Abstract
Although more than 100 genome sequences of Pasteurella multocida are available, comprehensive and complete genome sequence analysis is limited. This study describes the analysis of complete genome sequence and pathogenomics of P. multocida strain PMTB2.1. The genome of PMTB2.1 has 2176 genes with more than 40 coding sequences associated with iron regulation and 140 virulence genes including the complete tad locus. The tad locus includes several previously uncharacterized genes such as flp2, rcpC and tadV genes. A transposable phage resembling to Mu phages was identified in P. multocida that has not been identified in any other serotype yet. The multi-locus sequence typing analysis assigned the PMTB2.1 genome sequence as type ST101, while the comparative genome analysis showed that PMTB2.1 is closely related to other P. multocida strains with the genomic distance of less than 0.13. The expression profiling of iron regulating-genes of PMTB2.1 was characterized under iron-limited environment. Results showed significant changes in the expression profiles of iron-regulating genes (p < 0.05) whereas the highest expression of fecE gene (281 fold) at 30 min suggests utilization of the outer-membrane proteins system in iron acquisition at an early stage of growth. This study showed the phylogenomic relatedness of P. multocida and improved annotation of important genes and functional characterization of iron-regulating genes of importance to the bacterial growth.
1. Introduction
Pasteurella multocida (PM) species of the genus Pasteurella are Gram-negative facultative anaerobic bacteria belonging to the family Pasteurellaceae [1]. Based on its capsular antigen, the P. multocida is grouped into A, B, D, E and F capsular types and further classified into 16 serotypes (1–16) based on lipopolysaccharide (LPS) antigen [2,3]. P. multocida was earlier identified as a commensal in the upper respiratory tract of mammals and birds. However, P. multocida is often associated with acute as well as chronic infections in avian and bovine that can lead to significant morbidity and mortality [4,5]. Fowl cholera in avian, atrophic rhinitis in pigs and pasteurellosis characterized as respiratory diseases namely pneumonia and hemorrhagic septicemia (HS) in cattle and buffaloes are the most important veterinary diseases [6,7]. In general, pasteurellosis caused by P. multocida is an acute septicemic disease characterized by high morbidity and is a high-impact disease in livestock, according to the World Organization for Animal Health [7].
Pasteurella multocida serotype A:3 has been implicated in fatal pneumonia of cattle in India [8]. Furthermore, P. multocida serotype A:1 has also been reported to be responsible for respiratory diseases such as fatal pneumonia and septicemia of cattle and buffaloes [7,9]. Unlike pasteurellosis associated with pneumonia and septicemia induced by serotype A, HS is an acute septicemic disease caused by P. multocida serotypes B:2 and E:2 in cattle and buffaloes [6,7] that results in an outbreak with 100% mortality in infected animals [10]. Hence, HS and pneumonic septicemic pasteurellosis are considered to be the most important bacterial diseases economically in various countries in Asia and South Africa [10,11].
Currently, about 121 complete or draft P. multocida genomes are publicly available in the GenBank, National Center for Biotechnology Information (NCBI). However, only a few genomes have been studied and examined in detail such as PM70 (serotype F) [12], PM36950 (serotype A) [13], PMHB01 (serotype A) [14] and the genomes of P. multocida strains harboring the P. multocida toxin (PMT) gene such as PMHN06 (serotype D) [15]. The first complete genomic structure analysis of P. multocida PM70 revealed that it possesses some predicted virulence genes and iron-uptake genes and a complete set of the genes for metabolic pathways necessary for the metabolism of certain carbohydrates and synthesis of all 20 amino acids [12]. Subsequently, the first integrative conjugative element (ICE) harboring antibiotic resistance genes were identified from the complete genome sequence analysis of P. multocida PM36950 [13].
Iron is one of the most important nutrients for pathogenic bacteria like Pasteurella and the level of free iron available in the host is very limited [4,16,17]. It is possible that iron acquisition in P. multocida plays an important role in its survival and pathogenesis of the disease [4,16]. To this end, P. multocida strains have siderophore-independent iron acquisition systems homologous to the Actinobacillus AfeABCD system and the periplasmic binding protein-dependent iron transport systems homologous to Escherichia coli FecBCDE and Neisseria FbpABC systems [4]. The presence of multiple iron acquisition systems in the Pasteurella species may account for their ability to acquire iron under iron-limited conditions by utilizing a certain set of the genes under certain conditions. The gene expression patterns of P. multocida in iron-free chemically defined medium and in response to different iron sources have been studied by DNA microarray [17]. In another DNA microarray-based study [12], more than 150 genes showed more than two-fold altered expression when the P. multocida strain PM70 was grown with an iron chelating agent. In both of the above studies, the transcriptional responses of P. multocida to either low iron or defined iron sources have been studied using microarray-based technology. However, the expression profiles of these genes were not characterized further based on the real-time PCR assay.
Recently, the P. multocida strain HB01 complete genome sequence was produced using the next-generation sequencing (NGS) technology and primer walking Sanger sequencing [14], whereas, the P. multocida strain Razi 0001 (CP 017961.1) was produced as in silico complete genome sequence using the Pacific Biosciences (PacBio, Menlo Park, CA, USA) sequencing technology. The development of a third-generation sequencing technology such as PacBio, which is a long-read sequencing platform of relatively low cost and high throughput have greatly influenced the field of bacterial genome sequencing [18]. Recently we have isolated, P. multocida strain PMTB2.1 based on the Pasteurella multocida specific PCR (PM-PCR) [19], from buffalo that died of septicemia. However, the genomic characteristics of the isolate are not known. Thus, the aim of this study was to perform a comprehensive genome sequence analysis of PMTB2.1 and to study the gene expression pattern of selected iron-regulating genes namely fecE, yfeA, fbpB and fur as well as the colonization gene nanA (sialidase) to elucidate the actual response of these genes to the iron limiting condition.
2. Materials and Methods
2.1. Bacterial Strain and DNA Extraction
The P. multocida strain PMTB2.1 was isolated from a buffalo that died of septicemia in Malaysia [20] and was identified by species-specific PCR [7,19]. The bacteria were prepared from a pure stock culture that was grown in brain heart infusion (BHI) broth at 37 °C for 12–16 h. The PMTB2.1 genomic DNA was extracted using the DNeasy Mini spin column (Qiagen, Hilden, Germany). The extracted DNA was quantified and qualified using an Eppendorf Biospectrometer® (Eppendorf, Hamburg, Germany) and agarose gel electrophoresis stained with 1× RedSafeTM (iNTRon Biotechnology, Gyeonggi-do, Korea).
2.2. Genome Sequencing and Assembly
Genome sequencing was performed using the PacBio RS 2 sequencing platform (University of Malaya, Kuala Lumpur, Malaysia). The genomic DNA of PMTB2.1 was sequenced using the C2 chemistry [21] and primary filtering was performed on the RS Blade Centre server following which secondary analysis was performed using the SMRT analysis pipeline version 1.4 (PacBio). The raw sequencing data were used to extract circular consensus sequence (CCS) and continuous long read (CLR). Error correction of the CLR reads, based on the CCS reads was carried out by the PacBio ToCA module of Whole-Genome Shotgun Assembler v8.2 with 26× coverage. These error-corrected reads were then assembled by the Celera Assembler. The genome was assembled de novo by runCA (Celera Assembler) (http://wgs-assembler.sourceforge.net) using default parameters, which generated three contigs. The contigs were then aligned together and assembled into a single contig with minimum gaps, based on the overlapping layout method. Primers were designed to sequence the gaps between the contigs. The Sanger sequencing method was utilized to sequence amplicons by primer walking strategy. The first and last nucleotide position of the circular genome of PMTB2.1 was set based on homology to the reference genome of P. multocida strain PM36950 (accession no. CP003022.1) and circularity chromosome was confirmed by PCR.
