Abstract
The Himalayas are one of earth’s hotspots of biodiversity. Among its many cryptic and undiscovered organisms, including vertebrates, this complex high-mountain ecosystem is expected to harbour many species with adaptations to life in high altitudes. However, modern evolutionary genomic studies in Himalayan vertebrates are still at the beginning. Moreover, in organisms, like most amphibians with relatively high DNA content, whole genome sequencing remains bioinformatically challenging and no complete nuclear genomes are available for Himalayan amphibians. Here, we present the first well-annotated multi-tissue transcriptome of a Greater Himalayan species, the lazy toad Scutiger cf. sikimmensis (Anura: Megophryidae). Applying Illumina NextSeq 500 RNAseq to six tissues, we obtained 41.32 Gb of sequences, assembled to ~111,000 unigenes, translating into 54362 known genes as annotated in seven functional databases. We tested 19 genes, known to play roles in anuran and reptile adaptation to high elevations, and potentially detected diversifying selection for two (TGS1, SENP5) in Scutiger. Of a list of 37 genes, we also identify 27 candidate genes for sex determination or sexual development, all of which providing the first such data for this non-model megophryid species. These transcriptomes will serve as a valuable resource for further studies on amphibian evolution in the Greater Himalaya as a biodiversity hotspot.
1. Introduction
The Himalayas are a distinct biogeographic eco-region with high biodiversity and endemism due to great topographic and climatic variation [1] and isolation. The uplift of Tibet and the Himalayas since about 45 million years ago (Mya), with the Greater Himalayas starting to rise presumably the earliest in the post-Eocene, or even more recently (~20–10 Mya; for a review see the supplementary in Hofmann et al. [2]) appears to have resulted in a unique assemblage of species evolved under gradual high-altitude adaptation, as already shown for several Tibetan reptiles [3] and anurans [4,5] as well as interspecies comparisons with evidence for parallel evolution [6,7,8]. However, with respect to high-altitude adaptation in the Greater Himalayas, there is still a considerable knowledge deficiency as there is a general lack of knowledge about the biodiversity of this high-mountain range, causing its relatively large number of cryptic and undiscovered species [9], even among vertebrates. Despite its poorly accessible landscapes, the fragile and vulnerable Himalayan mountain ecosystems have not been spared from land use changes, anthropogenic habitat degradation, and other increasing pressures of growing human populations as well as tourism, and the region especially suffers from global climate change [10]. The associated loss and changing conditions for biodiversity are alarming and many species may be lost before they are known [11,12].
Molecular evolutionary studies of truly Greater Himalayan animal species are extremely rare (e.g., [13]) and, particularly, genomic data are lacking for most vertebrates (e.g., a PubMed search for “genomic” and “Himalaya” yielded 72 hits, only 18 when including “vertebrates” but zero for “amphibians”). To allow future studies in population genomics and phylogenetics, for example, using sequences capture approaches and to generally close this major data gap for regional amphibians, in the present paper, we have generated a new molecular marker set based on RNAseq for an anuran species from the Greater Himalaya.
Our target species is a megophryid toad, the Sikkim lazy toad (an alternative common name is “alpine toad”). Since there is great taxonomic uncertainty among Himalayan Scutiger [2], we refer to this specimen as Scutiger cf. sikimmensis. Importantly, so far, no well-annotated multi-tissue nuclear transcriptome or genome of a Scutiger species has been characterized, although transcriptomes of the skin [14] and pooled larval tissues [15] as well as brain, testes and skin [16] of related taxa from the family Megophryidae have recently been published. Megophryidae represent the sister group of Pelobatidae [17,18], and form a highly species-diverse family of oriental anurans with a basal phylogenetic position relative to the Neobatrachia [17,19,20,21,22,23]. All species comprise stream-breeding, forest ground-dwellers with toad-like morphology [24]. Lazy toads are a characteristic faunal element of the Himalaya-Tibet-Orogen. This genus comprises 23 recognized species, most of which distributed in the Hengduan Shan and adjacent regions to the east (Province Sichuan, China). Of these 23 taxa, at least six nominal species are only known from their type localities [2,25]. The Himalayan Scutiger group has been shown to be an excellent model for phylogeographic and evolutionary research due to its high local endemism (associated with many, so far undescribed lineages), the extremely limited distributional ranges and adaptation to high elevations of these species and the occurrence of strictly allopatric speciation [2,26,27]. During the last 15 years, four new Scutiger-species have been described, highlighting the potentially great number of undiscovered taxa. However, there is poor knowledge about species’ ranges in the Greater Himalaya and adjacent regions, as well as about their taxonomic diversity [28,29] and evolution [2], and this deficit is accompanied by a lack of high-quality molecular data.
