are in agreement with the earlier expression of the reporter gene detected at 16 h post-transfection, which point to a pool of DNA-gold NPs being internalized by clathrin-independent endocytosis (for instance, via caveolin-mediated vesicles [
7,
14]) and to cargo escaping lysosomal degradation. Nonetheless, we cannot discard the result that DNA-gold NPs used alternative endosomal trafficking or recycling routes [
24].
Our results are but a first step towards the potential use of non-cytotoxic gold NPs for in vivo gene therapy in retinal disorders. Indeed, several basic biological questions should be first addressed, such as the internalization routes and their intracellular endosome trafficking. According to our initial results, DNA-wrapped gold NPs might be considered for gene delivery in retinal tissues. However, before considering this system for in vivo applications, further work is required to fully understand the complexity of cell–NP interaction, which is crucial to increase their transfection efficiency, optimize the cargo delivery by avoiding lysosomal, autophagic or other degradative pathways, and ensure sustained expression of the therapeutic gene.