2.3. Genome Annotation and Genomic Organization of PMTB2.1
Genome annotation of P. multocida strain PMTB2.1 was carried out using the Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) provided by NCBI (http://ncbi.nlm.nih.gov). Genes were predicted using GeneMarks [22]; whereby the annotated sequences include CDs (coding sequences), ribosomal RNA (rRNA), transfer RNA (tRNA) and ncRNA (non-coding RNA). Functional annotations of the putative coding genes were predicted by PGAAP, NCBI using Basic Local Alignment Tools (BLAST) against non-redundant (nr) and Swiss-Prot (SP) protein databases [23] at a cut-off E-value ≤ 1 × 10−5 using Blast2GO [24]. Annotation was also carried out using the gene ontology Kyoto Encyclopedia of Genes and Genomes (KEGG) database [25]. Artemis [26] was used to visualize and analyze structural and functional annotation of the genome sequence of PMTB2.1. The origin of replication was determined using Ori-Finder [27], while the circular genomic plot of PMTB2.1 was plotted by Circos version 0.69 [28] and the genomic islands were identified using online software IslandViewer 3 [29].
2.4. Genome Sequence Analysis
The analysis of complete genome sequence of PMTB2.1 was performed to identify genes and sequence motifs of interest based on different databases. The potential virulence genes and antibiotic resistance genes were identified based on a homology search against Pathogen-Host Interaction Database (PHI-base) [30], Virulence Factors Database (VFDB) [31] and the Comprehensive Antibiotic Resistance Database (CARD) [32]. Phage and prophage-like sequences were predicted by PHAST [33]. Cut-off value of E-value ≤ 1 × 10−5 and subject coverage ≥ 70% and identity between two proteins ≥ 50% were applied to the blast p search. Multi-locus sequence typing (MLST) [34] was performed based on PubMLST (https://publst.org/multocida) and Centre of Genomic Epidemiology (CGE, https://cge.cbs.dtu.dk/services/MLST) against Rural Industries Research and Development Corporation (RIRDC) scheme.
2.5. Comparative Genomics of PMTB2.1
Comparative analysis was carried out on PMTB2.1 and another eight selected genomes. The selected genomes were comprised of five complete genomes of P. multocida belonging to different serotypes; A, D and F from different host origins and the remaining three included Actinobacillus pleuropneumoniae JL03 (CP000687), Haemophilus parasuis SH0165 (CP001321) and Escherichia coli K12 MG1655 (NC_000913). Genome distance matrix was obtained by Progressive Mauve v2.3.1 [35] and visualized in a heatmap using R v3.0.2. Pairwise coverage values are subtracted from one to yield a distance value. The matrix is used to infer a guide tree and to scale the break-point penalty during anchoring. The synteny analysis was carried out using Mauve v2.3.1 [35] to compare complete genome sequences to identify regions of similarity and rearrangements.
Identification of the orthology groups of the genome was carried out using OrthoMCL v2.0.9 [36] and information obtained was used to construct a phylogenomic tree and to compare the selected genomes with PMTB2.1, using the Venn diagram software [37].
2.6. Expression Profiling of Iron-Regulating Genes of PMTB2.1
The P. multocida strain PMTB2.1 cells were grown in 200 mL of fresh BHI broth with 2, 2’- Bipyridine (Merck, New Jersey, USA; 200 µM/mL). The cultures were incubated at 37 °C aerobically in a shaking incubator (Sartorius, Tagelswangen, Switzerland) and samples were removed from each cell culture at the time points of 30, 60, 120 and 180 min after incubation of the bacterial cell pellet. Bacterial cells were pelleted by centrifugation at 4000× rpm (Eppendorf Centrifuge 5415R, Hamburg, Germany) at 4 °C for 5–7 min, the supernatant was discarded and the resulted cell pellet was suspended in 2 mL of Bacterial RNAlater (Qiagen) and preserved at −80 °C for subsequent RNA extraction. The experiment was run in triplicate and bacterial cultures grown in BHI broth alone were maintained as controls.
Bacterial RNA extractions were performed with the RNeasy mini plus kit as recommended by the manufacturer (Qiagen). The quality of extracted bacterial RNA and their respective concentrations were checked using an Eppendorf Biospectrometer (Eppendorf, Germany). Reverse transcription PCR (RT-PCR) was carried out on a thermal cycler (Bio-Rad, California, USA) using the SensiFast cDNA synthesis kit (Bioline, London, UK) according to the manufacturer’s instructions. A total of four iron-regulating genes namely fecE, yfeA, fbpB and fur and a colonization gene nanA (sialidase) were selected to assess their relative mRNA expression using Taqman-based real time-PCR. Two housekeeping genes, glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) and DNA gyrase B (gyrB) were used to normalize the reactions. The primers and probes were designed using the PMTB2.1 complete genome sequence. The sequences of the primers and probes and their melting temperatures are shown in Table 1. Standard curves for the target and housekeeping genes were generated using extracted RNA converted to cDNA by Taqman Probe-based real-time PCR.

Table 1.
PMTB2.1 genes primer and probe sequences for real-time PCR assay.
The mean quantitative cycle value (Cq) for a selected gene at a time point was determined from three biological replicates each with three technical replicates. The relative fold changes of these genes from the treated samples at selected time points of treatment were compared to the control samples. Relative fold changes were calculated by the 2−ΔΔCt method. The data analysis, normalization with housekeeping genes and the quantification of fold changes were carried out as described by Bustin et al. [38] using Bio-Rad CFX manager software v3.1 (Bio-Rad). All quantitative data were expressed as a mean ± standard deviation. Statistical significance of relative fold change expressions was calculated by comparison between groups at p ≤ 0.05.
2.7. Data Availability
The complete genome sequence of P. multocida strain PMTB2.1 and eight selected genomes for comparative analysis are available on NCBI and can be accessed using the accession number listed in the table with title “Comparison of P. multocida strain PMTB2.1 and complete genome sequences of other P. multocida of different serotypes”, in the result section.
3. Results
3.1. Genome Assembly
A total of 49,844 CCS reads and 523,361 CLR were extracted from the PacBio raw sequencing data. Error correction of the CLR reads using CCS reads generated 13,231 reads with a mean read length of 2000 bp (total of 60,463,108 nucleotides) with 26× coverage. The genome was assembled de novo by runCA (Celera Assembler) using default parameters, which generated three contigs with the size of 1,057,336; 946,414; and 338,900 bp for each contig, respectively, to a total of 2,342,650 bp.
The unresolved gaps in the genome assembly were closed with the primer walking/Sanger sequencing reads by Staden v2.0.09. The circularity of the chromosome was confirmed by PCR where the trimmed sequences were successfully aligned with the complete genome sequence of PMTB2.1 in such a way that the first 1,538 bp at the end of circle aligned with the last nucleotides at position 2,326,603 to 2,328,141. Meanwhile, the remaining 112 bp (in a circle) aligned at the nucleotide positions of 1 to 112 of the complete genome sequence of P. multocida strain PMTB2.1. The final finished grade, complete genome sequences of P. multocida strain PMTB2.1 (2,315,138 bp) as a single contiguous circular chromosome was submitted to GenBank, NCBI under the accession number CP007205.2.
3.2. Genetic Organizations of the PMTB2.1 Complete Genome Sequence
The genome of P. multocida strain PMTB2.1 is composed of 2,315,138 base pairs with a GC content of approximately 40.32% (Figure 1), containing 2097 protein-coding sequences, 19 rRNA (including 6, 16S rRNA operon), 56 tRNA and 4 ncRNA (non-coding RNA) genes, making a total of 2176 genes (Table 2).
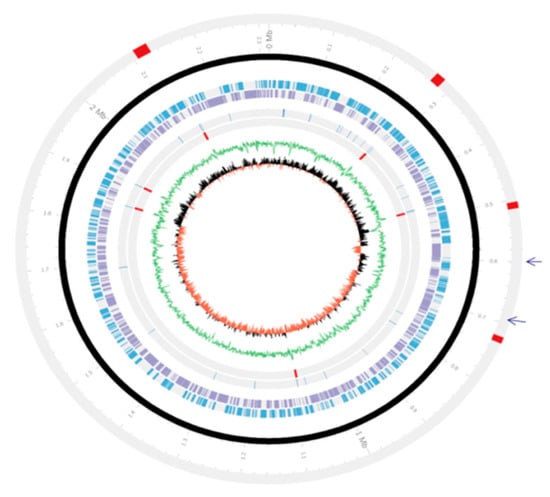
Figure 1.
