Identification of an Embryonic Cell-Specific Region within the Pineapple SERK1 Promoter
Abstract
1. Introduction
2. Materials and Methods
2.1. Vector Construction
2.2. Plant Materials
2.3. Transient Transfection
2.4. GUS Histochemical Staining
2.5. Promoter Activity Assay
2.6. Gene Expression Analysis
3. Results
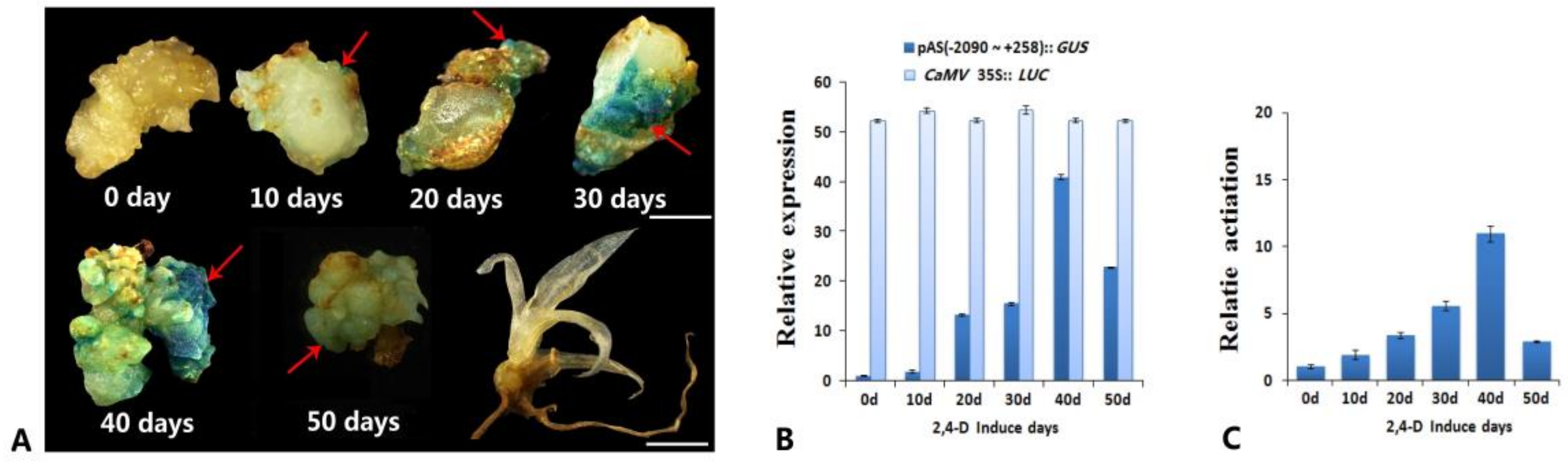
3.1. Analysis of the Promoter Activity of the Complete 5′ Upstream Regulatory Sequence of AcSERK1 during the Induction of Pineapple SE
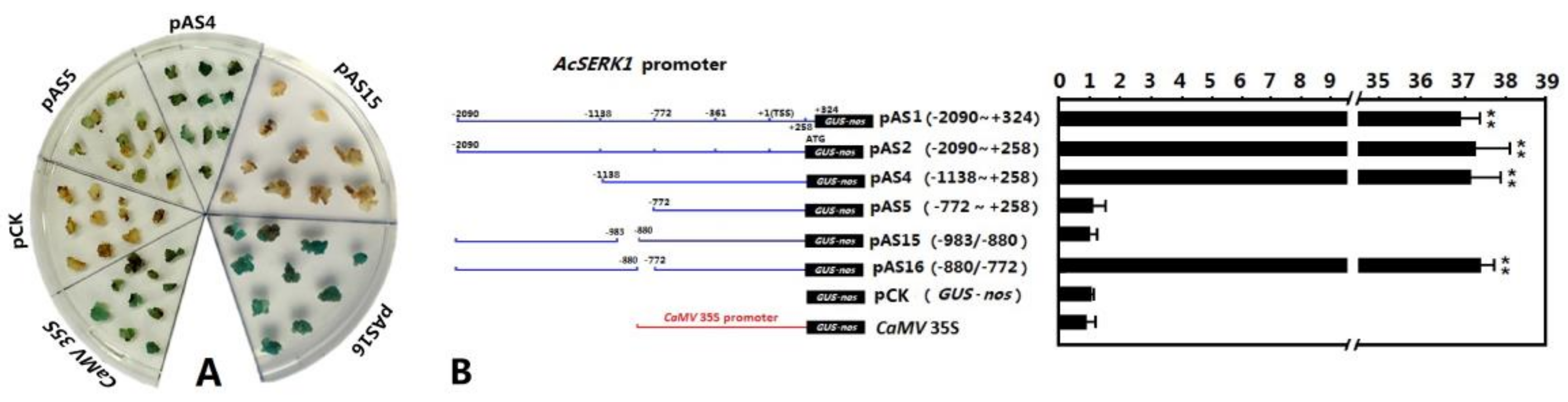
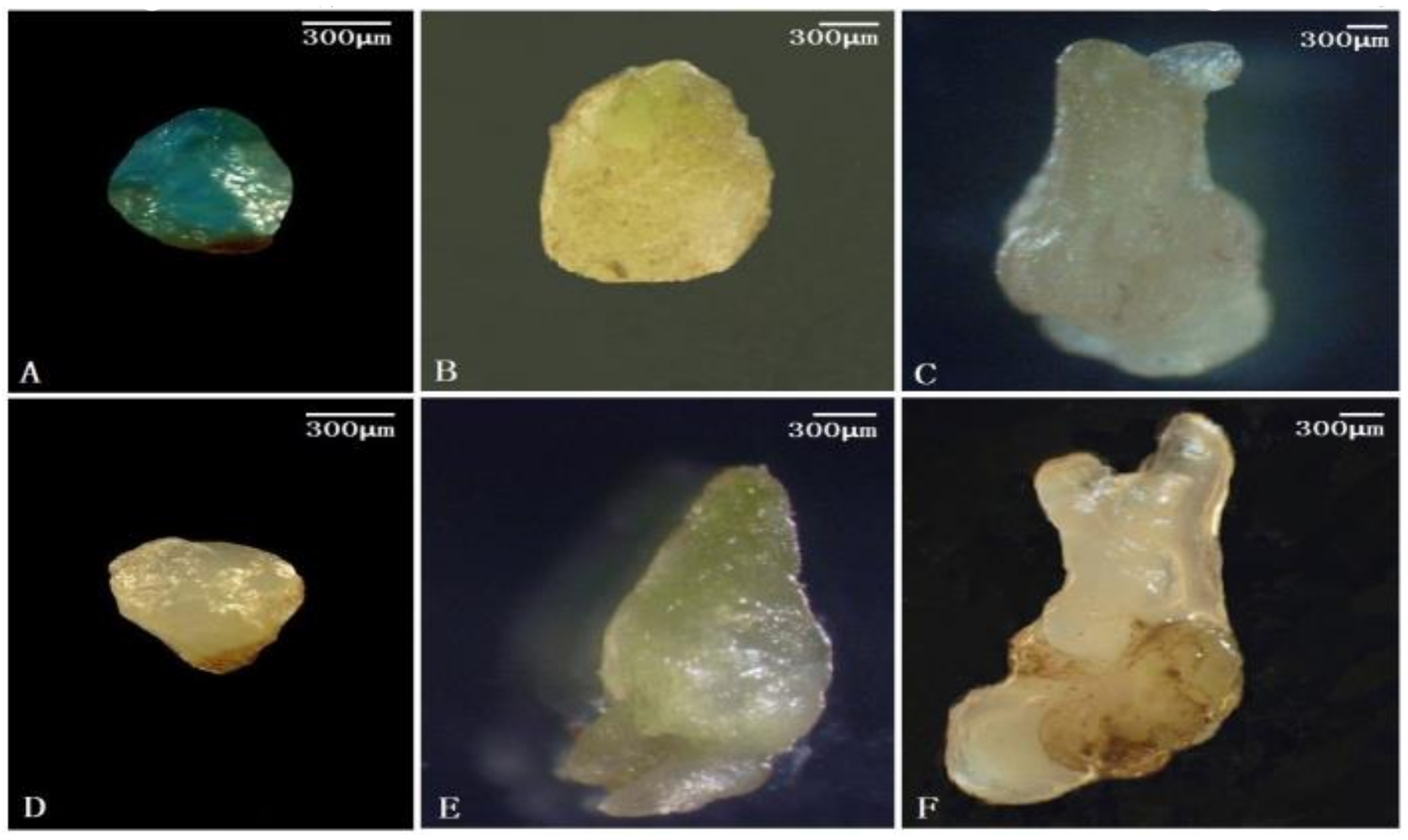
3.2. Identification of the Embryonic Cell-Specific Region in the 5′ Upstream Regulatory Region of AcSERK1
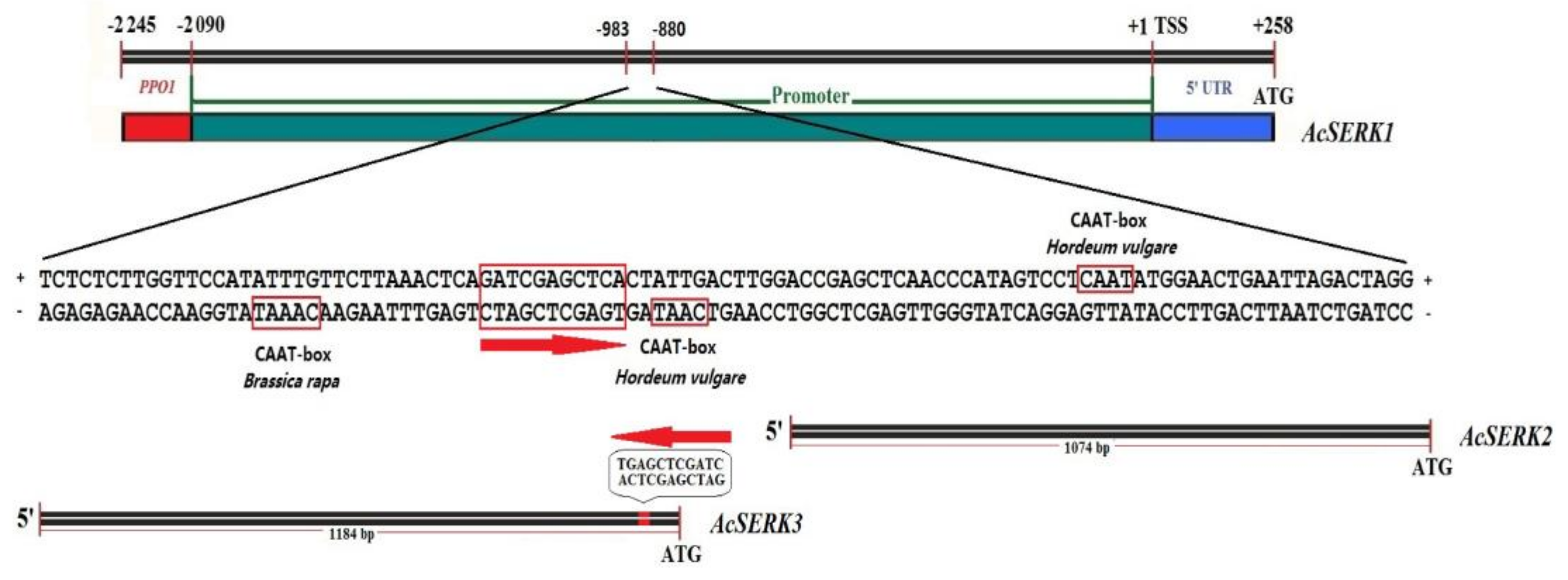
3.3. Bioinformatics Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sripaoraya, S.; Marchant, R.; Power, J.B.; Davey, M.R. Plant regeneration by somatic embryogenesis and organogenesis in commercial pineapple (Ananas comosus L.). Vitro. Cell. Dev. Plant 2003, 39, 450–454. [Google Scholar] [CrossRef]
- Firoozabady, E.; Gutterson, N. Cost-effective in vitro propagation methods for pineapple. Plant Cell Rep. 2003, 21, 844–850. [Google Scholar] [PubMed]
- Gonzalez-Olmedo, J.; Fundora, Z.; Molina, L.J.; Desjardins, Y.; Escalona, M. New contributions to propagation of pineapple (Ananas comosus L. Merr) in temporary immersion bioreactors. Vitro. Cell. Dev. Plant 2005, 41, 87–90. [Google Scholar] [CrossRef]
- Mhatre, M. Micropropagation of pineapple, Ananas comosus (L.) Merr. In Protocols for Micropropagation of Woody Trees and Fruits; Springer: Berlin, Germany, 2007; pp. 499–508. [Google Scholar] [CrossRef]
- Das, D.; Nirala, N.; Redoy, M.; Sopory, S.; Upadhyaya, K. Encapsulated somatic embryos of grape (Vitis vinifera L.): An efficient way for storage and propagation of pathogen-free plant material. VITIS-J. Grapevine Res. 2015, 45, 179. [Google Scholar]
- Gaj, M.D. Factors Influencing Somatic Embryogenesis Induction and Plant Regeneration with Particular Reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul. 2004, 43, 27–47. [Google Scholar] [CrossRef]
