Uncovering Missing Heritability in Rare Diseases
Abstract
:1. Introduction
2. Complexity of Rare Diseases
2.1. Heterogeneity
2.2. Mutation Spectrum
2.3. Phenotypic Variability
2.4. Unknowns
3. High Throughput Sequencing—Untangling Complexity
3.1. Exome Sequencing
3.2. Genome Sequencing
3.3. Genome and Phenome Resources
4. Uncovering Missing Heritability—“No Longer Just Looking under the Lamppost”
Funding
Acknowledgments
Conflicts of Interest
References
- Maher, B. Personal genomes: The case of the missing heritability. Nature 2008, 456, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Turkheimer, E. Still missing. Res. Hum. Dev. 2011, 8, 227–241. [Google Scholar] [CrossRef]
- Wright, C.F.; FitzPatrick, D.R.; Firth, H.V. Paediatric genomics: Diagnosing rare disease in children. Nat. Rev. Genet. 2018, 19, 253–268. [Google Scholar] [CrossRef]
- Montserrat Moliner, A.; Waligóra, J. The European Union policy in the field of rare diseases. Public Health Genomics 2013, 16, 268–277. [Google Scholar] [PubMed]
- Orphanet. Available online: https://www.orpha.net/consor/cgi-bin/index.php (accessed on 6 January 2019).
- OMIM—Online Mendelian Inheritance in Man. Available online: https://www.omim.org/ (accessed on 6 January 2019).
- Chakravorty, S.; Hegde, M. Gene and variant annotation for Mendelian disorders in the era of advanced sequencing technologies. Annu. Rev. Genomics Hum. Genet. 2017, 18, 229–256. [Google Scholar] [CrossRef] [PubMed]
- Caspar, S.M.; Dubacher, N.; Kopps, A.M.; Meienberg, J.; Henggeler, C.; Matyas, G. Clinical sequencing: From raw data to diagnosis with lifetime value. Clin. Genet. 2018, 93, 508–519. [Google Scholar] [CrossRef]
- Prokop, J.W.; May, T.; Strong, K.; Bilinovich, S.M.; Bupp, C.; Rajasekaran, S.; Worthey, E.A.; Lazar, J. Genome sequencing in the clinic: The past, present, and future of genomic medicine. Physiol. Genom. 2018, 50, 563–579. [Google Scholar] [CrossRef] [PubMed]
- Schindler, A.; Sumner, C.; Hoover-Fong, J.E. TRPV4-Associated Disorders. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Stephens, K., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, WA, USA, 1993. [Google Scholar]
- Schlingmann, K.P.; Bandulik, S.; Mammen, C.; Tarailo-Graovac, M.; Holm, R.; Baumann, M.; König, J.; Lee, J.J.Y.; Drögemöller, B.; Imminger, K.; et al. Germline de novo mutations in ATP1A1 cause renal hypomagnesemia, refractory seizures, and intellectual disability. Am. J. Hum. Genet. 2018, 103, 808–816. [Google Scholar] [CrossRef]
- Lassuthova, P.; Rebelo, A.P.; Ravenscroft, G.; Lamont, P.J.; Davis, M.R.; Manganelli, F.; Feely, S.M.; Bacon, C.; Brožková, D.Š.; Haberlova, J.; et al. Mutations in ATP1A1 cause dominant Charcot-Marie-Tooth type 2. Am. J. Hum. Genet. 2018, 102, 505–514. [Google Scholar] [CrossRef]
- Lynch, D.S.; Chelban, V.; Vandrovcova, J.; Pittman, A.; Wood, N.W.; Houlden, H. GLS loss of function causes autosomal recessive spastic ataxia and optic atrophy. Ann. Clin. Transl. Neurol. 2018, 5, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Rumping, L.; Büttner, B.; Maier, O.; Rehmann, H.; Lequin, M.; Schlump, J.-U.; Schmitt, B.; Schiebergen-Bronkhorst, B.; Prinsen, H.C.M.T.; Losa, M.; et al. Identification of a loss-of-function mutation in the context of glutaminase deficiency and neonatal epileptic encephalopathy. JAMA Neurol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Peron, A.; Au, K.S.; Northrup, H. Genetics, genomics, and genotype-phenotype correlations of TSC: Insights for clinical practice. Am. J. Med. Genet. C Semin. Med. Genet. 2018, 178, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Gil, N.; González-Del Pozo, M.; Martín-Sánchez, M.; Méndez-Vidal, C.; Rodríguez-de la Rúa, E.; Borrego, S.; Antiñolo, G. Unravelling the genetic basis of simplex Retinitis Pigmentosa cases. Sci. Rep. 2017, 7, 41937. [Google Scholar] [CrossRef] [PubMed]
- Chiurazzi, P.; Pirozzi, F. Advances in understanding - genetic basis of intellectual disability. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016, 44, D862–D868. [Google Scholar] [CrossRef]
- Sequence Ontology. Available online: http://www.sequenceontology.org/ (accessed on 7 January 2019).