Circular map of the complete genome of Pasteurella multocida strain PMTB2.1. The arrangement of genes within the chromosome of PMTB 2.1. Genomic islands = Red block in outer grey line; Circular chromosome = Black circle; Forward genes = Sky blue circle; Reverse genes = Light violet circle; tRNA = Innermost greyline; rRNA = Red line on inner grey line; GC plot = Green circle; GC skew = Red half inner circle figure, two arrows indicate the position of phages.

Table 2.
General features of the genomic organization of PMTB2.1.
The dnaA gene, designated AW43_0005 located between 101–1,456 nucleotides (nt), was selected as the first gene of the PMTB2.1 genome based on annotation of results of PGAAP. The putative replication origin (oriC) of the chromosome was identified by GC skew and DnaA-boxes immediately before the first putative coding sequence, gidA. The position of oriC is between 1,837,028–1,837,442 nt with 415 nucleotide length and closely situated to a first putative coding sequence gidA gene (1,837,443–1,839,332 nt).
3.3. Comparative Genomic Analysis and Genotyping of PMTB2.1
The PMTB2.1 genome was compared against five complete genomes of P. multocida based on different serotype and host origin and three different genera of the family Pasteurellaceae; Pasteurella sp., Actinobacillus sp. and Haemophilus sp.; while Escherichia sp. represents the family Enterobacteriaceae. Genomic distance analysis showed that PMTB2.1 is closely related to four other Pasteurella spp. with the distance of less than 0.12. The distance matrix of PMTB2.1 to PM70 is smaller (0.08) compared to that of PMTB2.1 to PM36950 (0.10, Table 3).

Table 3.
Genomic distance of PMTB2.1 compared to other selected genomes.
In contrast, synteny analysis of different P. multocida genomes showed the genome structure of PMTB2.1 resembles that of PM36950 and reveals large structural variations between PM3480 and PM70 (Figure 2).
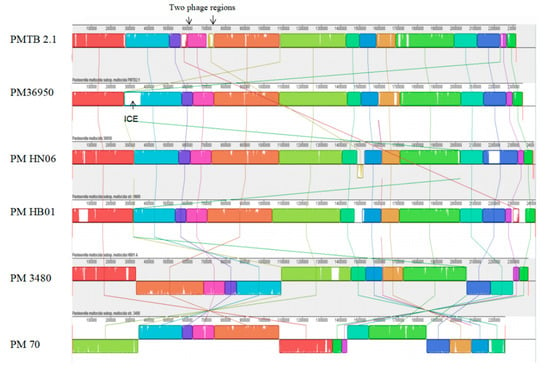
Figure 2.
Synteny analysis of P. multocida strain PMTB2.1 with other Pasteurella multocida complete genome sequences. ICE = integrative conjugative element.
The similar color box represents the similar genetic structure, white gaps representing area specific to each isolate/genome whereas cross lines represent the reorientation of similar genetic structure differently among the different P. multocida genomes.
Furthermore, a genomic comparison of six selected P. multocida complete sequences including PMTB2.1 is shown in Table 4, which revealed that P. multocida genomic size ranges between 2 and 2.4 Mbp in length, with a G+C content of between 40% to 41%. Moreover, the PMTB2.1 complete genome sequence comparison with P. multocida reference strains PM36950 and PM70 shows that the PMTB2.1 (2,315,138 bps) complete genome is approximately 34,380 kb smaller than PM36950 (2,349,518 bps) and 57,651 kb larger than PM70 (Table 4).

Table 4.
Comparison of P. multocida strain PMTB2.1 and complete genome sequences of other P. multocida of different serotypes.
A total of 20,630 protein sequences were retrieved from all nine genomes via the OrthoMCL analysis included in this study and clustered into 2831 orthologous clusters and 1025 single copy orthologous gene, which is used to determine the relatedness among Pasteurella strains from different host origins. The phylogenomic tree was constructed and rooted with E. coli, A. pleuropneumoniae and H. parasuis. The genome of PMTB2.1 was clustered with that of PM36950 and PMHB01, where both isolates are affecting bovine. They were separated from other P. multocida strains that infect avian and swine (Figure 3).
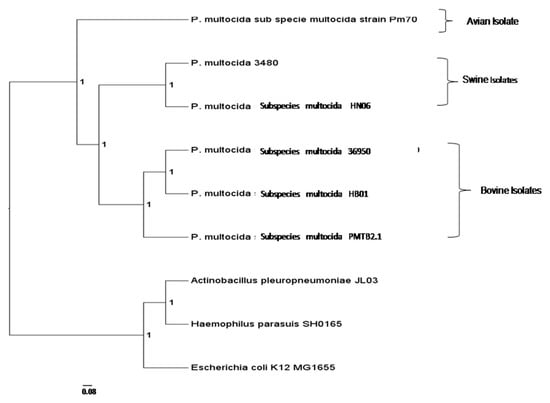
Figure 3.
Phylogenetic analysis of PMTB2.1 based on complete genome sequences. PMTB2.1 was clustered closely with other P. multocida strain 36950 and HB01 which were isolated from bovine.
The analysis of the PMTB2.1 sequence comparison based on the OrthoMCL analysis identified 282 CDs specific to P. multocida and 1,125 CDs with orthologs in all the Haemophilus SH0165 (Hi SH0165), E. coli (Ec.K12) and A. pleuropneumoniae JL03 (ActJL03) genomes (Figure 4).
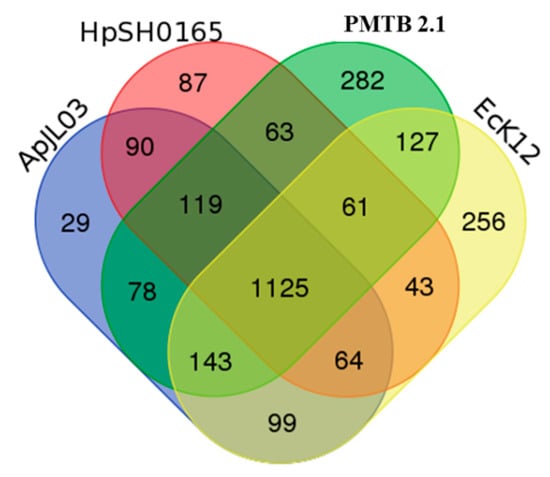
Figure 4.
Shared genes and strain-specific genes of PMTB2.1 compared to other selected genomes. strain-specific and shared genes among Pasteurella multocida strain PMTB2.1 and Haemophilus (SH0165), Escherichia coli (EcK12) and Actinobacillus pleuropneumoniae (APJL03).
Meanwhile, the genome comparison of PMTB2.1 with other Pasteurella indicated 1772 genes were common to serotypes A and D with each species showing their own strain-specific genes (Figure 5). Moreover, multi-locus sequence typing (MLST) against RIRDC scheme showed PMTB2.1 exactly matched the alleles of sequence type ST101 with 100% sequence similarity.
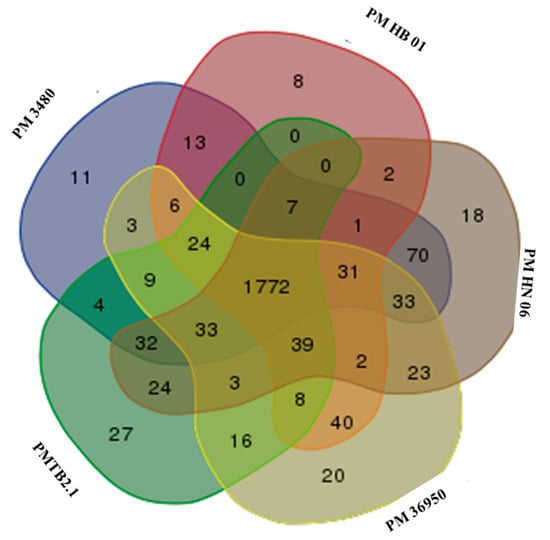
Figure 5.
