Comprehensive Analyses of Nitric Oxide-Induced Plant Stem Cell-Related Genes in Arabidopsis thaliana
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptome-Wide Identification and Characterization of Plant Stem Cell Genes
2.2. Correlation and Gene Ontology Analyses
2.3. Promoter Analysis
2.4. Phylogenetic Analysis
2.5. Plant Material and Growth Conditions
2.6. Redox Stress Assay
2.7. Pathogen Inoculation and Pathogenicity Assessment
2.8. q-RT PCR and Gene Expression
3. Results
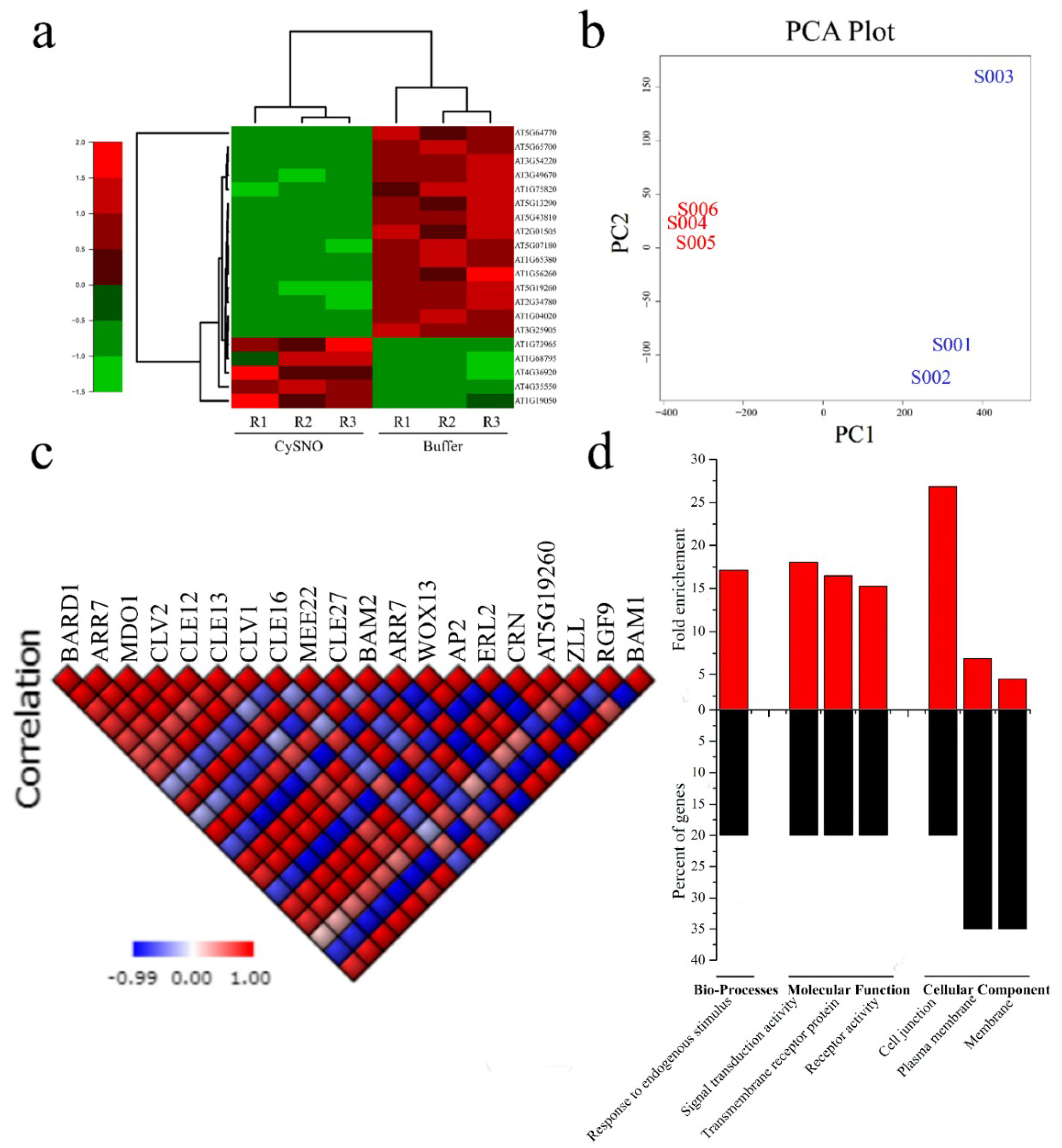
3.1. Transcriptome-Wide Identification and Characterization of Stem Cell-Related Genes in Response to Nitric Oxide
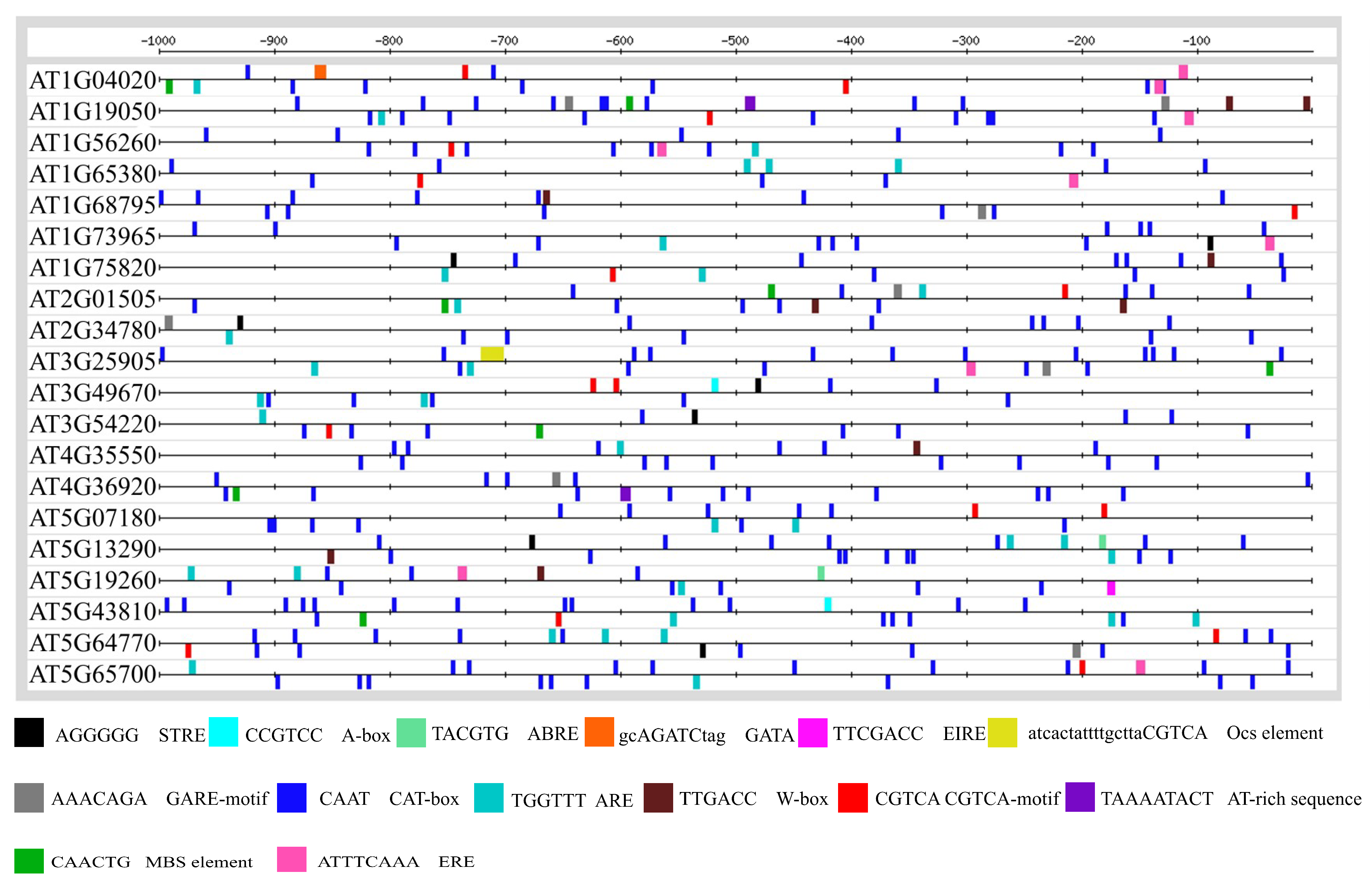
3.2. Promoter Anlaysis for Identification of Cis-Regulatory Elements
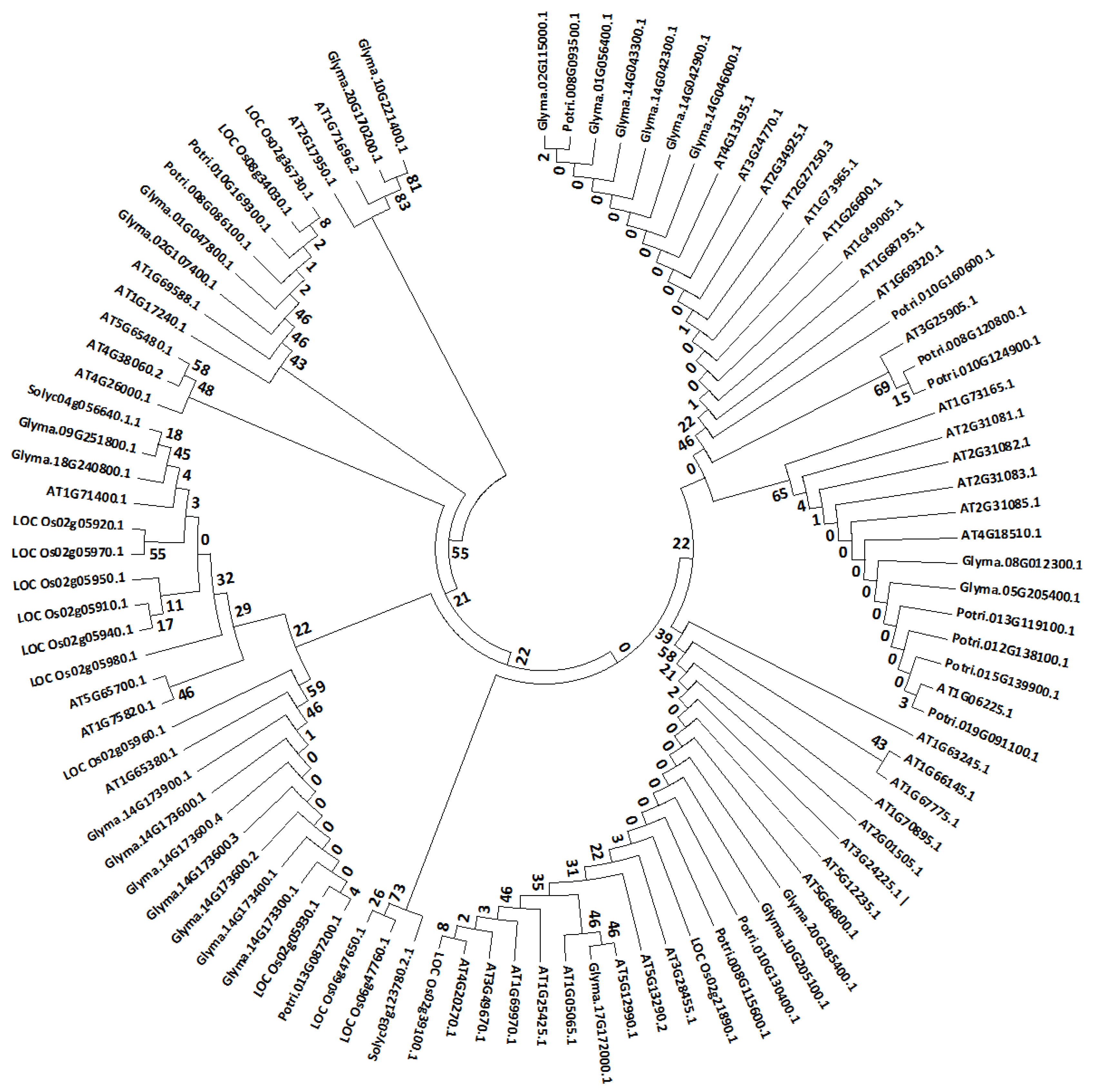
3.3. Phylogenetic Analsysis of Arabidopsis Stem Cells and their Orthologs in Other Species
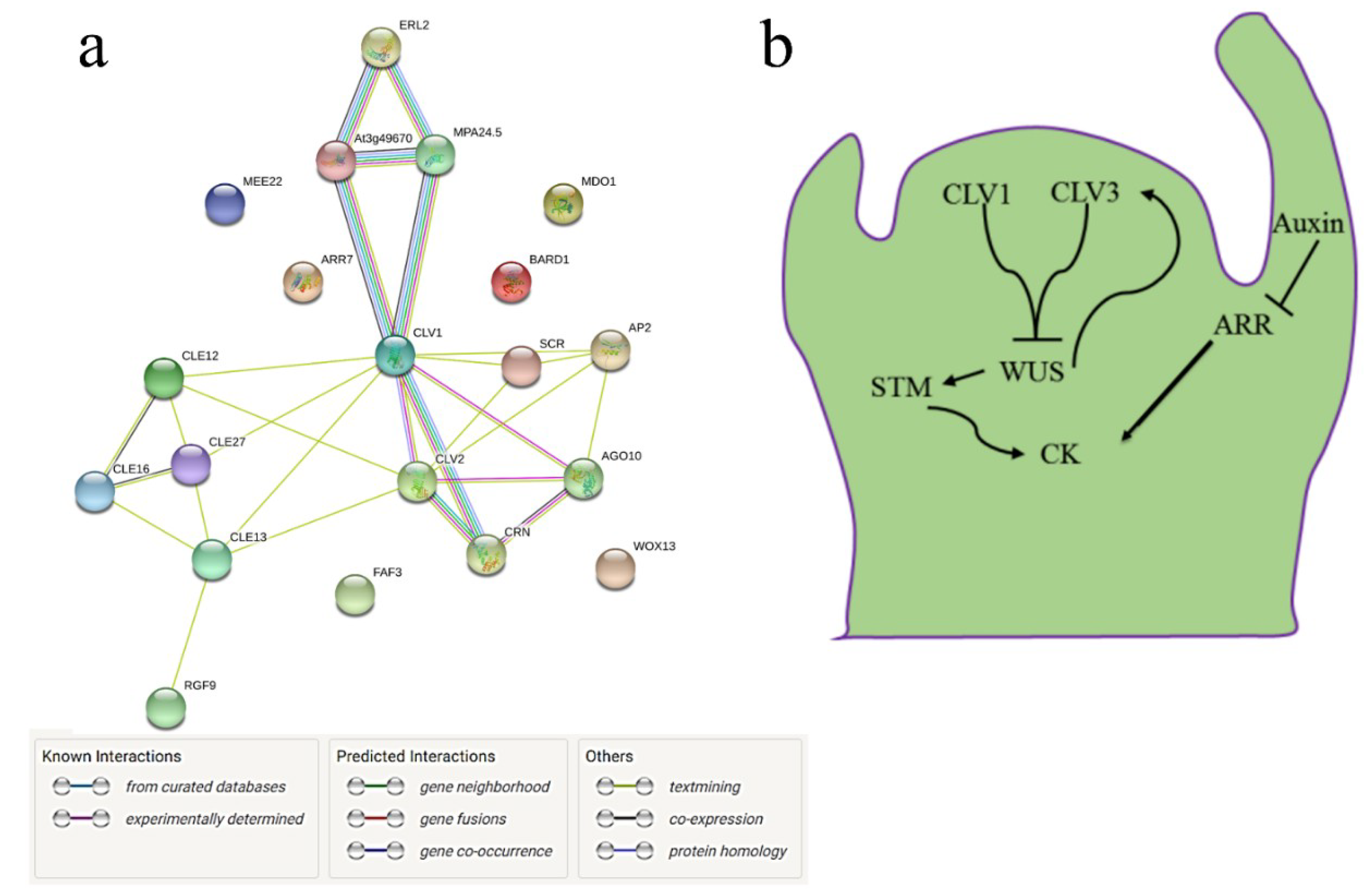
3.4. Interactome of CySNO-Induced Stem Cell-Related Genes
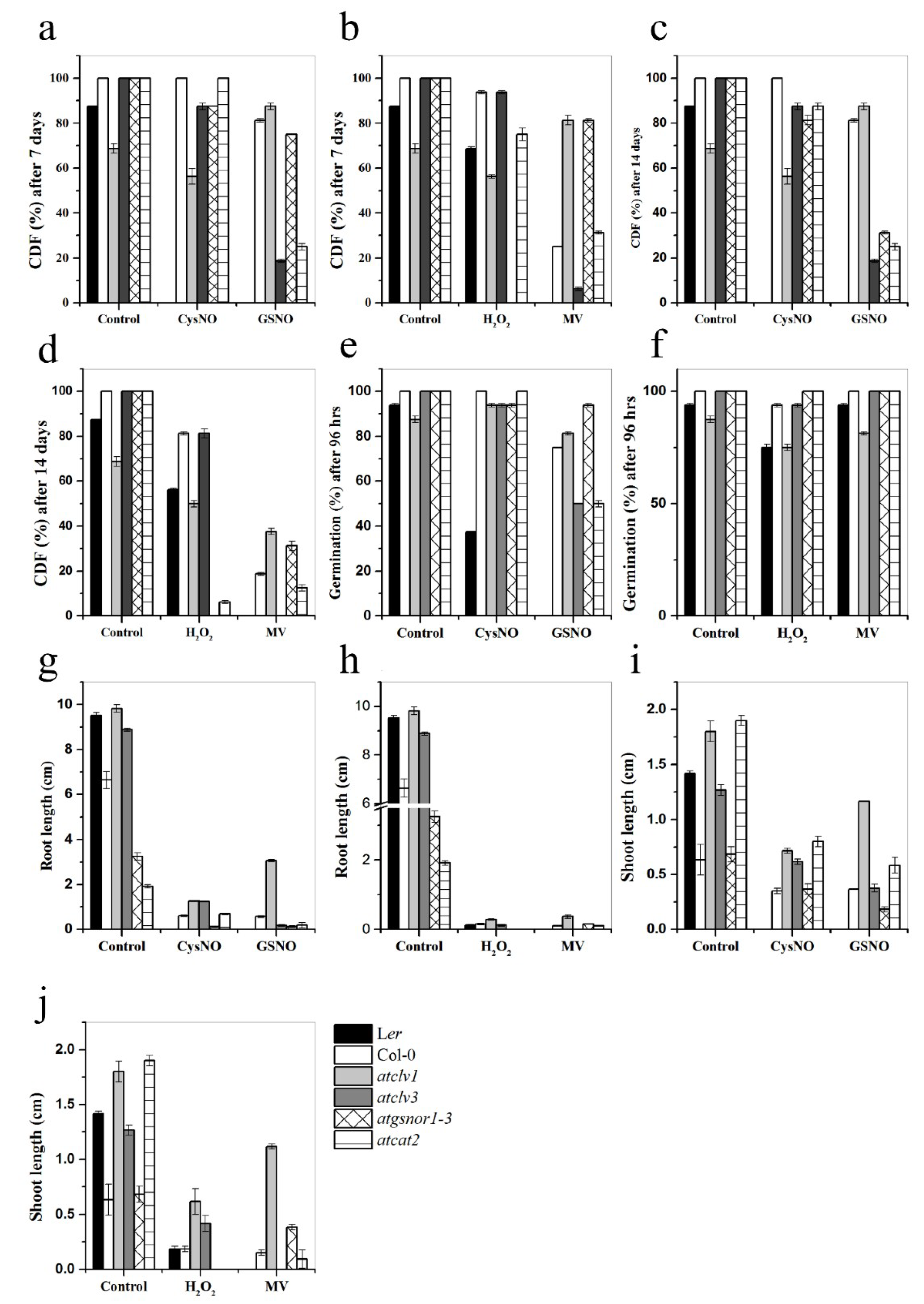
3.5. CLV1 and CLV3 Differentially Regulate Nitrosative and Oxidative Stresses
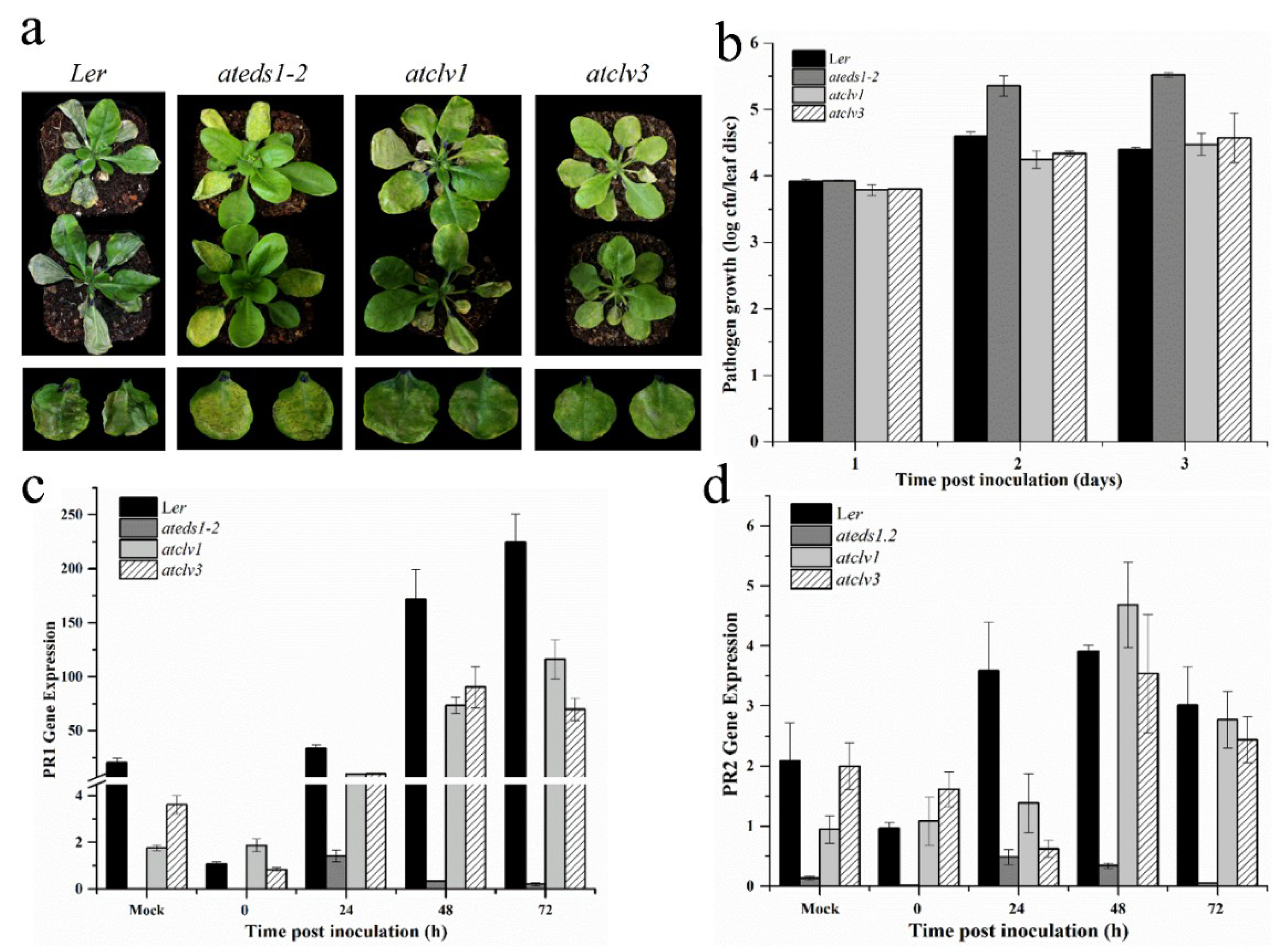
3.6. CLV1 and CLV3 Positively Regulates Basal Defense at Early Time Points
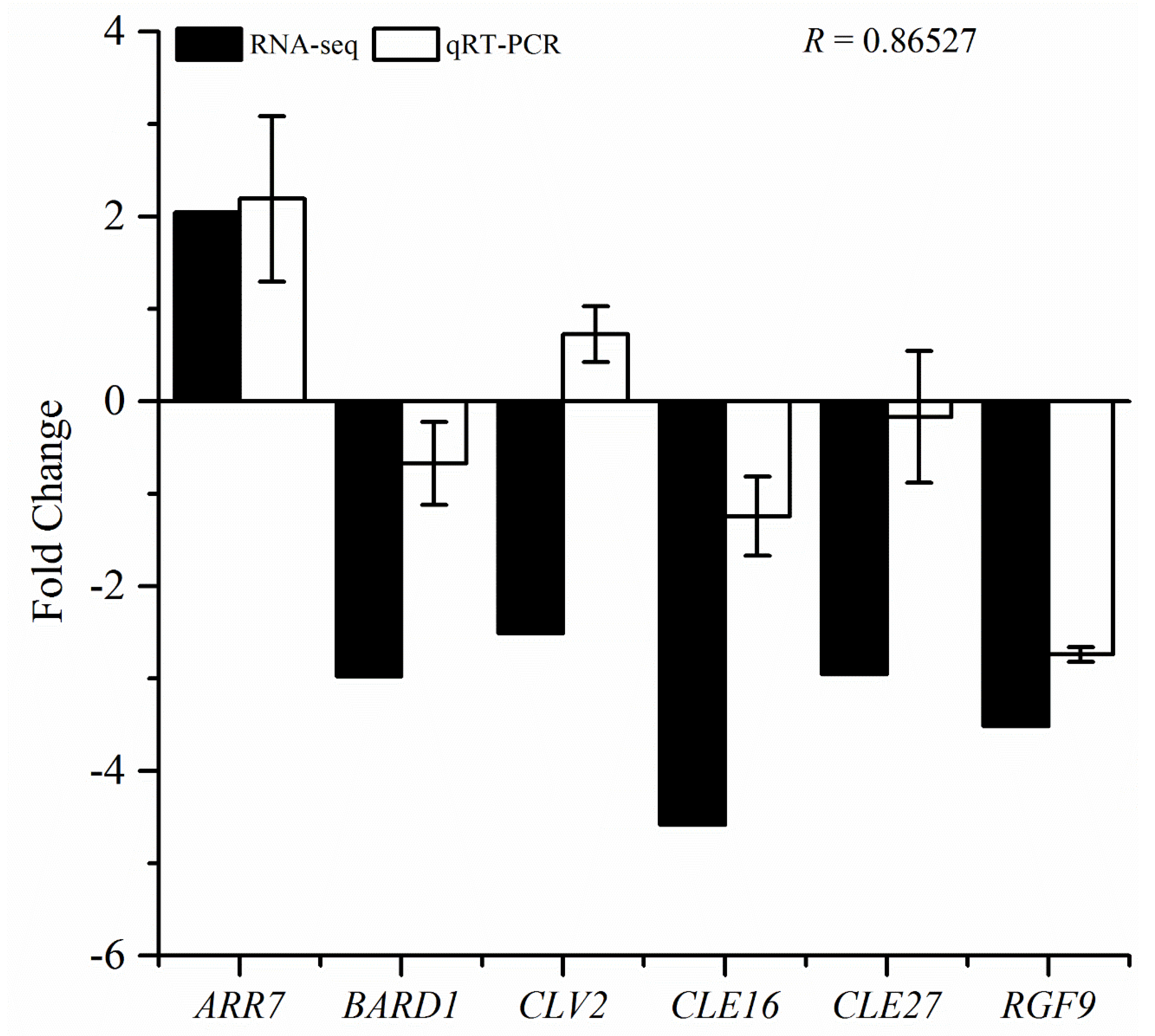
3.7. qRT-PCR Validation of RNA-Seq Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greb, T.; Lohmann, J.U. Plant Stem Cells. Curr. Biol. 2016, 26, R816–R821. [Google Scholar] [CrossRef] [PubMed]
- Gaillochet, C.; Lohmann, J.U. The never-ending story: From pluripotency to plant developmental plasticity. Development 2015, 142, 2237–2249. [Google Scholar] [CrossRef] [PubMed]