- Fokkema, I.F.A.C.; Taschner, P.E.M.; Schaafsma, G.C.P.; Celli, J.; Laros, J.F.J.; den Dunnen, J.T. LOVD v.2.0: The next generation in gene variant databases. Hum. Mutat. 2011, 32, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Ulirsch, J.C.; Verboon, J.M.; Kazerounian, S.; Guo, M.H.; Yuan, D.; Ludwig, L.S.; Handsaker, R.E.; Abdulhay, N.J.; Fiorini, C.; Genovese, G.; et al. The genetic landscape of diamond-blackfan anemia. Am. J. Hum. Genet. 2018, 103, 930–947. [Google Scholar] [CrossRef]
- Piazza, A.; Heyer, W.-D. Homologous recombination and the formation of complex genomic rearrangements. Trends Cell Biol. 2019, 29, 135–149. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Guryev, V.; van Roosmalen, M.; Duran, K.J.; de Bruijn, E.; Bakker, S.C.M.; Letteboer, T.; van Nesselrooij, B.; Hochstenbach, R.; Poot, M.; et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum. Mol. Genet. 2011, 20, 1916–1924. [Google Scholar] [CrossRef]
- McDermott, D.H.; Gao, J.-L.; Liu, Q.; Siwicki, M.; Martens, C.; Jacobs, P.; Velez, D.; Yim, E.; Bryke, C.R.; Hsu, N.; et al. Chromothriptic cure of WHIM syndrome. Cell 2015, 160, 686–699. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Shyr, C.; Ross, C.J.; Horvath, G.A.; Salvarinova, R.; Ye, X.C.; Zhang, L.-H.; Bhavsar, A.P.; Lee, J.J.Y.; Drögemöller, B.I.; et al. Exome Sequencing and the management of neurometabolic disorders. N. Engl. J. Med. 2016, 374, 2246–2255. [Google Scholar] [CrossRef]
- Posey, J.E.; Harel, T.; Liu, P.; Rosenfeld, J.A.; James, R.A.; Coban Akdemir, Z.H.; Walkiewicz, M.; Bi, W.; Xiao, R.; Ding, Y.; et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017, 376, 21–31. [Google Scholar] [CrossRef]
- Balci, T.B.; Hartley, T.; Xi, Y.; Dyment, D.A.; Beaulieu, C.L.; Bernier, F.P.; Dupuis, L.; Horvath, G.A.; Mendoza-Londono, R.; Prasad, C.; et al. Debunking Occam’s razor: Diagnosing multiple genetic diseases in families by whole-exome sequencing. Clin. Genet. 2017, 92, 281–289. [Google Scholar] [CrossRef]
- Wen, X.-Y.; Tarailo-Graovac, M.; Brand-Arzamendi, K.; Willems, A.; Rakic, B.; Huijben, K.; Da Silva, A.; Pan, X.; El-Rass, S.; Ng, R.; et al. Sialic acid catabolism by N-acetylneuraminate pyruvate lyase is essential for muscle function. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Pérez-Torras, S.; Mata-Ventosa, A.; Drögemöller, B.; Tarailo-Graovac, M.; Meijer, J.; Meinsma, R.; van Cruchten, A.G.; Kulik, W.; Viel-Oliva, A.; Bidon-Chanal, A.; et al. Deficiency of perforin and hCNT1, a novel inborn error of pyrimidine metabolism, associated with a rapidly developing lethal phenotype due to multi-organ failure. Biochim. Biophys. Acta Mol. Basis Dis. 2019. [Google Scholar] [CrossRef]
- Armour, C.M.; Smith, A.; Hartley, T.; Chardon, J.W.; Sawyer, S.; Schwartzentruber, J.; Hennekam, R.; Majewski, J.; Bulman, D.E.; FORGE Canada Consortium; et al. Syndrome disintegration: Exome sequencing reveals that Fitzsimmons syndrome is a co-occurrence of multiple events. Am. J. Med. Genet. A 2016, 170, 1820–1825. [Google Scholar] [CrossRef]
- Sass, J.O.; Gemperle-Britschgi, C.; Tarailo-Graovac, M.; Patel, N.; Walter, M.; Jordanova, A.; Alfadhel, M.; Barić, I.; Çoker, M.; Damli-Huber, A.; et al. Unravelling 5-oxoprolinuria (pyroglutamic aciduria) due to bi-allelic OPLAH mutations: 20 new mutations in 14 families. Mol. Genet. Metab. 2016, 119, 44–49. [Google Scholar] [CrossRef]
- DIDA | DIDA is a Novel Database that Provides for the First Time Detailed Information on Genes and Associated Genetic Variants Involved in Digenic Diseases, the Simplest form of Oligogenic Inheritance. Available online: http://dida.ibsquare.be/ (accessed on 21 February 2019).
- Gazzo, A.M.; Daneels, D.; Cilia, E.; Bonduelle, M.; Abramowicz, M.; Van Dooren, S.; Smits, G.; Lenaerts, T. DIDA: A curated and annotated digenic diseases database. Nucleic Acids Res. 2016, 44, D900–D907. [Google Scholar] [CrossRef]
- Lee, J.E.; Silhavy, J.L.; Zaki, M.S.; Schroth, J.; Bielas, S.L.; Marsh, S.E.; Olvera, J.; Brancati, F.; Iannicelli, M.; Ikegami, K.; et al. CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat. Genet. 2012, 44, 193–199. [Google Scholar] [CrossRef]
- Schäffer, A.A. Digenic inheritance in medical genetics. J. Med. Genet. 2013, 50, 641–652. [Google Scholar] [CrossRef]