Genomic comparison based on ortholog group of the genes among P. multocida. Venn diagram showing the number of shared and strain-specific genes of each of the analyzed Pasteurella genome with 1772 common genes; P. multocida strain PMHN06 and PM3480 have 18 and 11 strain-specific genes, respectively. PMTB 2.1 has more strain- specific genes, 27, as compared to PM36950 and PMHB01 which have 20 and 8 strain-specific genes, respectively.
3.4. Virulence Genes and Antibiotic Resistance Genes of PMTB2.1
Potential virulence genes and antibiotic resistance genes were identified based on a homology search against the Pathogen-Host Interaction Database (PHI-base), which identified 32 virulence-associated genes and the Virulence Factor Database (VFDB), which identified 128 genes (Supplementary Figure S1) in the PMTB2.1 genome. Among the virulence genes identified of particular importance are those which are involved in colonization such as the tad locus and capsular and LPS antigen (Supplementary Table S1). The presence of the tad locus gene in PMTB2.1 was identified from the VFDB analysis results and genome annotation at the positions of 1,010,863 to 1,011,243 (gene AW43_04840 to gene AW43_04895), which included all of the 14 genes of tad locus genes [39] (Table 5).

Table 5.
The tad locus of PMTB2.1.
The capsular (cap) locus in PMTB2.1, responsible for capsule biosynthesis and export is an approximately 14.9-kb region that encodes ten genes, genes AW43_04445 to gene AW43_04490 (911,463 to 926,403). In addition, the entire capsular sequence of the PMTB2.1, has 99% similarity to P. multocida serogroup A1 (accession no. AF067175.2). We have also identified the complete set of genes encoding enzymes for LPS biosynthesis in the genome of PMTB2.1 based on the KEGG pathway analysis. Included in these genes are those genes involved in lipid A biosynthetic process and were identified in PMTB2.1 from the VFDB analysis (Supplementary Table S1).
Furthermore, the Comprehensive Antibiotic Resistance Database (CARD) identified 12 genes (Supplementary Table S2) in PMTB2.1 that can bring about resistance against a specific group of antibiotics. Among these antibiotic resistance genes, genes AW43_04995 and AW43_08130 were similar to tetracycline resistance genes tet(35) and tet(34), respectively, whilst gene AW 43_04465 was similar to PmrE of polymyxin resistance operon (Pmr). However, the sensitivity tests against tetracycline and ampicillin (to which most Pasturella multocida strains are fully sensitive) using disk diffusion method found that PMTB2.1 was resistant to tetracycline and sensitive to ampicillin (data not shown) based on the zone size measured according to the British Society for Antimicrobial Chemotherapy (BSAC) guidelines [40].
3.5. Phage and Plasmid Sequences of PMTB2.1
Two apparently intact prophage sequences of approximately 62 kb designated as region 1 at positions 567,179–599,404 and region 2 at position 700,934–730,776 were detected in the PMTB2.1 genome (Figure 1). One of the phages (region 1) is similar to transposable Mu-like phage SfMu; however, the phage regions of PMTB2.1 were not associated with toxin-related genes, as detected in serotype D toxigenic strain of P. multocida. In addition, no plasmid sequence was detected in the complete genome sequence of PMTB2.1.
3.6. Iron Regulating Genes and Expression Profiling of Selected Iron-Regulating Genes in PMTB2.1
A total of five P. multocida strain PMTB2.1 genes were assessed for their expressions under iron-limited environment using TaqMan-based real-time PCR (qRT-PCR) analysis. The efficiency of the PCR and the co-efficient of the variance (R2) of the real-time PCR assays were between 90%–105% and ≥ 0.98, respectively (Supplementary Table S3). PMTB2.1 grown in iron-limited environment undergo significant changes (p < 0.05) in the expression profile of all of the four selected iron-regulating genes namely fecE, yfeA, fbpB and fur when compared to control (normal broth). Results of this study reflect that iron-limited environment has a significant effect on the expression profile of iron regulating-genes (p < 0.05) and all evaluated genes act differently in response to an iron limitation in the media (Table 6).

Table 6.
Relative fold change expressions of selected genes of PMTB2.1 at different time points.
The fur gene, a universal transcriptional regulator of iron-regulating genes, showed a significant increase in the relative fold changes expression in the iron-limited environment at all time points (2–5 fold) and started to be expressed early at 30 min of treatment (Table 6). The iron-regulating gene fecE (AW43_06150) of the FecABCDE periplasmic binding protein-dependent iron transport systems, was highly expressed (281 fold) after 30 min of treatment, which started to decrease remarkably, from 4.7 to −1.5-fold at 60 and 120 min, respectively, with elevated expression of other iron-regulating genes namely fbpB (AW43_01110) and yfeA (AW43_01910) of the FbpABC and yfeABCD system which are outer-membrane independent ABC iron transport system. The fbpB gene was significantly expressed at a higher level of relative fold change (25 fold) at 60 min (Table 6), whereas the yfeA gene was significantly expressed with the highest relative fold change expression (42 fold) at 120 min (Table 6). On the other hand, the relative fold change expression of nanA (sialidase) in the treated samples collected at all time points were low, that is, less than 2-fold (Table 6) that showed the treatment has no significant effect on the nanA gene expression.
4. Discussion
4.1. Complete Genome Sequencing
Long-read sequencing technologies such as PacBio RS can resolve repeat sequences that are found in most microbial genomes [21]. Hence, in this study, de novo assembly of the PMTB2.1 generated a near-complete genome with gaps that were closed by Sanger sequencing and a complete circular genome (2,315,138 bps) was produced based on the reference genome sequence of P. multocida strain PM36950. The genomic size of single circular genome sequences of P. multocida belonging to different serotypes available on NCBI, ranges from 2 and 2.4 Mbp with a G+C content of 40%–41% [16], which is also reflected in the genomic comparison of six selected P. multocida complete sequences including PMTB2.1 (Table 4).
4.2. Pathogenomics Analysis
Synteny analysis based on similarity of the genetic structure of different P. multocida genomes (Figure 2) revealed subtle differences in genetic structures indicating the dynamics of frequent gene transfer events among the pool of P. multocida strains. However, PM3480 and PM70 exhibited exceptionally large structural variations, as PM70 is an avian isolate (isolated from chicken) and PM3480 is a swine isolate, whereas PM36950, HB01 and PMTB2.1 are of bovine origin. However, the genomic contents of PMTB2.1 resemble more of the PM70 strain with a distance matrix of 0.08, while the genomic structure resembles more of the PM36950 strain with a distance matrix of 0.10, as compared to other Pasteurella strain (Table 3 and Figure 2). Furthermore, genomic distance analysis showed that PMTB2.1 is closely related to the other four Pasteurella spp. causing it to be grouped in the heatmap analysis with the distance of less than 0.12 (Figure 6).
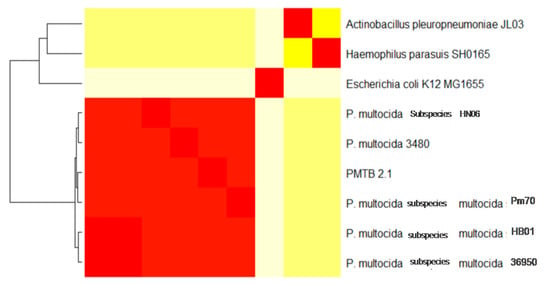
Figure 6.
Genomic distance based on heat map analysis of PMTB2.1 compared to other nine selected genomes. PMTB2.1 is grouped together with other P. multocida with genomic distance less than 0.13 (red color box), while Haemophilus and Actinobacillus are groups together with similar genomic distance 0.72 (lemon yellow color box) and E. coli with genomic distance 0.80 (off-white color box) Red color box in off-white E. coli bar indicating genomic region similar to Pasteurella, same for Haemophilus and Actinobacillus.