- Teyssier, C.; Maury, S.; Beaufour, M.; Grondin, C.; Delaunay, A.; Metté, C.L.; Ader, K.; Cadene, M.; Label, P. In search of markers for somatic embryo maturation in hybrid larch (Larix × eurolepis): Global DNA methylation and proteomic analyses. Physiol. Plant. 2013, 150, 271–291. [Google Scholar] [CrossRef]
- Fehér, A. Somatic embryogenesis-Stress-induced remodeling of plant cell fate. BBA Gene Regul. Mech. 2015, 1849, 385–402. [Google Scholar] [CrossRef]
- Thompson, D. Challenges for the large-scale propagation of forest trees by somatic embryogenesis-a review. In Proceedings of the Third International Conference of the IUFRO Unit, Vitoria-Gasteiz, Spain, 8–12 September 2014; pp. 81–91. [Google Scholar]
- Horstman, A.; Bemer, M.; Boutilier, K. A transcriptional view on somatic embryogenesis. Regeneration 2017, 4, 201–216. [Google Scholar] [CrossRef]
- Brandt, B.; Hothorn, M. SERK co-receptor kinases. Curr. Biol. 2016, 26, 225–226. [Google Scholar] [CrossRef]
- Fan, M.; Wang, M.; Bai, M.Y. Diverse roles of SERK family genes in plant growth, development and defense response. Sci. China Life Sci. 2016, 59, 889–896. [Google Scholar] [CrossRef]
- Hohmann, U.; Lau, K.; Hothorn, M. The Structural Basis of Ligand Perception and Signal Activation by Receptor Kinases. Annu. Rev. Plant Biol. 2017, 68, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; He, Y.; Wu, C.; Liu, H.; Hu, Z.; Sun, G. Cloning and Molecular Characterization of a SERK Gene Transcriptionally Induced during Somatic Embryogenesis in Ananas comosus cv. Shenwan. Plant Mol. Biol. Rep. 2012, 30, 195–203. [Google Scholar] [CrossRef]
