Global Vectors Representation of Protein Sequences and Its Application for Predicting Self-Interacting Proteins with Multi-Grained Cascade Forest Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Benchmark Datasets Preparation
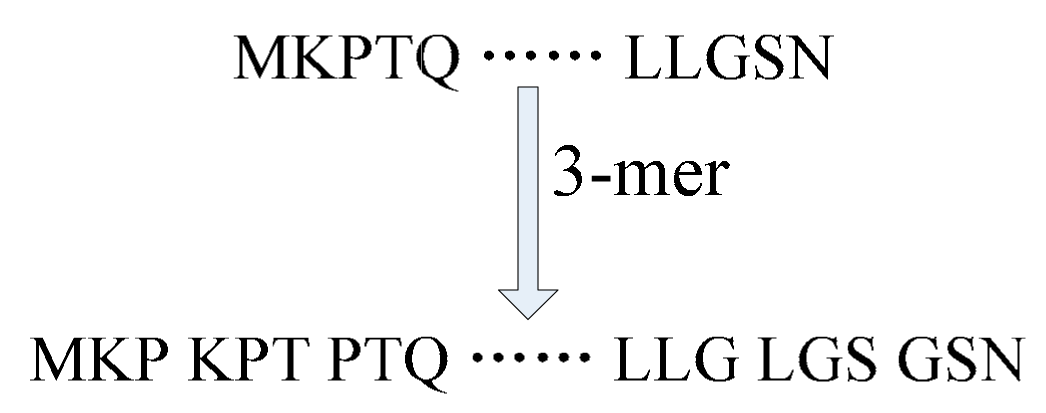
2.2. De Novo Assembly Protein Sequences
2.3. Global Vectors Representation of Protein Sequences
2.4. Multi-Grained Cascade Forest
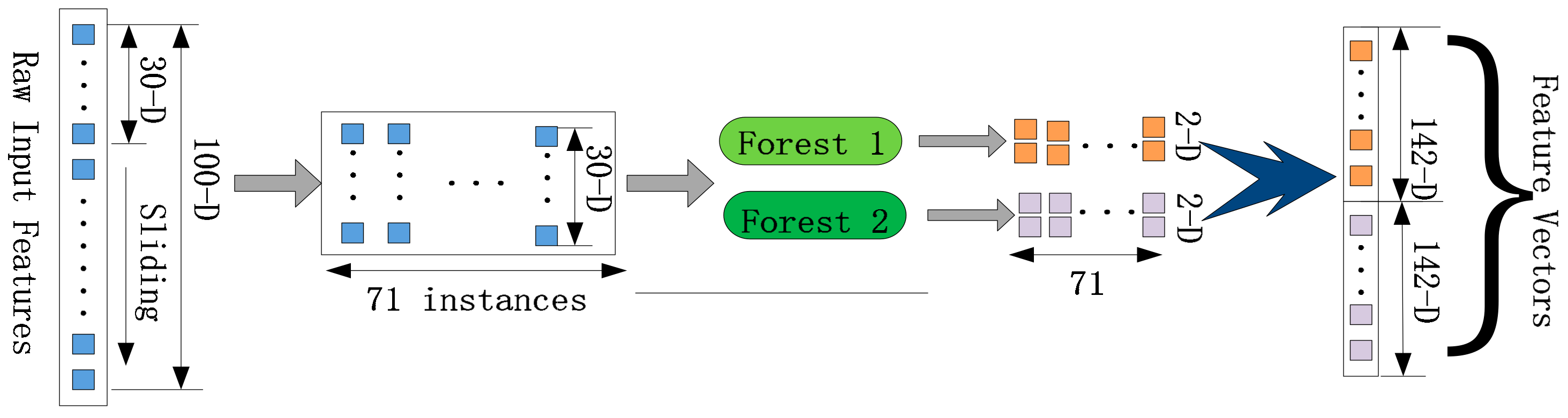
2.4.1. Multi-Grained Scanning
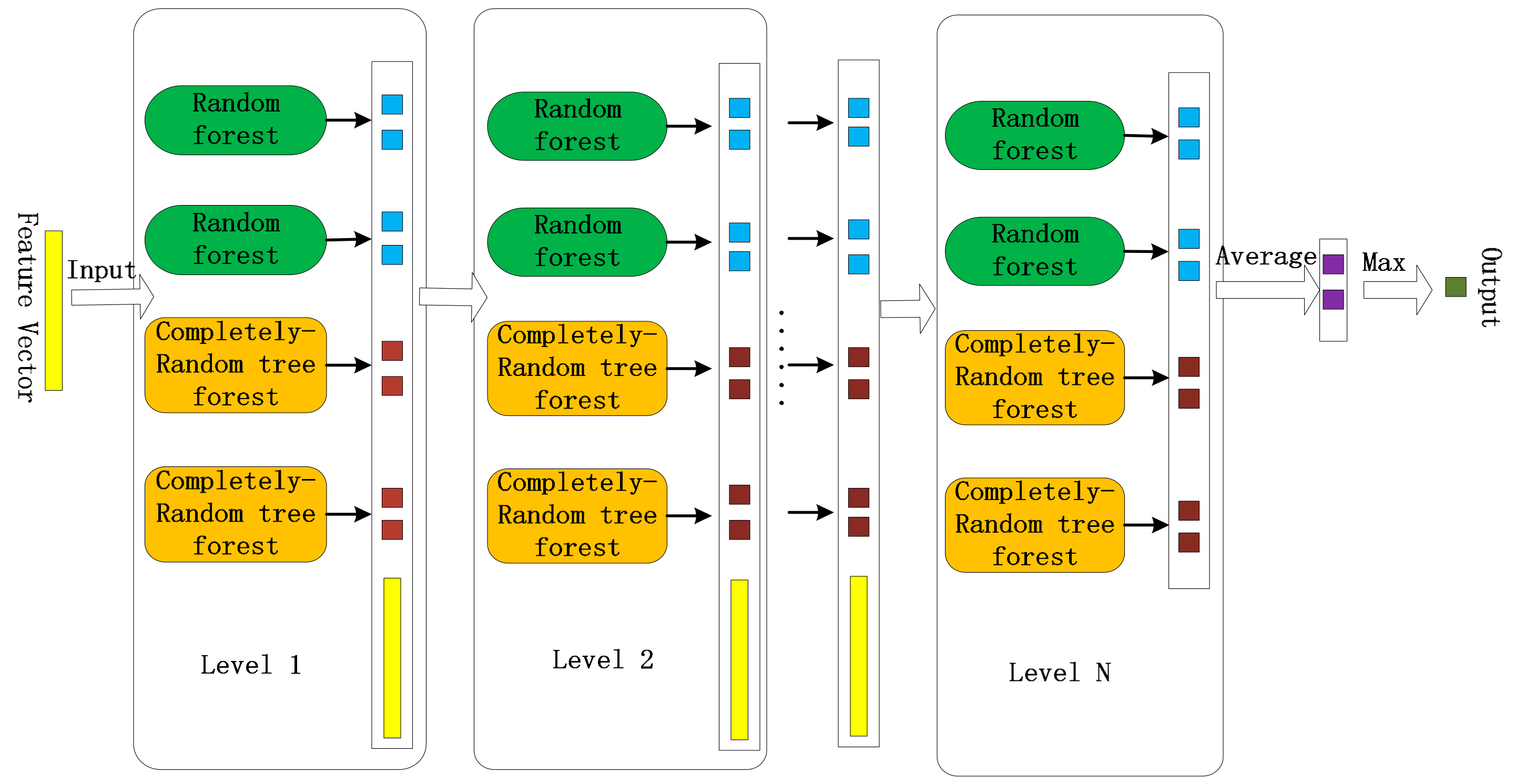
2.4.2. Cascade Forest
2.5. Performance Evaluation Indicators
- (1)
- TN: True negative, the number of true non-interacting pairs correctly predicted;
- (2)
- FN: False negative, the quantity of true non-interacting pairs falsely predicted;
- (3)
- FP: False positive, the count of true interacting pairs falsely predicted;
- (4)
- TP: True positive, the quantity of true interacting pairs correctly predicted.
- (1)
- Acc: Accuracy, the proportion of all judged correctly samples in the total observation values from the classification model;
- (2)
- TNR: True negative rate or specificity, the proportion of predicted correctly samples in the result with actual negative value;
- (3)
- F1-score: Measuring the overall performance of the classification model;
- (4)
- MCC: Matthews correlation coefficient, the geometric mean of the problem and dual regression coefficients; It is a better indicator for measuring unbalanced dataset and the most informative single fraction for assessing the quality of binary classifier from the CM.