The genome comparison of PMTB2.1 with five Pasteurella strains identified 1772 common genes among serotypes A and D (Figure 5). However, strain-specific genes belonging to each serotype and strains were present in all P. multocida genomes included in the analysis. P. multocida strain PMHN06 and PM3480 were found to have 18 and 11 strain-specific genes, respectively. As each strain has their own strain-specific genes, PMTB 2.1 has more strain-specific genes (27 genes) compared to another bovine serotype A, PM36950 with 8 and PMHB01 with 20 strain-specific genes; this probably reflects strain specificity of different Pasteurella even from the same serogroup of the similar host (bovine).
Strain-specific integrative conjugative element (ICE), ICEPmu1 of 82 kbp found to be present only in PM36950 [13], was absent in PMTB2.1 and PM70 (Figure 7). Meanwhile, 62,069 kb region (two prophage regions) was found to be present only in PMTB2.1 but was absent in PM36950. Moreover, although the PMTB2.1 genetic content more resembled PM70 compared to other PM strains, PMTB2.1 was found to have an extra 15-kb region (AW43_01320–AW43_01390), which was not present in the PM70 genome (Figure 7).
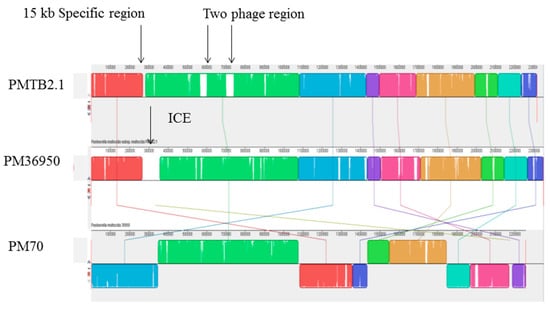
Figure 7.
Synteny analysis (co-linearity comparison) between PMTB2.1, PM70 and PM36950. Blocks with similar color represent similar sequences, while block with similar colors but with different orientation as in PM70 exhibit structural variation. PMTB2.1 specific two phage region and ICE region of PM36950 is represented by white color or gaps between nucleotide blocks indicated with arrows.
Remarkably, the number of orthologous genes/proteins shared between the bovine strains PMTB2.1 and the avian strain PM70 (65 genes) was also greater than those shared between the two bovine isolates PMTB2.1 and PM36950 (only 22 genes). In addition, the numbers of virulence-associated genes identified in the complete genome sequence of PMTB2.1 are 128, 32 and 12 based on the VFDB, PHI-base and CARD databases, respectively, are higher compared to previous studies [12]. One of the possible reasons is due to the high quality of genome sequences produced in this study.
Genome sequence comparison of P. multocida strain PMTB2.1 with Haemophilus SH0165, E. coli (Ec.K12) and A. pleuropneumoniae JL03 genomes identified 1125 ortholog CDs (Figure 4), reflecting overall close relationship and support the classification of these bacteria in the gamma subdivision of the Proteobacteria. Interestingly, the phylogenetic relationship within the Pasteurellaceae family based on 16S rRNA genes suggested that the Pasteurella species isolates are more closely related to the isolates of the genus Haemophilus [4], however the ortholog gene analysis shows that P. multocida PMTB2.1 shared a greater number of ortholog proteins with A. pleuropneumoniae JL03 (78 genes) compared to H. parasuis SH0165 (63 genes, Figure 4).
4.3. Phylogenomics Analysis
A total of 1025 single copy orthologous genes were used to determine the relatedness among different Pasteurella strains from different host origin that included PM70 avian isolate [12], PM3480 (unpublished) and HN06 [15] swine isolates and PM36950 [13], HB01 [14] and PMTB2.1 bovine isolates. The phylogenomic tree was constructed and rooted with E. coli, A. pleuropneumoniae and H. parasuis. The genome of PMTB2.1 was clustered with that of P. multocida strain PM36950 and HB01, where both isolates are of bovine origin (Figure 3). They were separated from other P. multocida strains that infect avian (PM70) and swine (P. multocida strain 3480 and HN06) and are distant from Actinobacillus and Haemophilus.
Hence, based on the complete genome sequences, a clear phylogenomic relatedness of P. multocida and its host can be established that was not possible in previous studies based on single nucleotide polymorphisms (SNPs) analysis [16] or MLST analysis [34]. In addition, a recent study based on genome-wide SNPs, at positions that were conserved across different P. multocida strains namely 17 HS-associated strains and 5-fowl cholera isolates including PM70 and X73, the swine isolates HN06 and 3480 and the bovine respiratory disease isolate PM36950, is still unable to show a clear correlation between strain relatedness and disease type other than for the HS strains [41]. This reflects that the complete genome sequence analysis proved useful to establish phylogenomic relatedness among the complete genomes of P. multocida.
4.4. Antibiotic Resistance Genes
Pasteurella multocida is an ampicillin-sensitive Gram-negative bacterium. Antimicrobial susceptibility study of P. multocida (108 avian isolates) by agar disc diffusion method showed that more than 90% of these isolates exhibited sensitivity to ampicillin [42]. However, it has been documented that P. multocida has developed resistance to commonly used antimicrobial agents. The antimicrobial sensitivity pattern of 56 P. multocida strains isolated from poultry reported exceptional resistance to sulphonamides, tetracyclines, first-generation quinolones and aminoglycosides [43]. The presence of tet(H) gene on the chromosome that belongs to the efflux-mediated class of gene for tetracycline resistance was detected in an avian strain of P. multocida [44] and later in other strains of P. multocida and P. hemolytic [45].
Pasteurella multocida strain PMTB2.1 possess genes tet(34) and gene tet(35) that are predicted to encode resistance to tetracycline. Tetracycline resistance is most often due to genes, which codes for the efflux of tetracycline or for a protein that protects the bacterial ribosome from the action of tetracycline. The genes tet(35) and tet(34) are mostly found in Gram-negative bacteria such as Vibrio, Pseudomonas, Serratia and Stenotrophomonas [46]. Moreover, the integrative conjugative element (ICEPmu1) of PM36950 also contains genes conferring resistance to tetracycline. Along with the gene responsible for tetracycline resistance, another gene AW43_04465 similar to PmrE has been identified in PMTB2.1. The protein product of PmrE gene is required for the synthesis of 4-amino-4-deoxy-l-arabinose (Ara4N) of lipid A, in Gram-negative bacteria. This, in turn, allows the bacterial cell to resist the antimicrobial activity of cationic antimicrobial peptides and antibiotic such as polymyxin. Resistance to polymyxin and cationic antimicrobial peptides in E. coli and Salmonella typhimurium is acquired by similar phenomenon [47].
4.5. Tad Locus Genes of PMTB2.1
The tad locus is generally a 14-gene locus (flp1-flp2-tadV-rcpCABtadZABCDEFG) (Table 5). The Tad locus cluster is widely present in many bacterial and archaeal species, encoding for a type IVb pilus and represents a subtype of type II secretion. The tad genes encode the machinery that is required for the assembly of adhesive Flp (fimbrial low-molecular-weight protein) pili [40]. Tad locus genes in PMTB2.1 are similar (51%–87% amino acid identity) to Aggregatibacter actinomycetemcomitans strain CU1000 tad locus gene [48]. Complete genome analysis of P. multocida strain PMTB2.1 was able to detect the complete tad loci in P. multocida genome with improved annotation (Supplementary Figure S3, Table 5) where genes such as flp2 was annotated as a hypothetical protein in previous studies but was annotated as pilus assembly gene in PMTB2.1. In addition, comparison of tad locus among different strains of P. multocida [14] found that the tad locus in HB01, PM70 and PM36950 is composed of only 13 genes and missing the flp2. The tad locus of strain 36950 is more truncated than other P. multocida due to the lack of rcpC gene. However, the tad locus in PMTB2.1 comprises all 14 genes including the flp2 and rcpC genes.