- Rocha, D.I.; Pinto, D.L.; Vieira, L.M.; Tanaka, F.A.; Dornelas, M.C.; Otoni, W.C. Cellular and molecular changes associated with competence acquisition during passion fruit somatic embryogenesis: Ultrastructural characterization and analysis of SERK gene expression. Protoplasma 2016, 253, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Vielle-Calzada, J.P.; Hartog, M.V.; Boutilier, K.; Grossniklaus, U.; Vries, S.C.D. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 Gene Is Expressed in Developing Ovules and Embryos and Enhances Embryogenic Competence in Culture. Plant Physiol. 2001, 127, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Nolan, K.E.; Irwanto, R.R.; Rose, R.J. Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol. 2003, 133, 218–230. [Google Scholar] [CrossRef]
- Ahmadi, B.; Masoomi-Aladizgeh, F.; Shariatpanahi, M.E.; Azadi, P.; Keshavarz-Alizadeh, M. Molecular characterization and expression analysis of SERK1 and SERK2 in Brassica napus L.: Implication for microspore embryogenesis and plant regeneration. Plant Cell Rep. 2016, 35, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Xiong, L.; Yang, Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta 2005, 222, 107–117. [Google Scholar] [CrossRef]
- Chen, H.Y.; Hsieh, E.J.; Cheng, M.C.; Chen, C.Y.; Hwang, S.Y.; Lin, T.P. ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 2016, 211, 599–613. [Google Scholar] [CrossRef]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef]
- Lin, W.Q.; Chen, C.J.; Luan, A.P.; Zhang, J.L.; Feng, J.T.; Zhang, Y.F.; Guo, C.H.; He, Y.H. Transcriptional activity of the 5′ upstream region(-499/+ 258 bp) of AcSERK1. J. Fruit Sci. 2016, 33, 1058–1064. [Google Scholar]
- Franssen, H.J.; Kulikova, O.; Willemsen, V.; Heidstra, R. Cis-regulatory PLETHORA promoter elements directing root and nodule expression are conserved between Arabidopsis thaliana and Medicago truncatula. Plant Signal. Behav. 2017, 12, 1278102. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, L.; Wang, Y.; Zhang, S.; He, R.; Li, W.; Han, J.; Cheng, X.G. Tomato SlDREB1 gene conferred the transcriptional activation of drought-induced gene and an enhanced tolerance of the transgenic Arabidopsis to drought stress. Plant Growth Regul. 2017, 81, 131–145. [Google Scholar] [CrossRef]