3. Results and Discussion
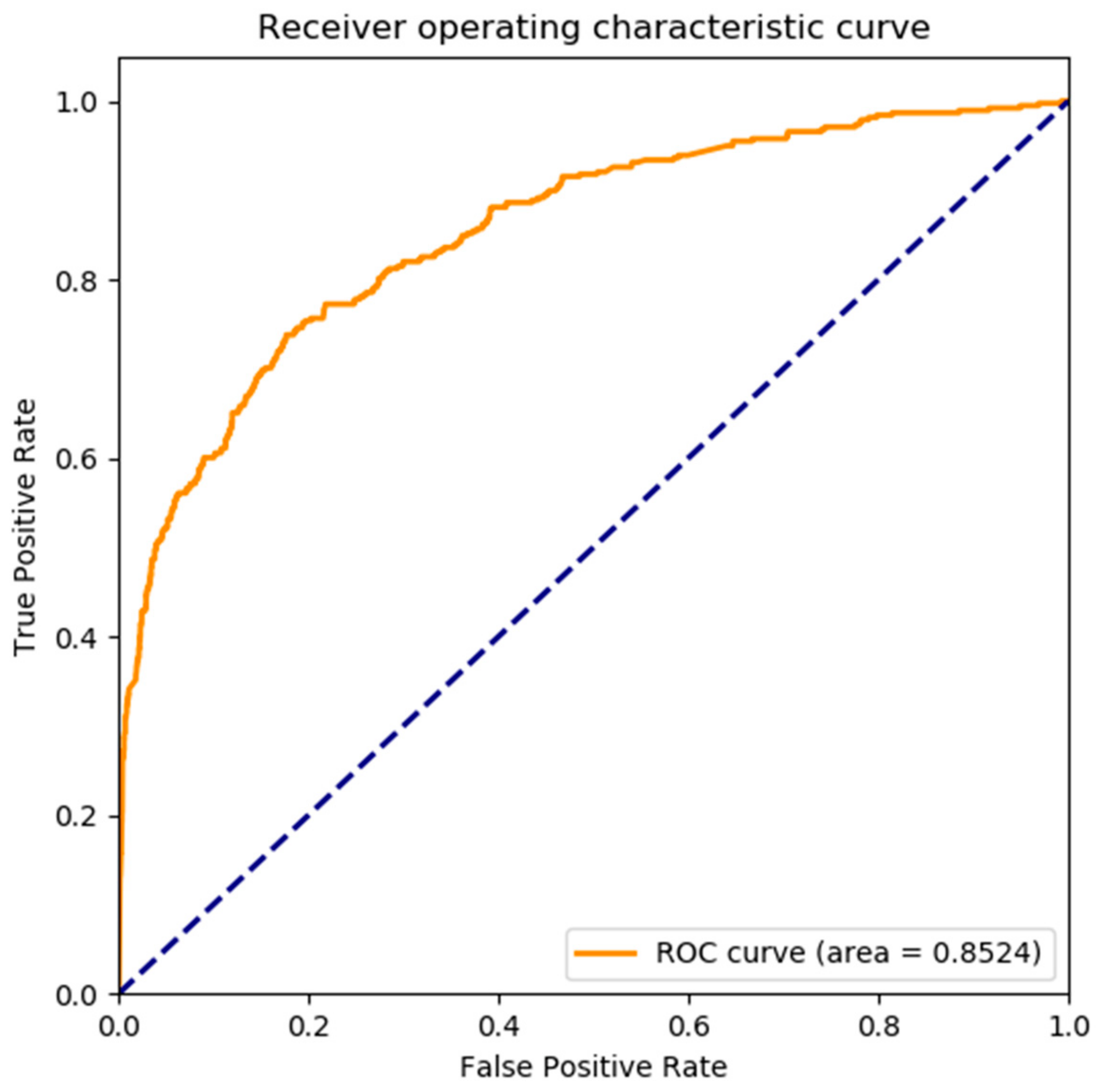
3.1. Performance Evaluation on Protein Self-Interaction
3.2. Comparison with Other Existing Methods for Predicting SIPs
4. Conclusions
5. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yıldırım, M.A.; Goh, K.-I.; Cusick, M.E.; Barabási, A.-L.; Vidal, M. Drug—Target network. Nat. Biotechnol. 2007, 25, 1119. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Clark, W.T.; Oron, T.R.; Schnoes, A.M.; Wittkop, T.; Sokolov, A.; Graim, K.; Funk, C.; Verspoor, K.; Ben-Hur, A.; et al. A large-scale evaluation of computational protein function prediction. Nat. Methods 2013, 10, 221. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Cheng, J. Integrated protein function prediction by mining function associations, sequences, and protein–protein and gene–gene interaction networks. Methods 2016, 93, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Ispolatov, I.; Yuryev, A.; Mazo, I.; Maslov, S. Binding properties and evolution of homodimers in protein–protein interaction networks. Nucleic Acids Res. 2005, 33, 3629–3635. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, B.; Panchenko, A. Deciphering protein-protein interactions. PLoS Comput. Biol. 2006, 3, e43. [Google Scholar]
- Reguly, T.; Breitkreutz, A.; Boucher, L.; Breitkreutz, B.-J.; Hon, G.C.; Myers, C.L.; Parsons, A.; Friesen, H.; Oughtred, R.; Tong, A.; et al. Comprehensive curation and analysis of global interaction networks in saccharomyces cerevisiae. J. Biol. 2006, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Salwinski, L.; Miller, C.S.; Smith, A.J.; Pettit, F.K.; Bowie, J.U.; Eisenberg, D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004, 32, D449–D451. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Oughtred, R.; Boucher, L.; Rust, J.; Chang, C.; Kolas, N.K.; O’Donnell, L.; Oster, S.; Theesfeld, C.; Sellam, A.; et al. The biogrid interaction database: 2017 update. Nucleic Acids Res. 2017, 45, D369–D379. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The string database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, S.; Li, C.; Zhang, Z.; Song, J. Spar: A random forest-based predictor for self-interacting proteins with fine-grained domain information. Amino Acids 2016, 48, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Deng, S.-P.; You, Z.-H.; Huang, D.