- Kim, A.; Savary, C.; Dubourg, C.; Carré, W.; Mouden, C.; Hamdi-Rozé, H.; Guyodo, H.; Douce, J.L.; FREX Consortium; GoNL Consortium; et al. Integrated clinical and omics approach to rare diseases: Novel genes and oligogenic inheritance in holoprosencephaly. Brain J. Neurol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Dewey, F.E.; Murray, M.F.; Overton, J.D.; Habegger, L.; Leader, J.B.; Fetterolf, S.N.; O’Dushlaine, C.; Van Hout, C.V.; Staples, J.; Gonzaga-Jauregui, C.; et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Shi, L.; Hakenberg, J.; Naughton, B.; Sklar, P.; Zhang, J.; Zhou, H.; Tian, L.; Prakash, O.; Lemire, M.; et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 2016, 34, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Tarailo-Graovac, M.; Zhu, J.Y.A.; Matthews, A.; van Karnebeek, C.D.M.; Wasserman, W.W. Assessment of the ExAC data set for the presence of individuals with pathogenic genotypes implicated in severe Mendelian pediatric disorders. Genet. Med. 2017, 12, 1300. [Google Scholar] [CrossRef] [PubMed]
- Wenger, A.M.; Guturu, H.; Bernstein, J.A.; Bejerano, G. Systematic reanalysis of clinical exome data yields additional diagnoses: Implications for providers. Genet. Med. 2017, 19, 209–214. [Google Scholar] [CrossRef]
- Short, P.J.; McRae, J.F.; Gallone, G.; Sifrim, A.; Won, H.; Geschwind, D.H.; Wright, C.F.; Firth, H.V.; FitzPatrick, D.R.; Barrett, J.C.; et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature 2018, 555, 611–616. [Google Scholar] [CrossRef]
- Guéant, J.-L.; Chéry, C.; Oussalah, A.; Nadaf, J.; Coelho, D.; Josse, T.; Flayac, J.; Robert, A.; Koscinski, I.; Gastin, I.; et al. APRDX1 mutant allele causes a MMACHC secondary epimutation in cblC patients. Nat. Commun. 2018, 9, 67. [Google Scholar] [CrossRef]
- Falkenberg, K.D.; Braverman, N.E.; Moser, A.B.; Steinberg, S.J.; Klouwer, F.C.C.; Schlüter, A.; Ruiz, M.; Pujol, A.; Engvall, M.; Naess, K.; et al. Allelic Expression imbalance promoting a mutant PEX6 allele causes Zellweger spectrum disorder. Am. J. Hum. Genet. 2017, 101, 965–976. [Google Scholar] [CrossRef]
- Ece Solmaz, A.; Onay, H.; Atik, T.; Aykut, A.; Cerrah Gunes, M.; Ozalp Yuregir, O.; Bas, V.N.; Hazan, F.; Kirbiyik, O.; Ozkinay, F. Targeted multi-gene panel testing for the diagnosis of Bardet Biedl syndrome: Identification of nine novel mutations across BBS1, BBS2, BBS4, BBS7, BBS9, BBS10 genes. Eur. J. Med. Genet. 2015, 58, 689–694. [Google Scholar] [CrossRef]
- Saudi Mendeliome Group. Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome Biol. 2015, 16, 134. [Google Scholar] [CrossRef]
- Matthews, A.M.; Tarailo-Graovac, M.; Price, E.M.; Blydt-Hansen, I.; Ghani, A.; Drögemöller, B.I.; Robinson, W.P.; Ross, C.J.; Wasserman, W.W.; Siden, H.; et al. A de novo mosaic mutation in SPAST with two novel alternative alleles and chromosomal copy number variant in a boy with spastic paraplegia and autism spectrum disorder. Eur. J. Med. Genet. 2017, 60, 548–552. [Google Scholar] [CrossRef]
- Ragotte, R.J.; Dhanrajani, A.; Pleydell-Pearce, J.; Del Bel, K.L.; Tarailo-Graovac, M.; van Karnebeek, C.; Terry, J.; Senger, C.; McKinnon, M.L.; Seear, M.; et al. The importance of considering monogenic causes of autoimmunity: A somatic mutation in KRAS causing pediatric Rosai-Dorfman syndrome and systemic lupus erythematosus. Clin. Immunol. 2017, 175, 143–146. [Google Scholar] [CrossRef]
- Boycott, K.M.; Rath, A.; Chong, J.X.; Hartley, T.; Alkuraya, F.S.; Baynam, G.; Brookes, A.J.; Brudno, M.; Carracedo, A.; den Dunnen, J.T.; et al. International cooperation to enable the diagnosis of all rare genetic diseases. Am. J. Hum. Genet. 2017, 100, 695–705. [Google Scholar] [CrossRef]
- Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature 2015, 519, 223–228. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Wasserman, W.W.; Van Karnebeek, C.D.M. Impact of next-generation sequencing on diagnosis and management of neurometabolic disorders: Current advances and future perspectives. Expert Rev. Mol. Diagn. 2017, 17, 307–309. [Google Scholar] [CrossRef]