With this study, we publish the first multi-tissue transcriptome of a high-altitude amphibian species from the Greater Himalaya and report genes known to play roles in adaptation of vertebrates to high elevations as well as in sex determination and sexual differentiation of this non-model amphibian. These data provide robust resources for future evolutionary investigations in Scutiger.
2. Materials and Methods
2.1. Animal Sampling and Ethics Statement
A single male Scutiger cf. sikimmensis was collected in Central Nepal, near Dunche (28.05 N, 85.26 E, 3289 m a.s.l.; Figure 1). Samples were collected in accordance with regulations for the protection of terrestrial wild animals under the permits of the Nepal expeditions of the Natural History Museum of Erfurt, Germany [30,31]. The male specimen was anesthetised in the field by immersion in tricaine methanesulfonate (MS 222; Sigma-Aldrich), sacrificed by decapitation, dissected and small pieces of organs transferred into RNAlater (Thermo Fisher), kept on ambient temperature during the time of the field work and later stored at −80 °C.

Figure 1.
Scutiger cf. sikimmensis found in the Central Himalaya at 3289 m.
2.2. RNA Extraction, cDNA Library Construction, and Illumina Sequencing
Total RNA was extracted from six different tissues (brain, heart, liver, lung, kidney, testes) and adjusted to equal concentrations. RNA integrity was assessed by RNA concentration, RIN value, 28S/18S and the fragment length distribution using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., CA, USA). Complementary DNA (cDNA) was synthesized and sequenced by BGI (BGI-Hongkong Co., Ltd.), using the Illumina NextSeq 500 sequencing system (Illumina, San Diego, USA).
2.3. Filtering, De Novo Assembly, Functional Annotation and Gene Expression
The raw Illumina reads were filtered by quality and for adaptor contamination, targeting reads containing adaptor, reads with more than 5% ambiguous bases, or reads with greater than 20% of bases with quality score below 15. The remaining high-quality, clean reads were de novo assembled to transcripts using Trinity v2.0.6 [32], based on the de Bruijn graph algorithm. Transcripts were clustered using Tgicl v2.0.6 [33] to obtain sequences that could no longer be extended. The resulting sequences were defined as unigenes. They represent expressed sequences, but are not characterized sufficiently to be denoted as a complete gene. The entire unigene set was functionally annotated by matching against seven frequently used databases, including Gene Orthology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), EuKaryotic Orthologous Groups (KOG), the non-redundant nucleotide (NT) database, the non-redundant (NR) protein database, SwissProt, and InterPro. We used Diamond v0.8.31 [34] or the BLASTx [35] algorithm with an E-value threshold of 1.0 × 10−5 to align unigenes to KEGG, KOG, NR, and NT and SwissProt for the annotation. Unigenes that matched to the NR database were annotated to the GO database with Blast2GOv2.5.0 [36] and classified into three categories: biological process, cellular components and molecular functions. BLAST2GO was also used to identify protein domains against the InterPro databases using the InterProScan5 v5.11-51.0 tool [37]. Candidate coding areas of these unigenes were predicted using TransDecoder v.3.0.1 (https://github.com/TransDecoder/TransDecoder) and the Pfam protein homologous sequences were searched by Blast to SwissProt and hmmscan v3.0 [38] to predict the coding region. For a coding sequence with more than two ORF, the longest one was identified as the sequence of interest. We mapped all unigenes to AnimalTFDB2.0 database and EMBOSS getorf v6.5.70 [39] to find ORF of each unigene and aligned the ORF to Transcription Factor domains using hmmsearch.
For each sample, expression profiles were obtained by mapping clean paired-end reads back to unigenes using Bowtie2 v2.2.5 [40] with default parameters. Raw expression counts were then calculated with RSEM v1.1.20 [41]. The raw counts were normalized into FPKM values to quantify transcript levels among the different tissue samples.
