A Weighted Genomic Relationship Matrix Based on Fixation Index (FST) Prioritized SNPs for Genomic Selection
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Simulation
2.2. SNPs Prioritization Based on FST Scores
2.3. Prioritized SNPs and Genomic Relationships
2.4. Data Analysis
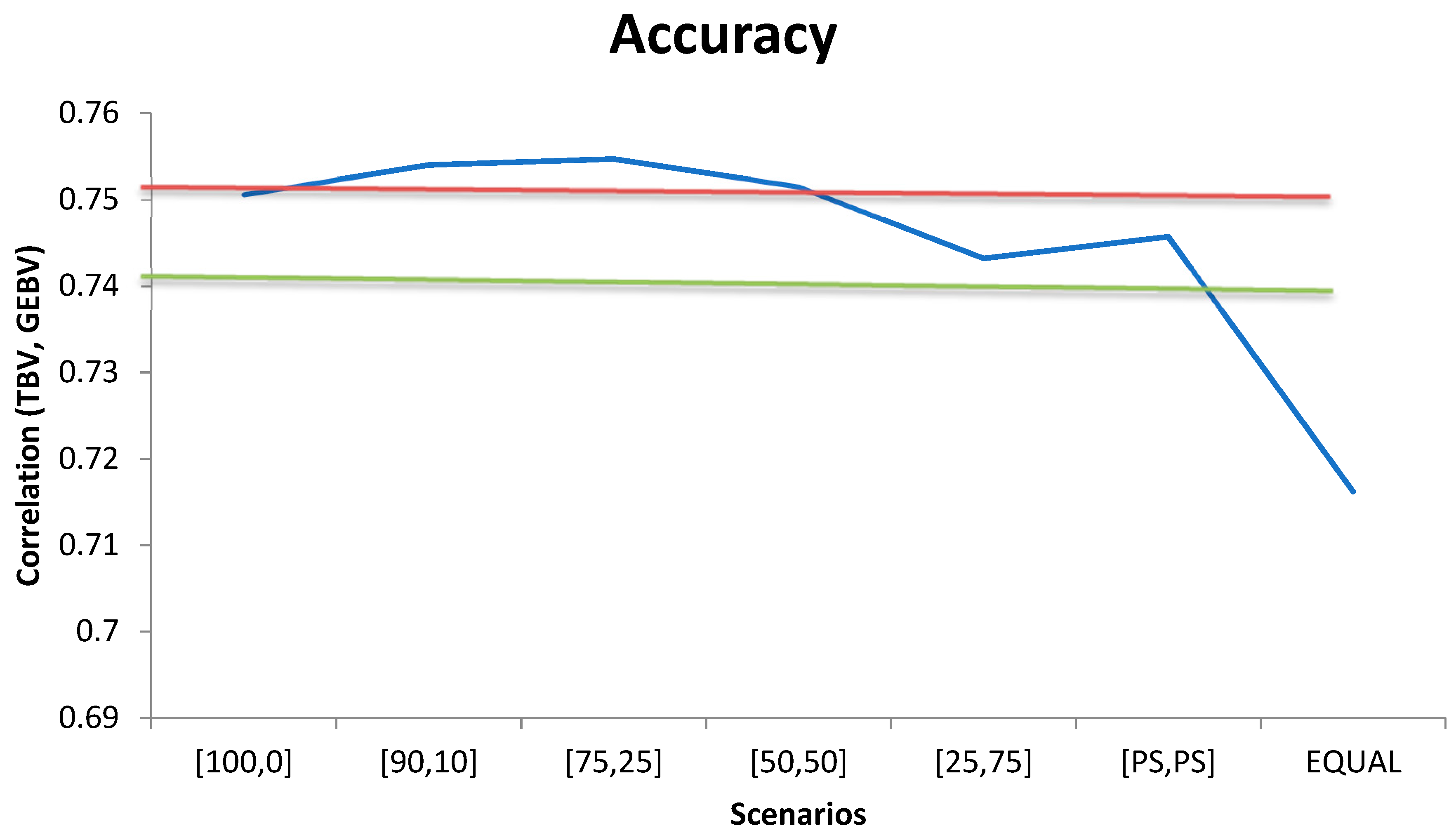
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Balloux, F.; Brunner, H.; Lugon-Moulin, N.; Hausser, J.; Goudet, J. Microsatellites can be misleading: An empirical and simulation study. Evolution 2000, 54, 1414–1422. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.; Van Tassell, C.; Wiggans, G.; Sonstegard, T.; Schnabel, R.; Taylor, J.; Schenkel, F.; Van Tassell, C.; Schnabel, R. Invited Review: Reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 2009, 92, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Guldbrandtsen, B.; Gregersen, V.; Lund, M. Preliminary investigation on reliability of genomic estimated breeding values in the Danish Holstein population. J. Dairy Sci. 2010, 93, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Madsen, P.; Nielsen, U.S.; Mäntysaari, E.A.; Aamand, G.P.; Christensen, O.F.; Lund, M.S. Genomic prediction for Nordic red cattle using one-step and selection index blending. J. Dairy Sci. 2012, 95, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Schefers, J.M.; Weigel, K.A. Genomic selection in dairy cattle: Integration of DNA testing into breeding programs. Anim. Front. 2012, 2, 4–9. [Google Scholar] [CrossRef]
- Zeng, J.; Toosi, A.; Fernando, R.L.; Dekkers, J.C.M.; Garrick, D.J. Genomic Selection of Purebred Animals for Crossbred Performance in the Presence of Dominant Gene Action. Genet. Sel. Evol. 2013, 45, 11. [Google Scholar] [CrossRef] [PubMed]
- Da, Y.; Wang, C.; Wang, S.; Hu, G. Mixed Model Methods for Genomic Prediction and Variance Component Estimation of Additive and Dominance Effects Using SNP Markers. PLoS ONE 2014, 9, e87666. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.A.; Hickey, J.M.; Van Der Werf, J.H. Different models of genetic variation and their effect on genomic evaluation. Genet. Sel. Evol. 2011, 43, 18. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J. Genomic selection. J. Anim. Breed. Genet. 2017, 124, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Pérez, P.; Campos, G.D.L. Genome-Wide Regression and Prediction with the BGLR Statistical Package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Pérez, P.; de los Campos, G.; Crossa, J.; Gianola, D. Genomic-enabled prediction based on molecular markers and pedigree using the Bayesian linear regression package in R. Plant Genome 2010, 3, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M.E.; Meuwissen, T.H. Prediction of identity by descent probabilities from marker-haplotypes. Genet. Sel. Evol. 2001, 33, 605–634. [Google Scholar]