- Van Kuilenburg, A.B.P.; Tarailo-Graovac, M.; Meijer, J.; Drogemoller, B.; Vockley, J.; Maurer, D.; Dobritzsch, D.; Ross, C.J.; Wasserman, W.; Meinsma, R.; et al. Genome sequencing reveals a novel genetic mechanism underlying dihydropyrimidine dehydrogenase deficiency: A novel missense variant c.1700G>A and a large intragenic inversion in DPYD spanning intron 8 to intron 12. Hum. Mutat. 2018, 39, 947–953. [Google Scholar] [CrossRef]
- Gilissen, C.; Hehir-Kwa, J.Y.; Thung, D.T.; van de Vorst, M.; van Bon, B.W.M.; Willemsen, M.H.; Kwint, M.; Janssen, I.M.; Hoischen, A.; Schenck, A.; et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014, 511, 344–347. [Google Scholar] [CrossRef]
- Alfares, A.; Aloraini, T.; Subaie, L.A.; Alissa, A.; Qudsi, A.A.; Alahmad, A.; Mutairi, F.A.; Alswaid, A.; Alothaim, A.; Eyaid, W.; et al. Whole-genome sequencing offers additional but limited clinical utility compared with reanalysis of whole-exome sequencing. Genet. Med. 2018, 20, 1328. [Google Scholar] [CrossRef]
- Lionel, A.C.; Costain, G.; Monfared, N.; Walker, S.; Reuter, M.S.; Hosseini, S.M.; Thiruvahindrapuram, B.; Merico, D.; Jobling, R.; Nalpathamkalam, T.; et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 2018, 20, 435–443. [Google Scholar] [CrossRef]
- Ostrander, B.E.P.; Butterfield, R.J.; Pedersen, B.S.; Farrell, A.J.; Layer, R.M.; Ward, A.; Miller, C.; DiSera, T.; Filloux, F.M.; Candee, M.S.; et al. Whole-genome analysis for effective clinical diagnosis and gene discovery in early infantile epileptic encephalopathy. NPJ Genom. Med. 2018, 3, 22. [Google Scholar] [CrossRef]
- Ishiura, H.; Doi, K.; Mitsui, J.; Yoshimura, J.; Matsukawa, M.K.; Fujiyama, A.; Toyoshima, Y.; Kakita, A.; Takahashi, H.; Suzuki, Y.; et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat. Genet. 2018, 50, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Fujita, M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 2018, 109, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Loose, M.W. The potential impact of nanopore sequencing on human genetics. Hum. Mol. Genet. 2017, 26, R202–R207. [Google Scholar] [CrossRef] [PubMed]
- Laver, T.; Harrison, J.; O’Neill, P.A.; Moore, K.; Farbos, A.; Paszkiewicz, K.; Studholme, D.J. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 2015, 3, 1–8. [Google Scholar] [CrossRef]
- Leggett, R.M.; Clark, M.D. A world of opportunities with nanopore sequencing. J. Exp. Bot. 2017, 68, 5419–5429. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.-Y.; Su, S.-C.; Fan, W.-L.; Lai, S.-H.; Tsai, M.-H.; Chen, S.-H.; Wong, K.-S.; Chung, W.-H. Whole-genome sequencing of a family with hereditary pulmonary alveolar proteinosis identifies a rare structural variant involving CSF2RA/CRLF2/IL3RA gene disruption. Sci. Rep. 2017, 7, 43469. [Google Scholar] [CrossRef] [PubMed]
- Borràs, D.M.; Vossen, R.H.A.M.; Liem, M.; Buermans, H.P.J.; Dauwerse, H.; van Heusden, D.; Gansevoort, R.T.; den Dunnen, J.T.; Janssen, B.; Peters, D.J.M.; et al. Detecting PKD1 variants in polycystic kidney disease patients by single-molecule long-read sequencing. Hum. Mutat. 2017, 38, 870–879. [Google Scholar] [CrossRef]
- Cretu Stancu, M.; van Roosmalen, M.J.; Renkens, I.; Nieboer, M.M.; Middelkamp, S.; de Ligt, J.; Pregno, G.; Giachino, D.; Mandrile, G.; Espejo Valle-Inclan, J.; et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017, 8, 1326. [Google Scholar] [CrossRef]
- Tavares, E.; Tang, C.Y.; Vig, A.; Li, S.; Billingsley, G.; Sung, W.; Vincent, A.; Thiruvahindrapuram, B.; Héon, E. Retrotransposon insertion as a novel mutational event in Bardet-Biedl syndrome. Mol. Genet. Genom. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Cowley, M.J.; Liu, Y.-C.; Oliver, K.L.; Carvill, G.; Myers, C.T.; Gayevskiy, V.; Delatycki, M.; Vlaskamp, D.R.M.; Zhu, Y.; Mefford, H.; et al. Reanalysis and optimisation of bioinformatic pipelines is critical for mutation detection. Hum. Mutat. 2018, 40, 374–379. [Google Scholar] [CrossRef]
- Miao, H.; Zhou, J.; Yang, Q.; Liang, F.; Wang, D.; Ma, N.; Gao, B.; Du, J.; Lin, G.; Wang, K.; et al. Long-read sequencing identified a causal structural variant in an exome-negative case and enabled preimplantation genetic diagnosis. Hereditas 2018, 155, 32. [Google Scholar] [CrossRef] [PubMed]