- Wang, L.W.; He, M.W.; Guo, S.R.; Zhong, M.; Shu, S.; Sun, J. NaCl stress induces CsSAMs gene expression in Cucumis sativus by mediating the binding of CsGT-3b to the GT-1 element within the CsSAMs promoter. Planta 2017, 245, 889–908. [Google Scholar] [CrossRef] [PubMed]
- Baudino, S.; Hansen, S.; Brettschneider, R.; Hecht, V.F.; Dresselhaus, T.; Lörz, H. Molecular characterisation of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta 2001, 213, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Takaya, K.; Kurata, N. Expression of SERK family receptor-like protein kinase genes in rice. BBA-Gene Struct. Expr. 2005, 1730, 253–258. [Google Scholar] [CrossRef]
- Sharma, S.K.; Millam, S.; Hein, I.; Bryan, G.J. Cloning and molecular characterisation of a potato SERK gene transcriptionally induced during initiation of somatic embryogenesis. Planta 2008, 228, 319–330. [Google Scholar] [CrossRef]
- Ma, J.; He, Y.H.; Hu, Z.Y.; Xu, W.T.; Xia, J.X.; Guo, C.H.; Lin, S.Q.; Chen, C.J.; Wu, C.H.; Zhang, J.L. Characterization of the third SERK gene in pineapple (Ananas comosus) and analysis of its expression and autophosphorylation activity in vitro. Genet. Mol. Biol. 2014, 37, 530–539. [Google Scholar] [CrossRef]
- Luan, A.P.; He, Y.H.; Lin, W.Q.; Chen, C.J.; Feng, J.T.; Xie, T.; Gong, X.; Xia, J.X. Identification of Transcription Start Site of AcSERK1 and Its Embryospecific Cell Promoter in Pineapple. J. Hortic. 2016, 43, 2251–2256. (In Chinese) [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- He, Y.H.; Luo, J.; Wu, H.T.; Wang, R.X.; Gao, A.P.; Zhao, C.X.; Yu, X.L.; Ye, Z.X.; Wang, Z.H.; Hang, J.Z.; et al. Somatic embryogenesis from leaf base callus of Ananas comosus. J. Fruit Sci. 2007, 24, 59–63. (In Chinese) [Google Scholar]