-S. Identifying spurious interactions in the protein-protein interaction networks using local similarity preserving embedding. IEEE/ACM Trans. Comput. Biol. Bioinform. (TCBB) 2017, 14, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.C.; Petrey, D.; Deng, L.; Qiang, L.; Shi, Y.; Thu, C.A.; Bisikirska, B.; Lefebvre, C.; Accili, D.; Hunter, T.; et al. Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature 2012, 490, 556. [Google Scholar] [CrossRef] [PubMed]
- You, Z.-H.; Zhou, M.; Luo, X.; Li, S. Highly efficient framework for predicting interactions between proteins. IEEE Trans. Cybern. 2017, 47, 731–743. [Google Scholar] [CrossRef] [PubMed]
- You, Z.-H.; Lei, Y.-K.; Gui, J.; Huang, D.-S.; Zhou, X. Using manifold embedding for assessing and predicting protein interactions from high-throughput experimental data. Bioinformatics 2010, 26, 2744–2751. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Yu, H.; Greenbaum, D.; Kluger, Y.; Krogan, N.J.; Chung, S.; Emili, A.; Snyder, M.; Greenblatt, J.F.; Gerstein, M. A bayesian networks approach for predicting protein-protein interactions from genomic data. Science 2003, 302, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Ofran, Y.; Rost, B. Protein–protein interaction hotspots carved into sequences. PLoS Comput. Biol. 2007, 3, e119. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhou, B.; Lai, L.; Pei, J. Sequence-based prediction of protein protein interaction using a deep-learning algorithm. BMC Bioinform. 2017, 18, 277. [Google Scholar] [CrossRef] [PubMed]
- Kovács, I.A.; Luck, K.; Spirohn, K.; Wang, Y.; Pollis, C.; Schlabach, S.; Bian, W.; Kim, D.-K.; Kishore, N.; Hao, T.; et al. Network-based prediction of protein interactions. Nat. Commun. 2019, 10, 1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-B.; You, Z.-H.; Li, X.; Jiang, T.-H.; Cheng, L.; Chen, Z.-H. Prediction of protein self-interactions using stacked long short-term memory from protein sequences information. BMC Syst. Biol. 2018, 12, 129. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-H.; You, Z.-H.; Li, L.-P.; Wang, Y.-B.; Wong, L.; Yi, H.-C. Prediction of self-interacting proteins from protein sequence information based on random projection model and fast fourier transform. Int. J. Mol. Sci. 2019, 20, 930. [Google Scholar] [CrossRef] [PubMed]
- George, A.; Ganesh, H.B.; Kumar, M.A.; Soman, K. Significance of Global Vectors Representation in Protein Sequences Analysis. In Computer Aided Intervention and Diagnostics in Clinical and Medical Images; Springer: Cham, Switzerland, 2019; pp. 261–269. [Google Scholar]