- Merker, J.D.; Wenger, A.M.; Sneddon, T.; Grove, M.; Zappala, Z.; Fresard, L.; Waggott, D.; Utiramerur, S.; Hou, Y.; Smith, K.S.; et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med. 2018, 20, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Sanchis-Juan, A.; Stephens, J.; French, C.E.; Gleadall, N.; Mégy, K.; Penkett, C.; Shamardina, O.; Stirrups, K.; Delon, I.; Dewhurst, E.; et al. Complex structural variants in Mendelian disorders: Identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med. 2018, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, T.; Toyota, T.; Adachi, H.; Miyake, N.; Matsumoto, N.; Miyatake, S. Detecting a long insertion variant in SAMD12 by SMRT sequencing: Implications of long-read whole-genome sequencing for repeat expansion diseases. J. Hum. Genet. 2019, 64, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Narzisi, G.; Schatz, M.C. The challenge of small-scale repeats for indel discovery. Front. Bioeng. Biotechnol. 2015, 3, 8. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Dolzhenko, E.; van Vugt, J.J.F.A.; Shaw, R.J.; Bekritsky, M.A.; van Blitterswijk, M.; Narzisi, G.; Ajay, S.S.; Rajan, V.; Lajoie, B.R.; Johnson, N.H.; et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017, 27, 1895–1903. [Google Scholar] [CrossRef]
- Hannan, A.J. Tandem repeats mediating genetic plasticity in health and disease. Nat. Rev. Genet. 2018, 19, 286–298. [Google Scholar] [CrossRef]
- De Koning, A.P.J.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 25, 4–10. [Google Scholar]
- Mousavi, N.; Shleizer-Burko, S.; Gymrek, M. Profiling the genome-wide landscape of tandem repeat expansions. bioRxiv 2018. [CrossRef]
- Sudmant, P.H.; Rausch, T.; Gardner, E.J.; Handsaker, R.E.; Abyzov, A.; Huddleston, J.; Zhang, Y.; Ye, K.; Jun, G.; Fritz, M.H.-Y.; et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015, 526, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Weischenfeldt, J.; Symmons, O.; Spitz, F.; Korbel, J.O. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat. Rev. Genet. 2013, 14, 125–138. [Google Scholar] [CrossRef]
- Bergman, C.M.; Quesneville, H. Discovering and detecting transposable elements in genome sequences. Brief. Bioinform. 2007, 8, 382–392. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Drögemöller, B.I.; Wasserman, W.W.; Ross, C.J.D.; van den Ouweland, A.M.W.; Darin, N.; Kollberg, G.; van Karnebeek, C.D.M.; Blomqvist, M. Identification of a large intronic transposal insertion in SLC17A5 causing sialic acid storage disease. Orphanet J. Rare Dis. 2017, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.M.; Ajay, S.S.; Rajan, V.; Brown, C.; Bluske, K.; Burns, N.J.; Chawla, A.; Coffey, A.J.; Malhotra, A.; Scocchia, A.; et al. Copy-number variants in clinical genome sequencing: Deployment and interpretation for rare and undiagnosed disease. Genet. Med. 2018. [Google Scholar] [CrossRef]
- Roller, E.; Ivakhno, S.; Lee, S.; Royce, T.; Tanner, S. Canvas: Versatile and scalable detection of copy number variants. Bioinformatics 2016, 32, 2375–2377. [Google Scholar] [CrossRef]
- Ivakhno, S.; Roller, E.; Colombo, C.; Tedder, P.; Cox, A.J. Canvas SPW: Calling de novo copy number variants in pedigrees. Bioinformatics 2018, 34, 516–518. [Google Scholar] [CrossRef]
- Becker, T.; Lee, W.-P.; Leone, J.; Zhu, Q.; Zhang, C.; Liu, S.; Sargent, J.; Shanker, K.; Mil-Homens, A.; Cerveira, E.; et al. FusorSV: An algorithm for optimally combining data from multiple structural variation detection methods. Genome Biol. 2018, 19, 38. [Google Scholar] [CrossRef]
- Antaki, D.; Brandler, W.M.; Sebat, J. SV2: Accurate structural variation genotyping and de novo mutation detection from whole genomes. Bioinformacs 2018, 34, 1774–1777. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef]
- Calabrese, C.; Simone, D.; Diroma, M.A.; Santorsola, M.; Guttà, C.; Gasparre, G.; Picardi, E.; Pesole, G.; Attimonelli, M. MToolBox: A highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics 2014, 30, 3115–3117. [Google Scholar] [CrossRef]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014, 15, R84. [Google Scholar] [CrossRef]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef]
- Ebler, J.; Schönhuth, A.; Marschall, T. Genotyping inversions and tandem duplications. Bioinformatics 2017, 33, 4015–4023. [Google Scholar] [CrossRef]