Data generated in this study are publicly available from the NCBI GenBank database under the Bioproject ID PRJNA532534. All clean sequence data were deposited in the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra/) under the accession numbers SRR9953599-SRR9953604, assembled sequences were transmitted to NCBI Transcriptome Shotgun Assembly Sequence Database (TSA, http://www.ncbi.nlm.nih.gov/genbank/tsa); the annotation dataset and statistics have been uploaded to figshare (doi 10.6084/m9.figshare.9913202).
2.4. Tests for Episodic Diversification in Selected Genes, Relevant for High Altitude Adaptation
We used the 21 most prominent genes, exhibiting convergent and continuous genetic adaptation to high elevations in Ranidae and Agamidae as provided by the authors of [6] upon request for assembled transcripts and annotated coding regions; thus, no whole-transcriptome selection analysis was feasible. The unigene annotation was screened for matches to the gene symbols of altitude adapted genes. Matching unigenes were filtered from the assembled transcriptome sequences. These candidate sequences were aligned with NR protein database to manually confirm the gene assignment. Coding sequence (CDS) of the transcripts was determined with TransDecoder. CDS were aligned codon based with the sequences from Sun et al. [6] using TranslatorX [42]. Finally, we removed stop codons and all codons that contained unaligned positions in at least one species. The alignments were submitted to aBSREL analysis at www.datamonkey.org [43,44], and therein were analyzed for potential diversifying selection at all branches, comparing selection through uncorrected p-values in the Nanorana-clade to those in Scutiger.
2.5. Identification of Genes Involved in Sex Determination or Sex Differentiation
Using a list of anuran genes presumably involved in male or female sex determination and sexual differentiation [45] to identify templates, we recovered such genes from the multiple tissues in Scutiger and studied their tissue-specific expression. Template protein sequences from the anuran model species Xenopus tropicalis or Xenopus laevis were obtained from Xenbase [46] and aligned with BLAT [47] (−t = dnax –q = prot) against Scutiger transcripts assembled from RNAseq data. Matching transcripts were identified and aligned to the NR database with BLASTx [35,48]. After manual inspection, we identified and removed paralogs or unspecific sequences. Unigenes were considered homologous, if the majority of the top scoring BLASTx hits were close to the gene of interest.
3. Results
3.1. Transcriptome
From separate cDNA libraries of six tissues (brain, heart, kidney, liver, lung, testis), a total of 425.4 million raw sequence data (41.32 Gb) were generated. After filtering and quality checks, 413.4 million clean reads were obtained, with an average of 68.9 million reads per tissue (Table 1). The GC-content of these clean reads ranged between 44.78% and 46.19%; 97% of all bases had a quality score of at least Q20. Over all samples, the assembly resulted in 110,889 non-redundant unigene sequences with a total length of 104,314,420 bp. Unigenes had an average length of 940 bp and a N50 of 1926 bp. A total of 103,396 unigenes ranged in length from 300 to 3000 bp (Table 2, Figure 2 and Supplementary Figure S1).

Table 1.
Summary of transcriptome sequencing based on RNA samples from six different tissues.

Table 2.
Summary of assembled unigenes per tissue.
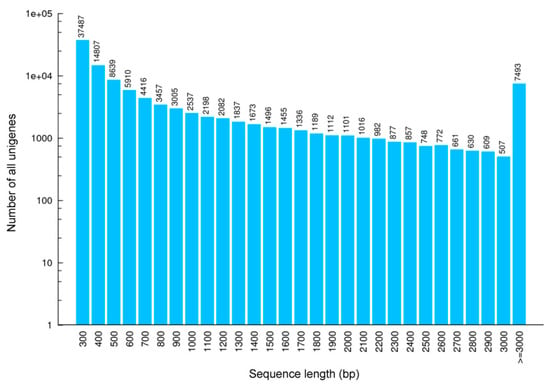
Figure 2.
Length distribution of all unigenes based on RNA samples from different tissues of Scutiger cf. sikimmensis. The x-axis represents the sequence length (base pairs), while the y-axis represents the number of transcripts.