- Wang, Y.; You, Z.-H.; Yang, S.; Li, X.; Jiang, T.-H.; Zhou, X. A high efficient biological language model for predicting protein–protein interactions. Cells 2019, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Wan, F.; Zeng, J. Deep learning with feature embedding for compound-protein interaction prediction. bioRxiv 2016, 086033. [Google Scholar]
- Luo, X.; Zhou, M.; Xia, Y.; Zhu, Q. An efficient non-negative matrix-factorization-based approach to collaborative filtering for recommender systems. IEEE Trans. Ind. Inform. 2014, 10, 1273–1284. [Google Scholar]
- Jin, L.; Li, S.; La, H.M.; Luo, X. Manipulability optimization of redundant manipulators using dynamic neural networks. IEEE Trans. Ind. Electron. 2017, 64, 4710–4720. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Liu, X.; Zeng, N.; Liu, Y.; Alsaadi, F.E. A survey of deep neural network architectures and their applications. Neurocomputing 2017, 234, 11–26. [Google Scholar] [CrossRef]
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.; Brinkman, F.S.; Lynn, D.J. Innatedb: Systems biology of innate immunity and beyond—Recent updates and continuing curation. Nucleic Acids Res. 2012, 41, D1228–D1233. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; Del-Toro, N.; et al. The mintact project—Intact as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2013, 42, D358–D363. [Google Scholar] [CrossRef] [PubMed]
- Clerc, O.; Deniaud, M.; Vallet, S.D.; Naba, A.; Rivet, A.; Perez, S.; Thierry-Mieg, N.; Ricard-Blum, S. Matrixdb: Integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 2018, 47, D376–D381. [Google Scholar] [CrossRef] [PubMed]
- Uniprot: The universal protein knowledgebase. Nucleic Acids Res. 2016, 45, D158–D169.
- Muppirala, U.K.; Honavar, V.G.; Dobbs, D. Predicting rna-protein interactions using only sequence information. BMC Bioinform. 2011, 12, 489. [Google Scholar] [CrossRef] [PubMed]
- Asgari, E.; Mofrad, M.R. Continuous distributed representation of biological sequences for deep proteomics and genomics. PLoS ONE 2015, 10, e0141287. [Google Scholar] [CrossRef] [PubMed]
- Pennington, J.; Socher, R.; Manning, C. Glove: Global vectors for word representation. In Proceedings of the 2014 conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Merchant, K.; Pande, Y. Nlp based latent semantic analysis for legal text summarization. In Proceedings of the 2018 International Conference on Advances in Computing, Communications and Informatics (ICACCI), Bangalore, India, 19–22 September 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1803–1807. [Google Scholar]