- Liang, Y.; Qiu, K.; Liao, B.; Zhu, W.; Huang, X.; Li, L.; Chen, X.; Li, K. Seeksv: An accurate tool for somatic structural variation and virus integration detection. Bioinformatics 2017, 33, 184–191. [Google Scholar] [CrossRef]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Källberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef]
- Ye, K.; Schulz, M.H.; Long, Q.; Apweiler, R.; Ning, Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009, 25, 2865–2871. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Guo, L.; Yang, X.; Lamijer, E.-W.; Raine, K.; Ning, Z. Split-read indel and structural variant calling using PINDEL. Methods Mol. Biol. 2018, 1833, 95–105. [Google Scholar]
- Wala, J.A.; Bandopadhayay, P.; Greenwald, N.F.; O’Rourke, R.; Sharpe, T.; Stewart, C.; Schumacher, S.; Li, Y.; Weischenfeldt, J.; Yao, X.; et al. SvABA: Genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018, 28, 581–591. [Google Scholar] [CrossRef] [PubMed]
- E pluribus unum. Nat. Methods 2010, 7, 331. [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Scherer, S. A Short Guide to the Human Genome; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2008; ISBN 978-0-87969-791-4. [Google Scholar]
- Ameur, A.; Che, H.; Martin, M.; Bunikis, I.; Dahlberg, J.; Höijer, I.; Häggqvist, S.; Vezzi, F.; Nordlund, J.; Olason, P.; et al. De novo assembly of two Swedish genomes reveals missing segments from the human grch38 reference and improves variant calling of population-scale sequencing data. Genes 2018, 9, 486. [Google Scholar] [CrossRef] [PubMed]
- Magi, A.; D’Aurizio, R.; Palombo, F.; Cifola, I.; Tattini, L.; Semeraro, R.; Pippucci, T.; Giusti, B.; Romeo, G.; Abbate, R.; et al. Characterization and identification of hidden rare variants in the human genome. BMC Genom. 2015, 16, 340. [Google Scholar] [CrossRef] [PubMed]
- Dewey, F.E.; Chen, R.; Cordero, S.P.; Ormond, K.E.; Caleshu, C.; Karczewski, K.J.; Whirl-Carrillo, M.; Wheeler, M.T.; Dudley, J.T.; Byrnes, J.K.; et al. Phased whole-genome genetic risk in a family quartet using a major allele reference sequence. PLoS Genet. 2011, 7, e1002280. [Google Scholar] [CrossRef]
- Novak, A.M.; Hickey, G.; Garrison, E.; Blum, S.; Connelly, A.; Dilthey, A.; Eizenga, J.; Elmohamed, M.A.S.; Guthrie, S.; Kahles, A.; et al. Genome Graphs. bioRxiv 2017. [Google Scholar] [CrossRef]
- Smigielski, E.M.; Sirotkin, K.; Ward, M.; Sherry, S.T. dbSNP: A database of single nucleotide polymorphisms. Nucleic Acids Res. 2000, 28, 352–355. [Google Scholar] [CrossRef]
- NHLBI Trans Omics for Precision Medicine. Available online: https://www.nhlbiwgs.org/ (accessed on 7 January 2019).
- Iranome. Available online: http://www.iranome.com/about (accessed on 7 January 2019).
- Lencz, T.; Yu, J.; Palmer, C.; Carmi, S.; Ben-Avraham, D.; Barzilai, N.; Bressman, S.; Darvasi, A.; Cho, J.H.; Clark, L.N.; et al. High-depth whole genome sequencing of an Ashkenazi Jewish reference panel: Enhancing sensitivity, accuracy, and imputation. Hum. Genet. 2018, 137, 343–355. [Google Scholar] [CrossRef]
- Oleksyk, T.K.; Brukhin, V.; O’Brien, S.J. Putting Russia on the genome map. Science 2015, 350, 747. [Google Scholar] [CrossRef]
- Oleksyk, T.K.; Brukhin, V.; O’Brien, S.J. The Genome Russia project: Closing the largest remaining omission on the world Genome map. GigaScience 2015, 4, 53. [Google Scholar] [CrossRef]
- Silent Genomes Project. Available online: https://www.bcchr.ca/silent-genomes-project (accessed on 7 January 2019).
- Stenson, P.D.; Mort, M.; Ball, E.V.; Evans, K.; Hayden, M.; Heywood, S.; Hussain, M.; Phillips, A.D.; Cooper, D.N. The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017, 136, 665–677. [Google Scholar] [CrossRef]
- Pawliczek, P.; Patel, R.Y.; Ashmore, L.R.; Jackson, A.R.; Bizon, C.; Nelson, T.; Powell, B.; Freimuth, R.R.; Strande, N.; Shah, N.; et al. ClinGen Allele Registry links information about genetic variants. Hum. Mutat. 2018, 39, 1690–1701. [Google Scholar] [CrossRef]
- Phan, L.; Hsu, J.; Tri, L.Q.M.; Willi, M.; Mansour, T.; Kai, Y.; Garner, J.; Lopez, J.; Busby, B. dbVar structural variant cluster set for data analysis and variant comparison. F1000Research 2016, 5, 673. [Google Scholar] [CrossRef]