- Erbe, M.; Hayes, B.; Matukumalli, L.; Goswami, S.; Bowman, P.; Reich, C.; Mason, B.; Goddard, M.; Goddard, M. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J. Dairy Sci. 2012, 95, 4114–4129. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.; Duran, C.; Berkman, P.J.; Lorenc, M.T.; Stiller, J.; Manoli, S.; Hayden, M.J.; Forrest, K.L.; Fleury, D.; Baumann, U.; et al. Single nucleotide polymorphism discovery from wheat next-generation sequence data. Plant Biotechnol. J. 2012, 10, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Farrer, R.A.; Henk, D.A.; MacLean, D.; Studholme, D.J.; Fisher, M.C. Using False Discovery Rates to Benchmark SNP-callers in next-generation sequencing projects. Sci. Rep. 2013, 3, 1512. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.; Golicz, A.; Hackett, C.A.; Milne, I.; Stephen, G.; Marshall, D.; Flavell, A.J.; Bayer, M. An investigation of causes of false positive single nucleotide polymorphisms using simulated reads from a small eukaryote genome. BMC Bioinform. 2015, 16, 382. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Brøndum, R.F.; Ma, P.; Guldbrandtsen, B.; Aamand, G.P.; Lund, M.S. Comparison of genomic predictions using medium-density (∼54,000) and high-density (∼777,000) single nucleotide polymorphism marker panels in Nordic Holstein and Red Dairy Cattle populations. J. Dairy Sci. 2012, 95, 4657–4665. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.Y.; Toghiani, S.; Ling, A.; Aggrey, S.E.; Rekaya, R. Correction to: High density marker panels, SNPs prioritizing and accuracy of genomic selection. BMC Genet. 2018, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [PubMed]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Christensen, O.F.; Lund, M.S. Genomic prediction when some animals are not genotyped. Genet. Sel. Evol. 2010, 42, 2. [Google Scholar] [CrossRef] [PubMed]
- Habier, D.; Fernando, R.L.; Dekkers, J.C.M. The impact of genetic relationship information on genome-assisted breeding values. Genetics 2008, 177, 2389–2397. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Fernando, R.L.; Garrick, D.J.; Dekkers, J.C.M. An iterative approach for efficient calculation of breeding values and genome-wide association analysis using weighted genomic BLUP. J. Anim. Sci. 2011, 89, 28. [Google Scholar]
- Aguilar, I.; Misztal, I.; Johnson, D.; Legarra, A.; Tsuruta, S.; Lawlor, T. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Fragomeni, B.O.; Lourenco, D.A.L.; Masuda, Y.; Legarra, A.; Misztal, I.; Masuda, Y. Incorporation of causative quantitative trait nucleotides in single-step GBLUP. Genet. Sel. Evol. 2017, 49, 59. [Google Scholar] [CrossRef] [PubMed]