- Gao, Y.; Zan, X.L.; Wu, X.F.; Yao, L.; Chen, Y.L.; Jia, S.W.; Zhao, K.J. Identification of fungus-responsive cis-acting element in the promoter of Brassica juncea chitinase gene, BjCHI1. Plant Sci. 2014, 215, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Mariashibu, T.S.; Subramanyam, K.; Arun, M.; Mayavan, S.; Rajesh, M.; Theboral, J.; Manickavasagam, M.; Ganapathi, A. Vacuum infiltration enhances the Agrobacterium-mediated genetic transformation in Indian soybean cultivars. Acta Physiol. Plant. 2013, 35, 41–54. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1988, 6, 3901–3907. [Google Scholar] [CrossRef]
- Wang, H.; Han, J.; Kanagarajan, S.; Lundgren, A.; Brodelius, P.E. Trichome-specific expression of the amorpha-4,11-diene 12-hydroxylase (cyp71av1) gene, encoding a key enzyme of artemisinin biosynthesis in Artemisia annua, as reported by a promoter-gus fusion. Plant Mol. Biol. 2013, 81, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.; Fu, C.; Habibi, G.; Mcmaster, W.R. Regions in the 3′ untranslated region confer stage-specific expression to the Leishmania mexicana a600-4 gene. Mol. Biol. Parasitol. 2007, 153, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Ure, M.E.; Heydari, E.; Pan, W.; Ramesh, A.; Rehman, S.; Morgan, C.; Pinsk, M.; Erickson, R.; Herrmann, J.M.; Dimke, H.; et al. A variant in a Cis-regulatory element enhances claudin-14 expression and is associated with pediatric-onset hypercalciuria and kidney stones. Hum. Mutat. 2017, 38, 649–657. [Google Scholar] [CrossRef]
- Villafuerte, B.C.; Barati, M.T.; Rane, M.J.; Isaacs, S.; Li, M.; Wilkey, D.W.; Merchant, M.L. Over-expression of insulin-response element binding protein-1 (IRE-BP1) in mouse pancreatic islets increases expression of RACK1 and TCTPP: Beta cell markers of high glucose sensitivity. BBA Proteins Proteom. 2017, 1865, 186–194. [Google Scholar] [CrossRef]
- Nishi-Tatsumi, M.; Yahagi, N.; Takeuchi, Y.; Toya, N.; Takarada, A.; Murayama, Y.; Aita, Y.; Sawada, Y.; Piao, X.; Oya, Y.; et al. A key role of nuclear factor Y in the refeeding response of fatty acid synthase in adipocytes. FEBS Lett. 2017, 591, 965–978. [Google Scholar] [CrossRef]