- Christmann, A.; Weiler, E.W.; Steudle, E.; Grill, E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007, 52, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Katayama, N.; Koi, S.; Kato, M. Expression of SHOOT MERISTEMLESS, WUSCHEL, and ASYMMETRIC LEAVES1 Homologs in the Shoots of Podostemaceae: Implications for the Evolution of Novel Shoot Organogenesis. Plant Cell 2010, 22, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.X.; Schoof, H.; Haecker, A.; Lenhard, M.; Jurgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Schuster, C.; Gaillochet, C.; Medzihradszky, A.; Busch, W.; Daum, G.; Krebs, M.; Kehle, A.; Lohmann, J.U. A Regulatory Framework for Shoot Stem Cell Control Integrating Metabolic, Transcriptional, and Phytohormone Signals. Dev. Cell 2014, 28, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.E.; Williams, R.W.; Meyerowitz, E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 1997, 89, 575–585. [Google Scholar] [CrossRef]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef] [PubMed]
- Strabala, T.J. CLE genes in plant development: Gain-of-function analyses, pleiotropy, hypermorphy and neomorphy. Plant Signal. Behav. 2008, 3, 457–459. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, A.; Nakamura, Y.; Sasaki, E.; Kyozuka, J.; Fukuda, H.; Sawa, S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007, 48, 1821–1825. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Replogle, A.; Hussey, R.; Baum, T.; Wang, X.H.; Davis, E.L.; Mitchum, M.G. Identification of potential host plant mimics of CLAVATA3/ESR (CLE)-like peptides from the plant-parasitic nematode Heterodera schachtii. Mol. Plant Pathol. 2011, 12, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Ramirez, J.; Fletcher, J.C. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol. Biol. 2003, 51, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Sawa, S.; Kinoshita, A.; Betsuyaku, S.; Fukuda, H. A large family of genes that share homology with CLE domain in Arabidopsis and rice. Plant Signal. Behav. 2008, 3, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 10328–10333. [Google Scholar] [CrossRef] [PubMed]
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant disease resistance. Nature 1998, 394, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Beligni, M.V.; Lamattina, L. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 2000, 210, 215–221. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tang, R.-H.; Hao, Y.; Stevens, R.D.; Cook, C.W.; Ahn, S.M.; Jing, L.; Yang, Z.; Chen, L.; Guo, F.; et al. Nitric Oxide Represses the Arabidopsis Floral Transition. Science 2004, 305, 1968–1971. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.; Yu, S.I.; Lee, H.; Yim, J.H.; Zhu, J.K.; Lee, B.H. Arabidopsis Serine Decarboxylase Mutants Implicate the Roles of Ethanolamine in Plant Growth and Development. Int. J. Mol. Sci. 2012, 13, 3176–3188. [Google Scholar] [CrossRef] [PubMed]
- Lee, U.; Wie, C.; Fernandez, B.O.; Feelisch, M.; Vierling, E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell Online 2008, 20, 786–802. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.D.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Pagnussat, G.C.; Simontacchi, M.; Puntarulo, S.; Lamattina, L. Nitric oxide is required for root organogenesis. Plant Physiol. 2002, 129, 954–956. [Google Scholar] [CrossRef] [PubMed]
- Gouvea, C.M.C.P.; Souza, J.F.; Magalhaes, A.C.N.; Martins, I.S. NO-releasing substances that induce growth elongation in maize root segments. Plant Growth Regul. 1997, 21, 183–187. [Google Scholar] [CrossRef]