Tad locus is essential for biofilm formation, colonization and pathogenesis in many members of the family Pasteurellaceae including the genera Actinobacillus, Haemophilus and Pasteurella [49]. The function and features of the products of several tad locus genes have been reviewed in various bacteria and it has been reported that at least 12 and probably 13 (excluding flp2), of the tad genes, are required for all adherence-related phenotypes [39], whereas rcpB and tadV are essential in adherence [48,49]. Studies have also revealed the flp1 encodes the Flp1 prepilin [50] and tadV encodes a protease that processes the Flp1 prepilin to the mature Flp1 pilin for assembly of pili. The product of rcpA has the properties of an outer membrane pore (secretin) and the product of rcpB is an outer-membrane protein that may gate the pore [48]. The tadZ encodes a protein that may localize the tad secretion machine to a pole, whereas the tadA product is an ATPase [51] and the tadE and tadF products are “pseudopilins,” whose functions are not known; but the pseudopilins are processed by tadV in the same way as the prepilin [49].
The presence of a homologous tad locus in the genomes of all five selected Pasteurella strains (PM70, 3480, HN06, 36950 and HB01) along with PMTB2.1, suggested that type IVb pili may play a role as a host colonization factor in the respiratory tract of ruminants and birds. Moreover, the presence of tad gene in P. multocida supports the idea of a pili-mediated host colonization and persistence mechanism for Pasteurella. In addition, tenacious adherence may be a property of Pasteurella because it must be able to colonize in the presence of extensive normal flora. Therefore, the strong phenotype of tenacious adherence may be a special property of the tad locus of Pasteurella. The analysis of tad loci in P. multocida genome is critically important to determine and understand the functions of the individual tad genes and whether and how the various proteins encoded by a tad locus acted to colonize a specific niche and the importance of the tad genes for P. multocida and other prokaryotes.
4.6. The Capsular and LPS Genes of PMTB2.1
The pathogenicity of P. multocida is associated with various virulence factors such as the capsule and the LPS [16]. NCBI blast revealed that the capsular region sequences of PMTB2.1 are similar to serotype A capsular region [52] with 99% identity. In addition, the cap locus operon comprises four genes, (AW43_04445 to AW43_04460) is associated with capsule biosynthesis and is similar to hyaEDCB, an operon in P. multocida serotype A:1. Downstream of hyaEDCB is another operon of four genes hexDCBA (AW43_04475 to AW43_04490), which form a complex involved in the export of capsule to the surface and three of these genes have a high level of similarity to hexDCA of Haemophilus influenzae strain 1007 while one gene is highly similar to Ctrc gene of Neisseria meningitidis strain MC58. Similar results have been obtained from a study carried out on bovine isolate HB01, a capsular serotype A strain, which has identified 10 genes within a region of approximately 14.9 kb [14].
Pasteurella multocida LPS plays a critical role in disease immunopathogenesis [53,54]. The typical structure of LPS produced by most P. multocida is primarily composed of lipid A, a highly hydrophobic lipid moiety and immunoreactive oligosaccharide that triggers defense-related responses and causes Gram-negative sepsis [14]. The biosynthesis of Kdo2-lipid A, the LPS substructure, involves nine enzymatic steps. However, not all Gram-negative bacteria have all nine enzymes. Previous studies have shown that most of the genes encodes for lipid A such as lpxA, lpxB, lpxC, lpxD, lpxH, lpxK, kdsA kdsB kdtA, lpxM, kdsC and htrB are conserved among Actinobacillus and Haemophilus [54] and are also highly similar among Pasteurella species [14]. The genome of PMTB2.1 based on VFDB analysis encode for lpxA, lpxB, lpxC, lpxD, lpxK, kdsA, kdsA, htrB, kdtA and kdkA; whereas recently sequenced genome of P. multocida strain HB01 was reported to have two genes namely kdtA and kdkA [14]. On the other hand, it has been reported that some Gram-negative bacteria have genes only for the first four enzymes lpxA, lpxC, lpxD and lpxB [55].
4.7. Bacteriophages in Pasteurella and Prophage Sequence in PMTB 2.1
Phage-related sequences have been reported from P. multocida serotype A strain, such as a temperate transducing phage F108 [56] and porcine toxigenic serotype D strain [15,57]. However, the only phage isolated from toxigenic strains were found to carry the gene for PMT toxin, a major virulence factor of atrophic rhinitis in pigs, which belongs to the Siphoviridae family of bacteriophage that includes lambda-like phage [57]. Recently, three regions of prophage sequences were detected in a bovine virulent isolate of P. multocida strain HB01 by complete genome sequencing [14]. In this study, a transposable temperate Mu-like prophage sequence similar (67%) to SfMu-like bacteriophage belonging to the family Myoviridae [58] was identified in Pasteurella strain PMTB 2.1. Two regions designated as region 1 and region 2 with prophage sequences were detected in PMTB2.1 using PHAST database (Supplementary Figure S4). Region 1 was predicted to be similar to SfMu accession no. NC_027382, which was isolated from Shigella flexneri [58], while region 2 did not have any significant similarity with the previously determined complete phage genome sequence.
4.8. Iron Regulating Genes and Expression Profiling of Selected Iron-Regulating Genes in PMTB2.1
Iron acts as a co-factor or prosthetic group in several essential enzymes [59] and is needed for amino acid, pyrimidine, DNA biosynthesis and participates in electron transport [60]. Results of this study reflect that iron-limited environment has a significant effect on the expression profile of iron-regulating genes (p < 0.05) and all evaluated genes act differently in response to an iron limitation in the media. The highest expression of fecE gene (281 fold) at an early stage of treatment (30 min, Table 6) suggests that in the initial stage of growth, Pasteurella utilizes the iron acquiring genes, which facilitate the direct import of ferrous iron to scavenge iron from the environment under iron-limited conditions. Utilization of the outer-membrane proteins system in iron acquisition at an early stage of infection may play an important role in the development of a vaccine against infection with P. multocida since many outer-membrane proteins involved in iron regulation are strong antigens and some of them have been used in protective experiments against bacterial pathogens [61].
Furthermore, the downregulation of relative expression of fecE gene in association with the increase in relative expression of fbpB and yfeA genes (Table 6) indicates that Pasteurella controls their iron requirements in response to iron availability by downregulating the expression of iron proteins during iron-restricted growth. This idea is supported by the high relative fold change expression of fur at 180 min (5.4 fold), when PMTB2.1 has started to decrease the expressions of fecE and fbpB, with only one gene yfeA being upregulated. The yfeABCD and FbpABC encodes an ABC transport system, whose expression is iron and Fur regulated. The fur gene has shown significantly higher relative fold changes expression at all-time points (2‒5 fold) (Table 6) and start to express early at 30 min of treatment (4 fold).These results show that like in other Gram-negative bacterial cells such as E. coli, the fur gene in P. multocida is significantly involved in the regulation of iron-regulating genes and controls the intracellular concentration of iron, which is toxic to the bacterial cell in high concentration [62]. Almost similar results were reported by May et al. [12] when they examined the pattern of gene expression of known iron-regulating genes of P. multocida strain PM70 under iron-depleted condition with microarray based study, with an overall increase in expression of yfeA and fecE genes at all time points tested (13).
Moreover, significantly higher relative fold change (25 fold) of the fbpB gene (ortholog of fbpB of FbpABC system) at 60 min is probably associated with the ability of P. multocida to directly use host iron complexes such as heme, hemoglobin, transferrin and lactoferrin. This suggestion is based on the previous study that showed that the FbpABC system in Neisseria spp. is involved in transporting iron, delivered by transferrin and lactoferrin across the cell membrane without the help of periplasmic protein and siderophore [63]. Besides these genes, the increase in relative expression (26 fold) of periplasmic chelated iron-binding protein gene, yfeA at an early time point (30 min) and highest relative expression (42 fold) at 120 min reflect the use of multiple iron acquiring systems in P. multocida as yfeA gene of PMTB2.1 is similar to one of the Yersinia pestis YfeABC iron transport system gene yfeA, [64].