- Toghiani, S.; Chang, L.Y.; Ling, A.; Aggrey, S.E.; Rekaya, R. Genomic differentiation as a tool for single nucleotide polymorphism prioritization for Genome wide association and phenotype prediction in livestock. Livest. Sci. 2017, 205, 24–30. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Schenkel, F.S. QMSim: A large-scale genome simulator for livestock. Bioinformatics 2009, 25, 680–681. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Amin, N.; Van Duijn, C.M.; Aulchenko, Y.S. A Genomic Background Based Method for Association Analysis in Related Individuals. PLoS ONE 2007, 2, e1274. [Google Scholar] [CrossRef] [PubMed]
- Gengler, N.; Mayeres, P.; Szydlowski, M. A simple method to approximate gene content in large pedigree populations: Application to the myostatin gene in dual-purpose Belgian Blue cattle. Animal 2007, 1, 21–28. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Weeks, D.; Chakravarti, A. Similarity of DNA Fingerprints Due to Chance and Relatedness. Hum. Hered. 1993, 43, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Blouin, M.S.; Lacaille, V.; Lotz, S.; Parsons, M. Use of microsatellite loci to classify individuals by relatedness. Mol. Ecol. 1996, 5, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Csilléry, K.; Johnson, T.; Beraldi, D.; Clutton-Brock, T.; Coltman, D.; Hansson, B.; Spong, G.; Pemberton, J.M. Performance of Marker-Based Relatedness Estimators in Natural Populations of Outbred Vertebrates. Genetics 2006, 173, 2091–2101. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M.; O’Connell, J.R.; Wiggans, G.R.; Weigel, K.A. Genomic evaluations with many more genotypes. Genet. Sel. Evol. 2011, 43, 10. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D. BLUPF90 and related programs (BGF90). In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002; pp. 27–28. [Google Scholar]
- Bird, C.E.; Karl, S.A.; Smouse, P.E.; Toonen, R.T. Detecting and measuring genetic differentiation. In Phylogeography and Population Genetics in Crustacea; Koenemann, S., Held, C., Schubart, C., Eds.; Crustacean Issues Series; CRC Press: Boca Raton, FL, USA, 2011; Volume 19, pp. 31–55. [Google Scholar]
| Scenario 2 | Weighting (%) | Genetic Variance | Residual Variance | Heritability | |
|---|---|---|---|---|---|
| 20K 1 | 380K | ||||
| 1 = (100,0) | 100 | 0 | 0.196 (0.026) | 0.671 (0.042) | 0.228 (0.033) |
| 2 = (90,10) | 90 | 10 | 0.213 (0.018) | 0.648 (0.032) | 0.247 (0.023) |
| 3 = (75,25) | 75 | 25 | 0.232 (0.015) | 0.633 (0.025) | 0.268 (0.018) |
| 4 = (50,50) | 50 | 50 | 0.257 (0.016) | 0.618 (0.021) | 0.294 (0.018) |
| 5 = (25,75) | 258 | 75 | 0.279 (0.021) | 0.619 (0.021) | 0.311 (0.023) |
| 6 = (PS 3,PS) | PS | PS | 0.251 (0.032) | 0.629 (0.037) | 0.285 (0.037) |