- Sharma, A.; Hussain, A.; Mun, B.-G.; Imran, Q.M.; Falak, N.; Lee, S.-U.; Kim, J.Y.; Hong, J.K.; Loake, G.J.; Ali, A. Comprehensive analysis of plant rapid alkalization factor (RALF) genes. Plant Physiol. Biochem. 2016, 106, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Kosack, K.E.; Jones, J.D. Resistance gene-dependent plant defense responses. Plant Cell 1996, 8, 1773–1791. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Mun, B.G.; Imran, Q.M.; Lee, S.U.; Adamu, T.A.; Shahid, M.; Kim, K.M.; Yun, B.W. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 975. [Google Scholar] [CrossRef] [PubMed]
- Imran, Q.M.; Hussain, A.; Mun, B.G.; Lee, S.U.; Asaf, S.; Ali, M.A.; Lee, I.J.; Yun, B.W. Transcriptome wide identification and characterization of NO-responsive WRKY transcription factors in Arabidopsis thaliana L. Environ. Exp. Bot. 2018, 148, 128–143. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Medina-Rivera, A.; Defrance, M.; Sand, O.; Herrmann, C.; Castro-Mondragon, J.A.; Delerce, J.; Jaeger, S.; Blanchet, C.; Vincens, P.; Caron, C.; et al. RSAT 2015: Regulatory Sequence Analysis Tools. Nucleic Acids Res. 2015, 43, W50–W56. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Feechan, A.; Kwon, E.; Yun, B.-W.; Wang, Y.; Pallas, J.A.; Loake, G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 8054–8059. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.Q.; Liu, S.; Yuan, H.M.; Li, J.; Yan, D.W.; Zhang, J.A.F.; Lu, Y.T. Functional comparison of catalase genes in the elimination of photorespiratory H2O2 using promoter- and 3’-untranslated region exchange experiments in the Arabidopsis cat2 photorespiratory mutant. Plant Cell Environ. 2010, 33, 1656–1670. [Google Scholar] [CrossRef] [PubMed]