The expression of nanA (sialidase) gene of PMTB2.1 was also assessed under iron-limited environment as this gene is involved in colonization. A previous study has shown that nanA encodes for sialidases that exhibits glycolytic activity on mucin and release terminal sialic acid residues that can then be used as a bacterial carbon source [65]. Compared to iron-regulating genes, the relative expression of nanA was low (Table 6), which shows that probably iron scarcity does not exhibit a significant effect on the colonization of P. multocida facilitated by the nanA gene. Although P. multocida strains producing sialidase have been reported, their role in the virulence of P. multocida has yet to be established [59,65].
5. Conclusions
Improved complete genome sequence analysis of P. multocida strain PMTB2.1 has provided valuable insights on the genomic structure with detection and identification of key important genes namely antibiotic resistance genes, a novel transposable Mu phage and various virulence genes including the Tad locus. In addition, clear phylogenomic relatedness for P. multocida can be established based on the complete genome analysis. Findings from this study has open up further studies on elucidating the mechanisms behind the molecular pathogenesis of P. multocida associated diseases in animals.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/10/2/81/s1, Table S1: List of virulence genes including the tad locus, LPS and capsular genes identified based on VFDB database; Table S2: Genes detected in PMTB2.1 based on CARD; Table S3: PCR efficiency and co-efficient of the variance of real-time PCR assays; Figure S1: Work flow for the identification of virulence-associated genes and antibiotic resistance gene in P. multocida strain PMTB2.1; Figure S2: Details of prophage sequence in PMTB2.1. The complete genome sequence of P. multocida strain PMTB2.1 was deposited in the National Center for Biotechnology Information (NCBI) accession number CP007205.2. The complete genome sequence of PMTB2.1 and eight selected genomes for comparative analysis is available on NCBI and can be accessed using the accession number mentioned in Table 4.
Author Contributions
S.J., F.J.F.A., Z.Z., N.M.I., Y.C.T., W.Y.Y. and A.R.O. conceived the research. Pasteurella multocida strain PMTB2.1 culture maintenance, sample handling, DNA and RNA extraction, PCR, real-time PCR and data analysis were conducted by S.J. and supervised by A.R.O. Sequencing data acquisition, data management and data analysis and interpretation, pathway analysis and comparative genomic were performed by S.J. with assistance from Y.C.T., W.Y.Y., Y.H.Y., D.A.S. and overseen by A.R.O. The manuscript was written by S.J., F.J.F.A., Z.Z., N.M.I., Y.C.T., W.Y.Y. and A.R.O. The manuscript was finalized and submitted by S.J.; all authors reviewed and approved the submitted manuscript.
Funding
This work was fully supported including the design of the current study, collection and analysis of the data by the FRGS Grant No. 02-02-13-1371FR from the Ministry of Higher Education, the Government of Malaysia.
Conflicts of Interest
The authors declare that they have no competing interest. Although we have used commercial software in this study supplied by the manufacturers, they had no influence on the experiments performed or on the results reported here.
References
- Kuhnert, P.; Christensen, H. Pasteurellaceae: Biology, Genomics and Molecular Aspects; Academic Press: Caister, UK, 2008; pp. 34–39. [Google Scholar]
- Carter, G.R. A haemagglutination test for the identification of serological types. Am. J. Vet. Res. 1955, 16, 481–484. [Google Scholar] [PubMed]
- Heddleston, K.L.; Gallagher, J.E.; Rebers, P.A. Fowl cholera: Gel diffusion precipitin test for serotyping Pasteruella multocida from avian species. Avian Dis. 1972, 16, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.A.; Ho, M. Pasteurella multocida: From zoonosis to cellular microbiology. Clin. Microbiol. Rev. 2013, 26, 631–655. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.D.; Adler, B. The capsule is a virulence determinant in pathogenesis of Pasteurella multocida M1404 (B:2). Infect. Immun. 2000, 68, 3463–3468. [Google Scholar] [CrossRef]
- De Alwis, M.C.L. Hemorrhagic Septicemia, Aciar Monograph Series; ACIAR: Canberra, Australia, 1999. [Google Scholar]
- OIE. Haemorrhagic Septicaemia. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; OIE: Paris, France, 2012; pp. 1–13. Available online: www.oie.int/en/international-standard-setting/terrestrial-manual/ (accessed on 11 December 2014).
- Dabo, S.M.; Taylor, J.D.; Confer, A.W. Pasteurella multocida and bovine respiratory disease. Anim. Health Res. Rev. 2007, 8, 129–150. [Google Scholar] [CrossRef]
- Praveena, P.E.; Periasamy, S.; Kumar, A.A.; Singh, N. Pathology of experimental infection by Pasteurella multocida serotype A:1. Vet. Pathol. 2014, 51, 1109–1112. [Google Scholar] [CrossRef]
- Shivachandra, S.B.; Viswas, K.N.; Kumar, A.A. A review of hemorrhagic septicemia in cattle and buffalo. Anim. Health Res. Rev. 2011, 12, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Zamri-Saad, M.; Annas, S. Vaccination against hemorrhagic septicemia of bovines: A review. Pak. Vet. J. 2015, 36, 1–5. [Google Scholar] [CrossRef]
- May, B.J.; Zhang, Q.; Li, L.L.; Paustian, M.L.; Whittam, T.S.; Kapur, V. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 2001, 98, 3460–3465. [Google Scholar] [CrossRef]
- Michael, G.B.; Kadlec, K.; Sweeney, M.T.; Brzuszkiewicz, E.; Liesegang, H.; Daniel, R.; Murray, R.W.; Watts, J.L.; Schwarz, S. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: Structure and transfer. J. Antimicrob. Chemother. 2012, 67, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Liang, W.; Liu, W.; Wu, B.; Tang, B.; Tan, C.; Zhou, R.; Chen, H. Genomic characterization of Pasteurella multocida HB01, a serotype A bovine isolate from China. Gene 2016, 581, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yang, M.; Xu, Z.; Zheng, H.; Liang, W.; Zhou, R.; Wu, B.; Chen, H. Complete genome sequence of Pasteurella multocida HN06, a toxigenic strain of serogroup D. J. Bacteriol. 2012, 194, 3292–3293. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.D.; Seemann, T.; Adler, B.; Harper, M. Pathogenomics of Pasteurella multocida. Curr. Top. Microbiol. Immunol. 2012, 361, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Paustian, M.L.; May, B.J.; Kapur, V. Transcriptional response of Pasteurella multocida to nutrient limitation. J. Bacteriol. 2002, 184, 3734–3739. [Google Scholar] [CrossRef] [PubMed]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 2012, 13, 341. [Google Scholar] [CrossRef] [PubMed]
- Townsend, K.M.; Frost, A.J.; Lee, C.W.; John, M.; Dawkins, H.J.S.; Papadimitriou, J.M. Development of PCR assays for species- and type-specific identification of Pasteurella multocida isolates. J. Clin. Microbiol. 1998, 36, 1096–1100. [Google Scholar] [PubMed]
- Jabeen, S.; Yap, H.Y.; Jesse Abdullah, F.F.; Zakaria, Z.; Mat Isa, N.; Tan, Y.C.; Hoh, C.C.; Yee, W.Y.; Omar, A.R. Complete genome sequence of Pasteurella multocida serotype A strain PMTB2.1 isolated from buffaloes died of septicemia in Malaysia. Genome Announc. 2017, 5, e01190-17. [Google Scholar] [CrossRef] [PubMed]
- English, A.C.; Richards, S.; Han, Y.; Wang, M.; Vee, V.; Qu, J.; Qin, X.; Muzny, D.M.; Reid, J.G.; Worley, K.C.; et al. Mind the gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE 2012, 7, e47768. [Google Scholar] [CrossRef]