- Preste, R.; Vitale, O.; Clima, R.; Gasparre, G.; Attimonelli, M. HmtVar: A new resource for human mitochondrial variations and pathogenicity data. Nucleic Acids Res. 2018, 47, D1202–D1210. [Google Scholar] [CrossRef]
- Köhler, S.; Carmody, L.; Vasilevsky, N.; Jacobsen, J.O.B.; Danis, D.; Gourdine, J.-P.; Gargano, M.; Harris, N.L.; Matentzoglu, N.; McMurry, J.A.; et al. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef]
- Haendel, M.A.; Chute, C.G.; Robinson, P.N. Classification, ontology, and precision medicine. N. Engl. J. Med. 2018, 379, 1452–1462. [Google Scholar] [CrossRef]
- Sobreira, N.; Schiettecatte, F.; Valle, D.; Hamosh, A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015, 36, 928–930. [Google Scholar] [CrossRef]
- Buske, O.J.; Girdea, M.; Dumitriu, S.; Gallinger, B.; Hartley, T.; Trang, H.; Misyura, A.; Friedman, T.; Beaulieu, C.; Bone, W.P.; et al. PhenomeCentral: A portal for phenotypic and genotypic matchmaking of patients with rare genetic diseases. Hum. Mutat. 2015, 36, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Philippakis, A.A.; Azzariti, D.R.; Beltran, S.; Brookes, A.J.; Brownstein, C.A.; Brudno, M.; Brunner, H.G.; Buske, O.J.; Carey, K.; Doll, C.; et al. The Matchmaker Exchange: A platform for rare disease gene discovery. Hum. Mutat. 2015, 36, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U.; Walter, K.; Min, J.L.; Huang, J.; Crooks, L.; Memari, Y.; McCarthy, S.; Perry, J.R.B.; Xu, C.; Futema, M.; et al. The UK10K project identifies rare variants in health and disease. Nature 2015, 526, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Splinter, K.; Adams, D.R.; Bacino, C.A.; Bellen, H.J.; Bernstein, J.A.; Cheatle-Jarvela, A.M.; Eng, C.M.; Esteves, C.; Gahl, W.A.; Hamid, R.; et al. Effect of genetic diagnosis on patients with previously undiagnosed disease. N. Engl. J. Med. 2018, 379, 2131–2139. [Google Scholar] [CrossRef]
- Collins, F.S. 2005 William Allan Award address. No longer just looking under the lamppost. Am. J. Hum. Genet. 2006, 79, 421–426. [Google Scholar] [CrossRef]
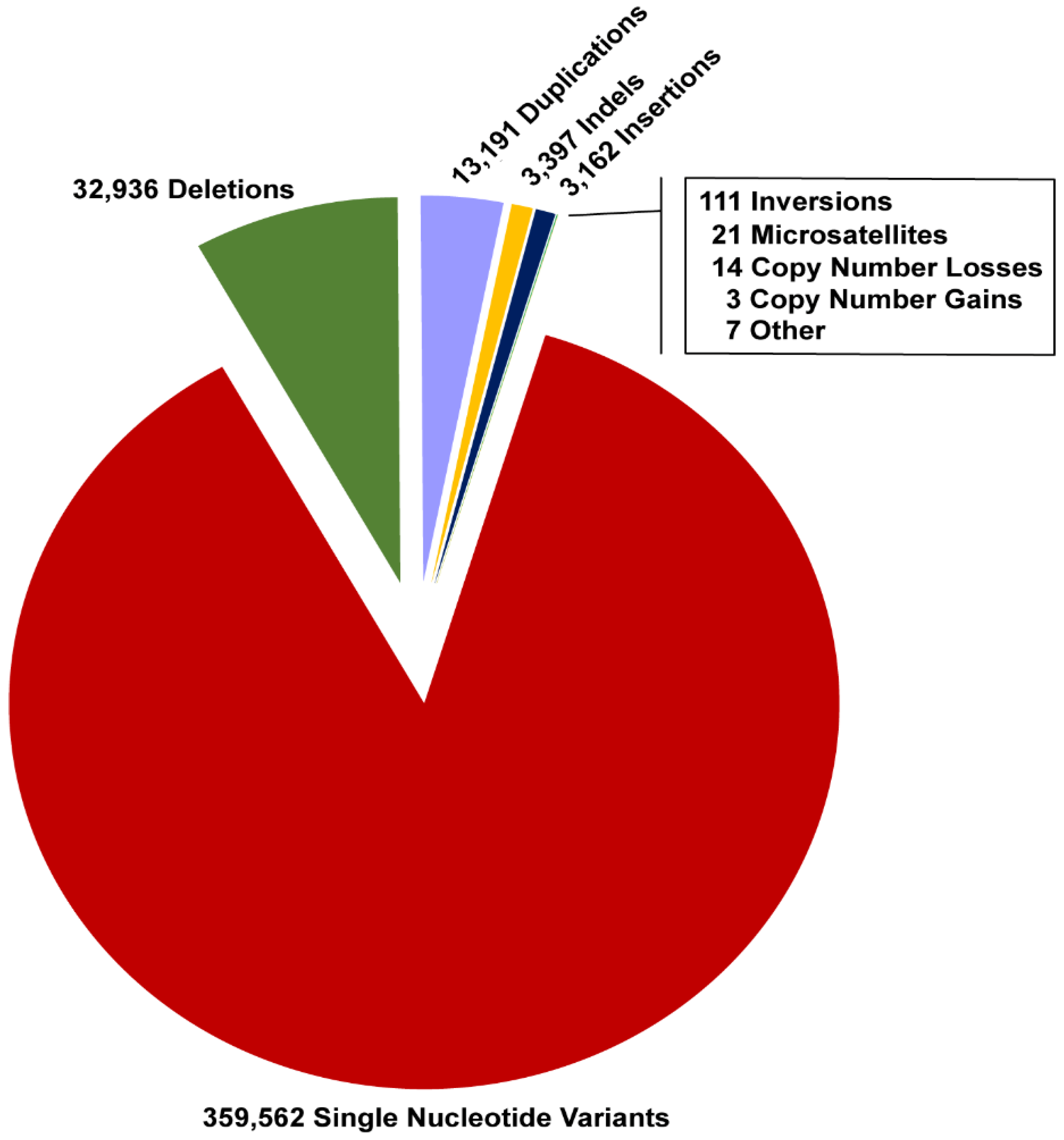
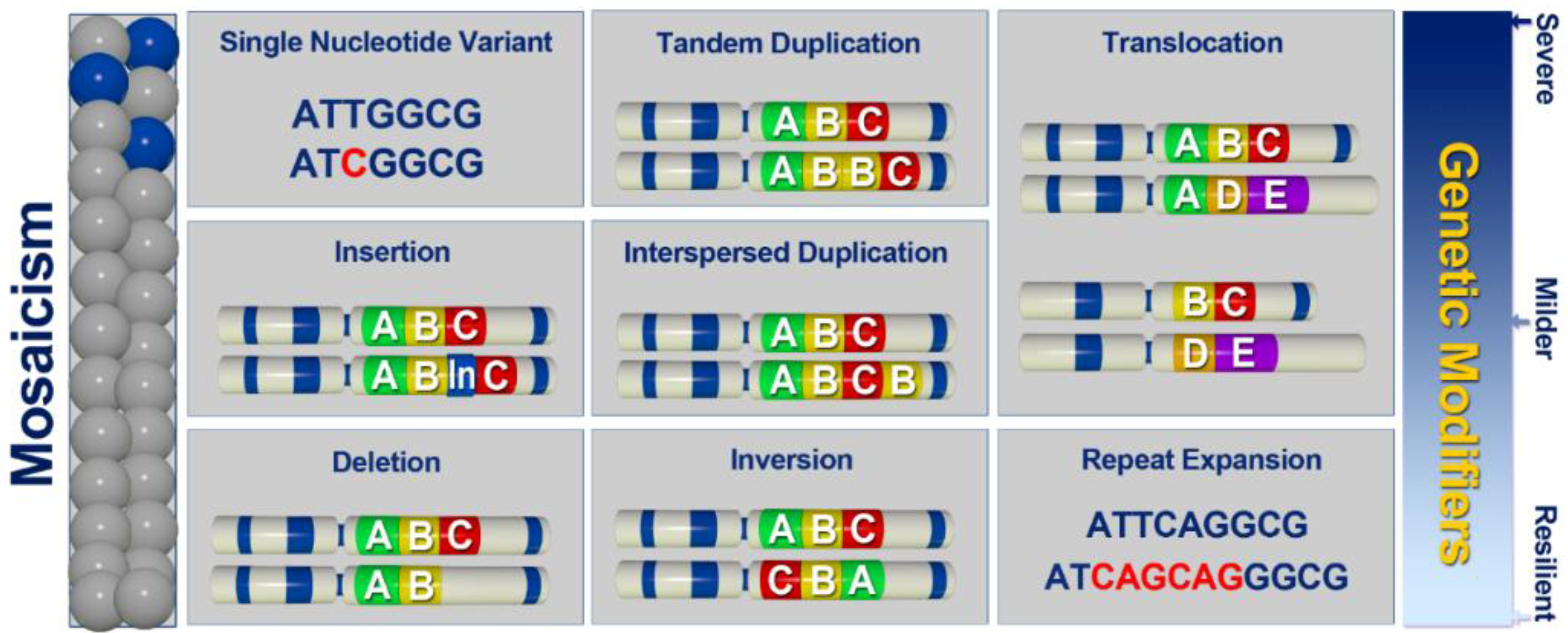
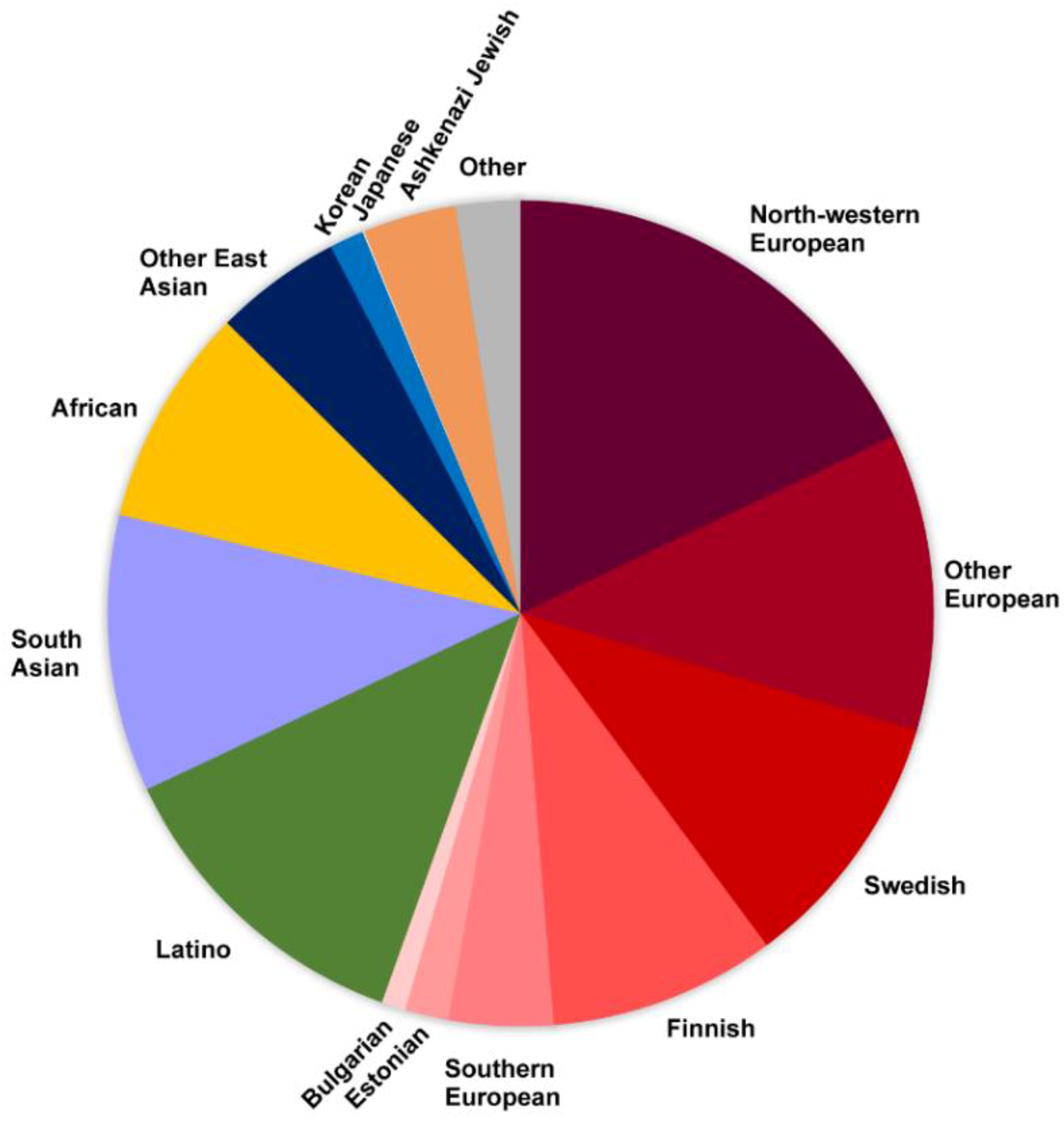
| Authors | Year | Gene | Disease | Type of Variation | Type of WGS | Ref. |
|---|---|---|---|---|---|---|
| Kloosterman et al. | 2011 | Multiple | Severe congenital abnormalities | De novo SV (chromothripsis) | SOLiD | [23] |
| Gilissen et al. | 2014 | SHANK3 | Phelan-McDermid syndrome | De novo 66 kb deletion | Complete Genomics | [53] |
| Gilissen et al. | 2014 | VPS13B | Cohen syndrome | 1.7 kb and 122 kb deletions | Complete Genomics | [53] |
| Gilissen et al. | 2014 | MECP2 | Rett syndrome | De novo 0.6 kb deletion | Complete Genomics | [53] |
| Gilissen et al. | 2014 | IQSEC2 | Intellectual disability | De novo 62 kb interspersed duplication | Complete Genomics | [53] |
| Gilissen et al. | 2014 | SMC1A | Cornelia de Lange syndrome | De novo 2.1 kb deletion | Complete Genomics | [53] |
| Gilissen et al. | 2014 | Multiple | 16p11.2 deletion syndrome | De novo 611 kb deletion | Complete Genomics | [53] |
| Gilissen et al. | 2014 | STAG1 | Intellectual Disability | De novo 382 kb deletion | Complete Genomics | [53] |
| van Kuilenburg et al. | 2017 | DPYD | DPDD | Large intragenic inversion | Illumina | [52] |
| Chiu et al. | 2017 | Multiple | Pulmonary alveolar proteinosis | 425 kb deletion | Illumina | [63] |
| Borràs et al. | 2017 | PKD1 | Polycystic kidney disease | Various, 18/19 probands | PacBio | [64] |
| Cretu Stancu et al. | 2017 | Multiple | Severe congenital abnormalities | De novo SV (chromothripsis) | ONT 1 + Illumina | [65] |
| Alfares et al. | 2018 | PHOX2B | Central hypoventilation syndrome | GCN (25) repeat expansion [+25] | Illumina | [54] |
| Alfares et al. | 2018 | TPM3 | Nemaline myopathy 1 | Large deletion | Illumina | [54] |
| Alfares et al. | 2018 | TSC2 | Tuberous sclerosis type 2 | De novo deep intronic SNV | Illumina | [54] |
| Lionel et al. 2 | 2018 | GPR143 | Ocular albinism | Deep intronic variant | Illumina | [55] |
| Lionel et al. 2 | 2018 | OTC | Ornithine transcarbamylase deficiency | Deep intronic variant | Illumina | [55] |