- Liu, P.; Qiu, X.; Huang, X. Learning context-sensitive word embeddings with neural tensor skip-gram model. In Proceedings of the Twenty-Fourth International Joint Conference on Artificial Intelligence, Buenos Aires, Argentina, 25–31 July 2015. [Google Scholar]
- Zhou, Z.-H.; Feng, J. Deep forest: Towards an alternative to deep neural networks. arXiv 2017, arXiv:1702.08835. [Google Scholar]
- Chen, Z.-H.; Li, L.-P.; He, Z.; Zhou, J.-R.; Li, Y.; Wong, L. An improved deep forest model for predicting self-interacting proteins from protein sequence using wavelet transformation. Front. Genet. 2019, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.; Deng, L.; Yu, D.; Dahl, G.; Mohamed, A.-r.; Jaitly, N.; Senior, A.; Vanhoucke, V.; Nguyen, P.; Kingsbury, B.; et al. Deep neural networks for acoustic modeling in speech recognition. IEEE Signal Process. Mag. 2012, 29, 82–97. [Google Scholar] [CrossRef]
- Liu, X.; Liu, W.; Ma, H.; Fu, H. Large-scale vehicle re-identification in urban surveillance videos. In Proceedings of the 2016 IEEE International Conference on Multimedia and Expo (ICME), Seattle, WA, USA, 11–15 July 2016; pp. 1–6. [Google Scholar]
- Liu, Z.; Guo, F.; Zhang, J.; Wang, J.; Lu, L.; Li, D.; He, F. Proteome-wide prediction of self-interacting proteins based on multiple properties. Mol. Cell. Proteom. 2013, 12, 1689–1700. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Cheng, J.; Zheng, T.; Duan, Z.; Qian, F. A novel feature extraction scheme with ensemble coding for protein–protein interaction prediction. Int. J. Mol. Sci. 2014, 15, 12731–12749. [Google Scholar] [CrossRef] [PubMed]
- Zahiri, J.; Yaghoubi, O.; Mohammad-Noori, M.; Ebrahimpour, R.; Masoudi-Nejad, A. Ppievo: Protein–protein interaction prediction from pssm based evolutionary information. Genomics 2013, 102, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Zahiri, J.; Mohammad-Noori, M.; Ebrahimpour, R.; Saadat, S.; Bozorgmehr, J.H.; Goldberg, T.; Masoudi-Nejad, A. Locfuse: Human protein–protein interaction prediction via classifier fusion using protein localization information. Genomics 2014, 104, 496–503. [Google Scholar] [CrossRef] [PubMed]
| Predict | |||
|---|---|---|---|
| Negative | Positive | ||
| Actual | Negative | TN | FN |
| Positive | FP | TP | |