| 7 = Equal weights | Equal weights | Equal weights | 0.247 (0.027) | 0.692 (0.016) | 0.263 (0.025) |
| OD | ||
|---|---|---|
| OD < −0.05 | 2.32 | 1.61 |
| 0.05 < OD < −0.03 | 9.85 | 8.39 |
| 0.03 < OD < −0.01 | 28.18 | 29.35 |
| −0.01 < OD < 0.01 | 33.48 | 36.14 |
| 0.01 < OD < 0.03 | 17.21 | 16.52 |
| 0.03 < OD < 0.05 | 5.52 | 4.86 |
| OD > 0.05 | 3.46 | 4.86 |
| OD | Weights | No Weight | Scenario | ||||
|---|---|---|---|---|---|---|---|
| (100,0) 2 | (90,10) | (75,25) | (50,50) | (25,75) | |||
| OD < −0.05 | 0.92 | 0.00 | 2.32 | 1.66 | 0.92 | 0.25 | 0.03 |
| −0.05 < OD < −0.03 | 4.60 | 0.77 | 9.85 | 8.72 | 6.81 | 3.67 | 1.42 |
| −0.03 < OD < −0.01 | 30.26 | 29.77 | 28.18 | 29.16 | 30.45 | 31.72 | 31.22 |
| −0.01 < OD < 0.01 | 43.93 | 52.43 | 33.49 | 35.54 | 38.86 | 44.57 | 49.81 |
| 0.01 < OD < 0.03 | 13.52 | 11.52 | 17.21 | 16.74 | 15.74 | 13.64 | 11.89 |
| 0.03 < OD < 0.05 | 4.04 | 3.30 | 5.52 | 5.01 | 4.36 | 3.68 | 3.36 |
| OD > 0.05 | 2.73 | 2.21 | 3.46 | 3.19 | 2.86 | 2.49 | 2.27 |
| Scenario | Residual Variance | Intercept | Slope | −2LogL |
|---|---|---|---|---|
| 1 = (100,0) | 0.671 (0.04) | −1.208 (0.04) | 0.664 (0.03) | 26,409.13 (310.78) |
| 2 = (90,10) | 0.648 (0.03) | −1.228 (0.04) | 0.675 (0.02) | 26,397.40 (275.55) |
| 3 = (75,25) | 0.633 (0.03) | −1.244 (0.04) | 0.683 (0.02) | 26,404.90 (233.44) |
| 4 = (50,50) | 0.618 (0.02) | −1.240 (0.05) | 0.682 (0.02) | 26,489.84 (180.68) |
| 5 = (25,75) | 0.619 (0.02) | −1.185 (0.06) | 0.651 (0.02) | 26,746.99 (106.85) |
| 6 = (PS,PS) | 0.629 (0.04) | −1.205 (0.04) | 0.662 (0.03) | 26,619.76 (232.62) |
| 7 = Equal weight | 0.692 (0.02) | −0.921 (0.13) | 0.505 (0.05) | 27,378.30 (82.38) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, L.-Y.; Toghiani, S.; Hay, E.H.; Aggrey, S.E.; Rekaya, R. A Weighted Genomic Relationship Matrix Based on Fixation Index (FST) Prioritized SNPs for Genomic Selection. Genes 2019, 10, 922. https://doi.org/10.3390/genes10110922
Chang L-Y, Toghiani S, Hay EH, Aggrey SE, Rekaya R. A Weighted Genomic Relationship Matrix Based on Fixation Index (FST) Prioritized SNPs for Genomic Selection. Genes. 2019; 10(11):922. https://doi.org/10.3390/genes10110922
Chicago/Turabian StyleChang, Ling-Yun, Sajjad Toghiani, El Hamidi Hay, Samuel E. Aggrey, and Romdhane Rekaya. 2019. "A Weighted Genomic Relationship Matrix Based on Fixation Index (FST) Prioritized SNPs for Genomic Selection" Genes 10, no. 11: 922. https://doi.org/10.3390/genes10110922
APA StyleChang, L.-Y., Toghiani, S., Hay, E. H., Aggrey, S. E., & Rekaya, R. (2019). A Weighted Genomic Relationship Matrix Based on Fixation Index (FST) Prioritized SNPs for Genomic Selection. Genes, 10(11), 922. https://doi.org/10.3390/genes10110922