- Imran, Q.M.; Hussain, A.; Lee, S.-U.; Mun, B.-G.; Falak, N.; Loake, G.J.; Yun, B.-W. Transcriptome profile of NO-induced Arabidopsis transcription factor genes suggests their putative regulatory role in multiple biological processes. Sci. Rep. 2018, 8, 771. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.W.; Feechan, A.; Yin, M.H.; Saidi, N.B.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.G.; Kwon, E.; Spoel, S.H.; et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011, 478, 264. [Google Scholar] [CrossRef] [PubMed]
- Imran, Q.M.; Falak, N.; Hussain, A.; Mun, B.-G.; Sharma, A.; Lee, S.-U.; Kim, K.-M.; Yun, B.-W. Nitric oxide responsive heavy metal-associated gene AtHMAD1 contributes to development and disease resistance in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1712. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Li, Q.; Zhu, Y.X. Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell 2008, 20, 1482–1493. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Jiao, Y.; Liu, Z.H.; Zhu, Y.X. ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat. Commun. 2015, 6, 6003. [Google Scholar] [CrossRef] [PubMed]
- Hashimura, Y.; Ueguchi, C. The Arabidopsis MERISTEM DISORGANIZATION 1 gene is required for the maintenance of stem cells through the reduction of DNA damage. Plant J. 2011, 68, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Somssich, M.; Je, B.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Trotochaud, A.E.; Clark, S.E. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 1999, 11, 1925–1933. [Google Scholar] [CrossRef] [PubMed]