- Peters, B.; Casey, J.; Aidley, J.; Zohrab, S.; Borg, M.; Twell, D.; Brownfield, L. A Conserved cis-Regulatory Module Determines Germline Fate through Activation of the Transcription Factor DUO1 Promoter. Plant Physiol. 2017, 173, 280–293. [Google Scholar] [CrossRef]
- Nolan, K.E. Expression of the Somatic Embryogenesis Receptor-Like Kinase1 (SERK1) gene is associated with developmental change in the life cycle of the model legume Medicago truncatula. J. Exp. Bot. 2009, 60, 1759–1771. [Google Scholar] [CrossRef]
| Primer Name | Primer Sequence | References |
|---|---|---|
| GUSF | 5′-AACCGTTCTACTTTACTGGCTTTGG-3′ | Wang et al., 2013 [36] |
| GUSR | 5′-GCATCTCTTCAGCGTAAGGGTAAT-3′ | |
| Fluc-F | 5′-TGCACATATCGAGGTGGACATC-3′ | Murray et al., 2017 [37] |
| Fluc-R | 5′-AGAACCAGAAGAATTTGCAGCAT-3′ | |
| β-actinF | 5′-CTGGCCTACGTGGCACTTGACTT-3′ | Ma et al., 2012 [14] |
| β-actinR | 5′-CACTTCTGGGCAGCGGAACCTTT-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luan, A.; He, Y.; Xie, T.; Chen, C.; Mao, Q.; Wang, X.; Li, C.; Ding, Y.; Lin, W.; Liu, C.; et al. Identification of an Embryonic Cell-Specific Region within the Pineapple SERK1 Promoter. Genes 2019, 10, 883. https://doi.org/10.3390/genes10110883
Luan A, He Y, Xie T, Chen C, Mao Q, Wang X, Li C, Ding Y, Lin W, Liu C, et al. Identification of an Embryonic Cell-Specific Region within the Pineapple SERK1 Promoter. Genes. 2019; 10(11):883. https://doi.org/10.3390/genes10110883
Chicago/Turabian StyleLuan, Aiping, Yehua He, Tao Xie, Chengjie Chen, Qi Mao, Xiaoshuang Wang, Chuhao Li, Yaqi Ding, Wenqiu Lin, Chaoyang Liu, and et al. 2019. "Identification of an Embryonic Cell-Specific Region within the Pineapple SERK1 Promoter" Genes 10, no. 11: 883. https://doi.org/10.3390/genes10110883
APA StyleLuan, A., He, Y., Xie, T., Chen, C., Mao, Q., Wang, X., Li, C., Ding, Y., Lin, W., Liu, C., Xia, J., & He, J. (2019). Identification of an Embryonic Cell-Specific Region within the Pineapple SERK1 Promoter. Genes, 10(11), 883. https://doi.org/10.3390/genes10110883