- Besemer, J.; Borodovsky, M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005, 33, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Got, S.; Garcia-gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Mackiewicz, P.; Zakrzewska-Czerwinska, J.; Zawilak, A.; Dudek, M.R.; Cebrat, S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 2004, 32, 3781–3791. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Li, C.; Han, S.; Lopez-Baltazar, J.; Zhang, X.; Wang, X. Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci. Rep. 2016, 6, 35245. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, B.K.; Laird, M.R.; Shay, J.A.; Winsor, G.L.; Lo, R.; Nizam, F.; Pereira, S.K.; Waglechner, N.; McArthur, A.G.; Langille, M.G.I.; et al. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 2015, 43, 104–108. [Google Scholar] [CrossRef]
- Winnenburg, R.; Baldwin, T.K.; Urban, M.; Rawlings, C.; Kohler, J.; Hammond-Kosack, K.E. PHI-base: A new database for pathogen host interactions. Nucleic Acids Res. 2006, 34, D459–D464. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB: Hierarchical and refined dataset for big data analysis 10-years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef]
- Subaaharan, S.; Blackall, L.L.; Blackall, P.J. Development of a multi-locus sequence typing scheme for avian isolates of Pasteurella multocida. Vet. Microbiol. 2010, 141, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T.; Batzoglou, S.; Zhong, Y. Progressive mauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Creevey, C.J.; Muller, J.; Doerks, T.; Thompson, J.D.; Arendt, D.; Bork, P. Identifying single copy orthologs in metazoa. PLoS Comput. Biol. 2011, 7, e1002269. [Google Scholar] [CrossRef] [PubMed]
- Heydari, M.; Miclotte, G.; Demeester, P.; Van de peer, Y.; Fostier, J. Evaluation of the impact of Illumina error correction tools on de novo genome assembly. BMC Bioinformatics 2017, 18, 374. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaff, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Figurski, D.H.; Fine, D.H.; Perez-Cheeks, B.A.; Grosso, V.W.; Karin, E.; Kram, J.H.; Xu, K.; Hedhli, J. Targeted mutagenesis in the study of the tight adherence (tad) locus of Aggregatibacter actinomycetemcomitans. In Genetic Manipulation of DNA and Protein—Examples from Current Research; Figurski, D., Ed.; InTech Publisher: London, UK, 2013; pp. 953–978. [Google Scholar] [CrossRef]
- Wootton, M. BSAC Methods for Antimicrobial Susceptibility Testing; Version 12; BSAC: Birmingham, UK, 2013. [Google Scholar]
- Moustafa, A.M.; Seemann, T.; Gladman, S.; Adler, B.; Harper, M.; Boyce, J.D.; Bennett, M.D. Comparative genomic analysis of Asian haemorrhagic septicaemia-associated strains of Pasteurella multocida identifies more than 90 haemorrhagic septicaemia- specific genes. PLoS ONE 2015, 10, e130296. [Google Scholar] [CrossRef] [PubMed]
- Habrun, B.; Kompes, G.; Cvetni, Ž.; Špi, S.; Beni, M.; Mitak, M. Antimicrobial sensitivity of Escherichia coli, Salmonella spp., Pasteurella multocida, Streptococcus suis and Actinobacillus pleuropneumoniae isolated from diagnostic samples from large pig breeding farms in Croatia. Vet. Arh. 2010, 80, 571–583. [Google Scholar]
- Sellyei, B.; Varga, Z.; Szentesi-Samu, K.; Kaszanyitzky, E.; Magyar, T. Antimicrobial susceptibility of Pasteurella multocida isolated from swine and poultry. Acta Vet. Hung. 2009, 57, 357–367. [Google Scholar] [CrossRef]
- Hansen, L.M.; McMurry, L.M.; Levy, S.B.; Hirsh, D.C. A new tetracycline resistance determinant, tet(H), from Pasteurella multocida specifying active efflux of tetracycline. Antimicrob. Agents Chemother. 1993, 37, 2699–2705. [Google Scholar] [CrossRef]
- Hansen, L.M.; Blanchard, P.C.; Hirsh, D.C. Distribution of tet(H) among Pasteurella isolates from the United States and Canada. Antimicrob. Agents Chemother. 1996, 40, 1558–1560. [Google Scholar] [CrossRef]
- Miranda, C.D.; Kehrenberg, C.; Ulep, C.; Schwarz, S.; Roberts, M.C. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob. Agents Chemother. 2003, 47, 883–888. [Google Scholar] [CrossRef]
- Falagas, M.E.; Rafailidis, P.I.; Matthaiou, D.K. Resistance to polymyxins: Mechanisms, frequency and treatment options. Drug Resist. Updates 2010, 13, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Clock, S.A.; Planet, P.J.; Perez, B.A.; Figureurski, D.H. Outer membrane components of the Tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 2008, 190, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Tomich, M.; Planet, P.J.; Figurski, D.H. The tad locus: Postcards from the widespread colonization island. Nat. Rev. Microbiol. 2007, 5, 363–375. [Google Scholar] [CrossRef]
- Kachlany, S.C.; Planet, P.J.; Desalle, R.; Fine, D.H.; Figurski, D.H.; Kaplan, J.B. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 2001, 40, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, M.K.; Kachlany, S.C.; Fine, D.H.; Figurski, D.H. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 2001, 20, 5927–5936. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.Y.; Zhang, Y.; Adler, B. The capsule biosynthetic locus of Pasteurella multocida A:1. FEMS Microbiol. Lett. 1998, 166, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Faez, F.J.A.; Lawan, A.; Abdinasir, Y.O.; Zunita, Z.; Rasedee, A. Clinicopathological responses of calves associated with infection of Pasteurella multocida type B and the bacterial lipopolysaccharide and outer membrane protein immunogens. Int. J. Anim. Vet. Adv. 2013, 5, 190–198. [Google Scholar]
- Xu, Z.; Yue, M.; Zhou, R.; Jin, Q.; Fan, Y.; Bei, W.; Chen, H. Genomic characterization of Haemophilus parasuis SH0165, a highly virulent strain of serovar 5 prevalent in China. PLoS ONE 2011, 6, e19631. [Google Scholar] [CrossRef] [PubMed]
- Opiyo, S.O.; Pardy, R.L.; Moriyama, H.; Moriyama, E.N. Evolution of the Kdo2-lipid A biosynthesis in bacteria. BMC Evol. Biol. 2010, 10, 362. [Google Scholar] [CrossRef] [PubMed]
- Campoy, S.; Aranda, J.; Alvarez, G.; Barbe, J.; Llagostera, M. Isolation and sequencing of a temperate transducing phage for Pasteurella multocida. Appl. Environ. Microbiol. 2006, 72, 3154–3160. [Google Scholar] [CrossRef] [PubMed]
- Pullinger, G.D.; Bevir, T.; Lax, A.J. The Pasteurella multocida toxin is encoded within a lysogenic bacteriophage. Mol. Microbiol. 2004, 51, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Jakhetia, R.; Verma, N.K. Identification and molecular characterisation of a novel Mu-like bacteriophage, SfMu, of Shigella flexneri. PLoS ONE 2015, 10, e0124053. [Google Scholar] [CrossRef] [PubMed]
- Harper, M.; Boyce, J.D.; Adler, B. Pasteurella multocida pathogenesis: 125 years after Pasteur. FEMS Microbiol. Lett. 2006, 265, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, D.; Henestrosa, A.R.; Badiola, I.; Saco, M.; Perez Rozas, D.A.M.; Campoy Frawley, E.R.; Fang, F.C. The ins and outs of bacterial iron metabolism. Mol. Microbiol. 2014, 93, 609–616. [Google Scholar] [CrossRef]
- Hatfaludi, T.; Al-Hasani, K.; Boyce, J.D.; Adler, B. Outer membrane proteins of Pasteurella multocida. Vet. Microbiol. 2010, 144, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Adhikari, P.; Berish, S.A.; Nowalk, A.J.; Veraldi, K.L.; Morse, S.A.; Mietzner, T.A. The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J. Bacteriol. 1996, 178, 2145–2149. [Google Scholar] [CrossRef]
- Bearden, S.W.; Perry, R.D. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 1999, 32, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.; Mizan, S.; Quist, C.; Schroder, P.; Juneau, M.; Dawe, D.; Ritchie, B.; Lee, M.D. Serological response to Pasteurella multocida NanH sialidase in persistently colonized rabbits. Clin. Diagn. Lab. Immunol. 2004, 11, 825–834. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).