- Bolle, C.; Koncz, C.; Chua, N.H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Gene Dev. 2000, 14, 1269–1278. [Google Scholar] [PubMed]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Drews, G.N.; Bowman, J.L.; Meyerowitz, E.M. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 1991, 65, 991–1002. [Google Scholar] [CrossRef]
- Müller, R.; Bleckmann, A.; Simon, R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 2008, 20, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Wahl, V.; Brand, L.H.; Guo, Y.L.; Schmid, M. The FANTASTIC FOUR proteins influence shoot meristem size in Arabidopsis thaliana. Bmc Plant Biol. 2010, 10, 285. [Google Scholar] [CrossRef] [PubMed]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2012, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Caarls, L.; Pieterse, C.M.; Van Wees, S.C. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.; Bussey, H.; Andrews, B.J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007, 8, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 1995, 121, 2057–2067. [Google Scholar]
- Koornneef, M.; Van Eden, J.; Hanhart, C.; Stam, P.; Braaksma, F.; Feenstra, W. Linkage map of Arabidopsis thaliana. J. Hered. 1983, 74, 265–272. [Google Scholar] [CrossRef]
- Wendehenne, D.; Pugin, A.; Klessig, D.F.; Durner, J. Nitric oxide: Comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 2001, 6, 177–183. [Google Scholar] [CrossRef]
- Garcia-Mata, C.; Lamattina, L. Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiol. 2002, 128, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, F.; Guo, J.; Yang, Y.; Li, B.; Zhang, L. Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of Reed. Plant Physiol. 2004, 134, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R.; Erdjument-Bromage, H.; Ferris, C.D.; Tempst, P.; Snyder, S.H. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001, 3, 193. [Google Scholar] [CrossRef] [PubMed]