| Ostrander et al. | 2018 | Multiple | Global developmental delay | Balanced inverted translocation | Illumina | [56] |
| Ostrander et al. | 2018 | CDKL5 | Global developmental delay | De novo 63 kb tandem duplication | Illumina | [56] |
| Tavares et al. | 2018 | BBS1 | Bardet-Biedl syndrome | Retrotransposon insertion | Illumina | [66] |
| Cowley et al. | 2018 | SYNGAP1 | Epileptic encephalopathy | De novo 13 bp duplication | Illumina | [67] |
| Miao et al. | 2018 | G6PC | Glycogen storage disease type Ia | 7.1 kb deletion | ONT 1 | [68] |
| Merker et al. | 2018 | PRKAR1A | Carney complex | De novo 2184 bp deletion | PacBio | [69] |
| Sanchis-Juan et al. | 2018 | ARID1B | Coffin-Siris syndrome | De novo complex SV dupINVinvDEL | Illumina | [70] |
| Sanchis-Juan et al. | 2018 | HNRNPU | Seizures; Intellectual disability | De novo complex SV delINVdup | Illumina | [70] |
| Sanchis-Juan et al. | 2018 | CEP78 | Cone-rod dystrophy; Hearing loss | complex homozygous SV delINVdel | Illumina | [70] |
| Sanchis-Juan et al. | 2018 | CDKL5 | Birth asphyxia; Fetal distress | De novo complex SV dupINVdup | Illumina + ONT 1 | [70] |
| Ishiura et al. | 2018 | SAMD12 | BAFME 3 | TTTCA and TTTTA repeat expansions | PacBio +ONT | [57] |
| Ishiura et al. | 2018 | TNRC6A | BAFME 3 | TTTCA and TTTTA repeat expansions | PacBio + ONT | [57] |
| Ishiura et al. | 2018 | RAPGEF2 | BAFME 3 | TTTCA and TTTTA repeat expansions | PacBio + ONT | [57] |
| Mizuguchi et al. | 2019 | SAMD12 | BAFME 3 | 4.6 kb intronic repeat insertion | PacBio | [71] |
| Authors | Year | Tool | Method | Input 1 | Variants Detected | Reference |
|---|---|---|---|---|---|---|
| Abyzov et al. | 2011 | CNVnator | Read Depth | PE2 Short read WGS | Copy Number Variants | [88] |
| Rausch et al. | 2012 | DELLY | Paired-ends, Read depth, Split-reads | Short read WGS | Structural Variants | [89] |
| Calabrese et al. | 2014 | MToolBox | Read re-alignment | WGS or WES | Mitochondrial Variants | [90] |
| Layer et al. | 2014 | LUMPY | Paired-ends, Read depth, Split-reads | PE short read WGS | Structural Variants | [91] |
| Roller et al. | 2016 | Canvas | Read Depth | WGS or WES | Copy Number Variations | [84,85] |
| Chen et al. | 2016 | Manta | Pair Read, Split Read | PE short read WGS | Indels, Structural Variants | [92] |
| Dolzhenko et al. | 2017 | Expansion-Hunter | Sequence-graph | PE short read WGS | Large Expansion of Short Tandem Repeats | [74] |
| Ebler et al. | 2017 | DIGTYPER | Breakpoint-Spanning, Split Alignments | PE short read WGS | Inversions, Tandem Duplications | [93] |
| Liang et al. | 2017 | Seeksv | Split Read, Discordant Paired-End, Read Depth, 2 Ends Unmapped | SE/PE 2 short read WGS | Structural Variants + Virus Integration | [94] |
| Mousavi et al. | 2018 | GangSTR | Enclosing, Fully Repetitive, Spanning and Off-target Fully Repetitive Read Pairs | PE short read WGS | Tandem Repeat expansions | [78] |
| Kim et al. | 2018 | Strelka2 | Mixture-model | PE short read WGS | Single Nucleotide Variants, Indels | [95] |
| Ye et al. | 2018 | Pindel | Split-reads | PE short read WGS | Indels, Structural Variants (small and medium-size) | [96,97] |
| Wala et al. | 2018 | SvABA | Local assembly | PE short read WGS | Indels, Structural Variants (20–300 bp) | [98] |
| Becker et al. | 2018 | SVE/FusorSV | 8 SV callers combination + Data mining | PE short read WGS | Deletions + Duplications + Inversions 3 | [86] |
| Antaki et al. | 2018 | SV2 | Supervised support vector machine classifiers | PE short read WGS | Deletions + Duplications | [87] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maroilley, T.; Tarailo-Graovac, M. Uncovering Missing Heritability in Rare Diseases. Genes 2019, 10, 275. https://doi.org/10.3390/genes10040275
Maroilley T, Tarailo-Graovac M. Uncovering Missing Heritability in Rare Diseases. Genes. 2019; 10(4):275. https://doi.org/10.3390/genes10040275
Chicago/Turabian StyleMaroilley, Tatiana, and Maja Tarailo-Graovac. 2019. "Uncovering Missing Heritability in Rare Diseases" Genes 10, no. 4: 275. https://doi.org/10.3390/genes10040275
APA StyleMaroilley, T., & Tarailo-Graovac, M. (2019). Uncovering Missing Heritability in Rare Diseases. Genes, 10(4), 275. https://doi.org/10.3390/genes10040275