| Datasets | Acc (%) | TNR (%) | F1-Score (%) | MCC |
|---|---|---|---|---|
| yeast | 91.45 | 99.71 | 37.56 | 0.4389 |
| human | 93.12 | 99.57 | 39.10 | 0.4421 |
| Model | Acc (%) | TNR (%) | F1-Score (%) | MCC | AUC |
|---|---|---|---|---|---|
| SLIPPER [40] | 71.90 | 72.18 | 36.16 | 0.2842 | 0.7723 |
| DXECPPI [41] | 87.46 | 94.93 | 34.89 | 0.2825 | 0.6934 |
| PPIevo [42] | 66.28 | 87.46 | 28.92 | 0.1801 | 0.6728 |
| LocFuse [43] | 66.66 | 68.10 | 27.53 | 0.1577 | 0.7087 |
| CRS [10] | 72.69 | 74.37 | 33.05 | 0.2368 | 0.7115 |
| SPAR [10] | 76.96 | 80.02 | 34.54 | 0.2484 | 0.7455 |
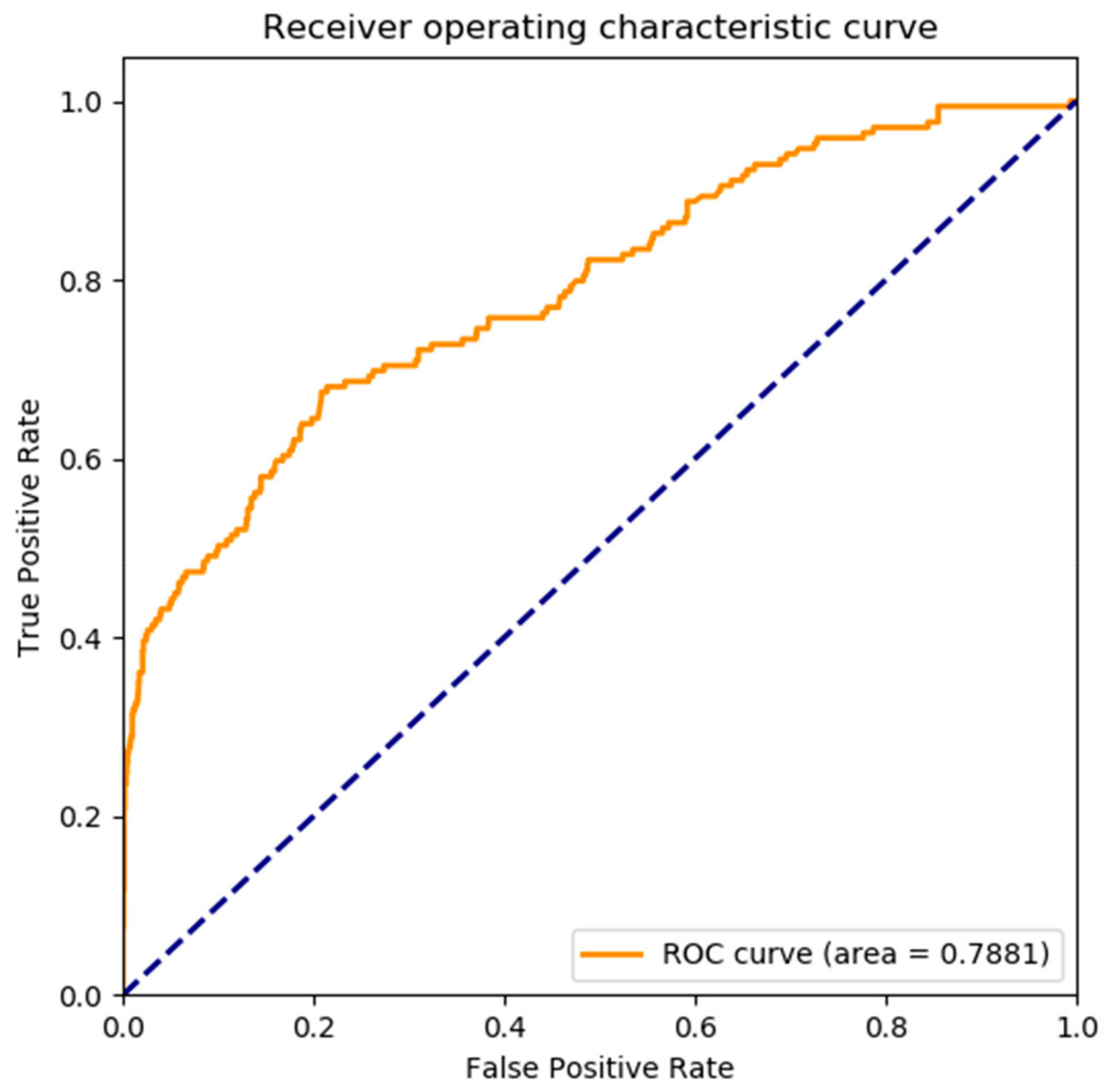
| Proposed method | 91.45 | 99.71 | 37.56 | 0.4389 | 0.7881 |
| Model | Acc (%) | TNR (%) | F1-score (%) | MCC | AUC |
|---|---|---|---|---|---|
| SLIPPER [40] | 91.10 | 95.06 | 46.82 | 0.4197 | 0.8723 |
| DXECPPI [41] | 30.90 | 25.83 | 17.28 | 0.0825 | 0.5806 |
| PPIevo [42] | 78.04 | 25.82 | 27.73 | 0.2082 | 0.7329 |
| LocFuse [43] | 80.66 | 80.50 | 27.65 | 0.2026 | 0.7087 |
| CRS [10] | 91.54 | 96.72 | 36.83 | 0.3633 | 0.8196 |
| SPAR [10] | 92.09 | 97.40 | 41.13 | 0.3836 | 0.8229 |
| Proposed method | 93.12 | 99.57 | 39.10 | 0.4421 | 0.8524 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.-H.; You, Z.-H.; Zhang, W.-B.; Wang, Y.-B.; Cheng, L.; Alghazzawi, D. Global Vectors Representation of Protein Sequences and Its Application for Predicting Self-Interacting Proteins with Multi-Grained Cascade Forest Model. Genes 2019, 10, 924. https://doi.org/10.3390/genes10110924
Chen Z-H, You Z-H, Zhang W-B, Wang Y-B, Cheng L, Alghazzawi D. Global Vectors Representation of Protein Sequences and Its Application for Predicting Self-Interacting Proteins with Multi-Grained Cascade Forest Model. Genes. 2019; 10(11):924. https://doi.org/10.3390/genes10110924
Chicago/Turabian StyleChen, Zhan-Heng, Zhu-Hong You, Wen-Bo Zhang, Yan-Bin Wang, Li Cheng, and Daniyal Alghazzawi. 2019. "Global Vectors Representation of Protein Sequences and Its Application for Predicting Self-Interacting Proteins with Multi-Grained Cascade Forest Model" Genes 10, no. 11: 924. https://doi.org/10.3390/genes10110924
APA StyleChen, Z.-H., You, Z.-H., Zhang, W.-B., Wang, Y.-B., Cheng, L., & Alghazzawi, D. (2019). Global Vectors Representation of Protein Sequences and Its Application for Predicting Self-Interacting Proteins with Multi-Grained Cascade Forest Model. Genes, 10(11), 924. https://doi.org/10.3390/genes10110924