- Parani, M.; Rudrabhatla, S.; Myers, R.; Weirich, H.; Smith, B.; Leaman, D.W.; Goldman, S.L. Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol. J. 2004, 2, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Polverari, A.; Molesini, B.; Pezzotti, M.; Buonaurio, R.; Marte, M.; Delledonne, M. Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol. Plant Microbe 2003, 16, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; von Rad, U.; Durner, J. Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 2002, 215, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Luque, F.; Leyva-Pérez, M.O.; Leterrier, M.; Corpas, F.J.; Barroso, J.B. Differential transcriptomic analysis by RNA-Seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant Cell Physiol. 2014, 55, 1080–1095. [Google Scholar] [CrossRef] [PubMed]
- Misra, A.N.; Misra, M.; Singh, R. Nitric oxide ameliorates stress responses in plants. Plant Soil Environ. 2011, 57, 95–100. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.X.; Jurgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Kim, J.S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K.; et al. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A Gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 2011, 52, 2136–2146. [Google Scholar] [CrossRef] [PubMed]
- Mukumoto, F.; Hirose, S.; Imaseki, H.; Yamazaki, K. DNA-sequence requirement of a Tata element-binding protein from Arabidopsis for transcription in-Vitro. Plant Mol. Biol. 1993, 23, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y. Interaction of tobacco nuclear protein with an elicitor-responsive element in the promoter of a basic class I chitinase gene. Plant Mol. Biol. 1997, 34, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Laxmi, A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 2015, 6, 895. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, M.C.; Sell, S.; Huang, X.; Scherf, M.; Werner, T.; Durner, J.; Lindermayr, C. Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: A bioinformatics approach. J. Exp. Bot. 2008, 59, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; Sheen, J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 2008, 453, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahid, M.; Imran, Q.M.; Hussain, A.; Khan, M.; Lee, S.U.; Mun, B.G.; Yun, B.-W. Comprehensive Analyses of Nitric Oxide-Induced Plant Stem Cell-Related Genes in Arabidopsis thaliana. Genes 2019, 10, 190. https://doi.org/10.3390/genes10030190
Shahid M, Imran QM, Hussain A, Khan M, Lee SU, Mun BG, Yun B-W. Comprehensive Analyses of Nitric Oxide-Induced Plant Stem Cell-Related Genes in Arabidopsis thaliana. Genes. 2019; 10(3):190. https://doi.org/10.3390/genes10030190
Chicago/Turabian StyleShahid, Muhammad, Qari Muhammad Imran, Adil Hussain, Murtaza Khan, Sang Uk Lee, Bong Gyu Mun, and Byung-Wook Yun. 2019. "Comprehensive Analyses of Nitric Oxide-Induced Plant Stem Cell-Related Genes in Arabidopsis thaliana" Genes 10, no. 3: 190. https://doi.org/10.3390/genes10030190
APA StyleShahid, M., Imran, Q. M., Hussain, A., Khan, M., Lee, S. U., Mun, B. G., & Yun, B.-W. (2019). Comprehensive Analyses of Nitric Oxide-Induced Plant Stem Cell-Related Genes in Arabidopsis thaliana. Genes, 10(3), 190. https://doi.org/10.3390/genes10030190