3.2. Functional Annotation
Altogether, 54362 (49.02%) unigenes showed homology to entries in at least one of the seven functional databases (Figure 3, Supplementary Table S1); 11,933 unigenes were annotated by all these databases (Supplementary File S1). Most matches were obtained in the NR database, with 48805 unigenes (44.01%), of which 26,071 (23.51%) could be assigned to gene ontology terms. Regarding the biological process ontology, the most common classifications were cellular processes (13,065), metabolic processes (9499) and biological regulation (6523). The most common categories for the cellular component ontology were cellular (13,125), cell compartments (13,004) and organelles (9276). In terms of the molecular function, most common categories involved binding (13,958), catalytic activity (9871) and molecular transporter activity (1479) (Supplementary Figure S2).
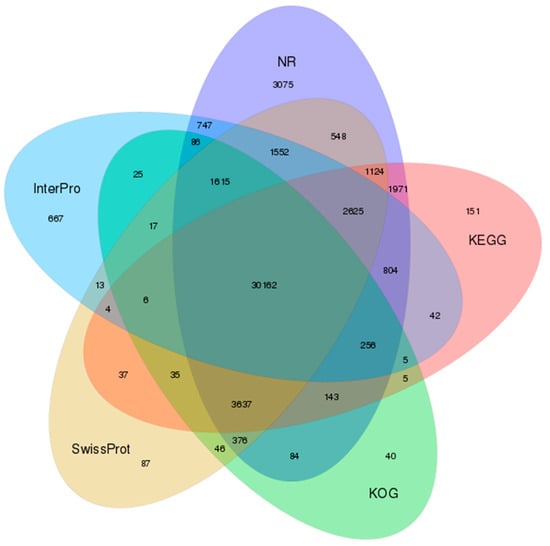
Figure 3.
Venn diagram of shared and unique unigenes in Scutiger cf. sikimmensis among the five most informative databases used for annotation. GO = gene orthology; InterPro = integrative protein signature database; KEGG = Kyoto Encyclopedia of Genes and Genomes; KOG = EuKaryotic Orthologous Groups; NR = non-redundant protein database; NT = non-redundant nucleotide database; SwissProt = Swiss Protein Sequence Database.
The KOG functional classification revealed general function (8788), signal transduction mechanisms (8017 unigenes), posttranslational modification (3845 unigenes), unknown function (3430 unigenes) and transcription (3113) as top five categories (Supplementary Figure S3).
We also performed a search of all unigenes against the KEGG database to identify the active biological pathways in Scutiger cf. sikimmensis. We found 41,007 unigenes that shared homology to entries, mapping to pathways related to cellular processes (8951), environmental information processing (8646), genetic information processing (6562), human diseases (20053), metabolism (13259) and organismal systems (15435). Predominantly, the unigenes were enriched in “signal transduction” (6573), followed by “global and overview maps” (5042), “cancers: overview” (4111), “infectious diseases: viral” (3725), and “immune system” (3633) categories (Figure S4).
3.3. Testing Potential Genetic Adaptation to High Elevations
From the Scutiger transcriptome-based gene set, we were able to successfully align 19 out of 21 anuran genes, exhibiting convergent and continuous genetic adaptation to high elevations in Ranidae and Agamidae (Table in Figure 1 in Sun et al. 2018 [6]). We did not recover the genes XPS and TINAGL1 from Scutiger. Using aBSREL, we then examined each of the 19 alignments for the occurrence of episodic diversification. We detected evidence of diversifying selection in two Scutiger genes (TGS1: uncorr. p = 0.0339, corr. p = 0.2373; SENP5: uncorr. p = 0.0053, corr. p = 0.0475). In all analyses, the test statistics confirmed the selection shown by Sun et al. in Nanorana, as reported by these authors (Supplementary File S2: note that corrected p-values are provided therein as “Test p-value”) [6].
3.4. Genes Related to Sex Determination or Sex Differentiation and Their Expression Levels
Of a list of 37 genes from the clawed frog genomes, about 73% (27) had significant blast hits in the Scutiger transcriptomes, all of which provide the first sequence data for these genes in this non-model anuran species. In comparison to the length of Xenopus mRNAs, which were used as BLAST templates, the majority of the Himalayan Scutiger unigenes is shorter (between 4% and 98%) and may thus represent fragments. However, unigenes are also up to 3.5 times longer, which may be indicative of insertions (Table 3). The majority (∼49%) of the target genes were expressed in all tissues and only ∼5% were expressed solely in a single one.

Table 3.
Sex-determining and sex differentiation gene inventory and homologous Scutiger cf. sikimmensis transcripts. Gene expression is calculated as FPKM based on RNA-seq data from six tissues; the highest gene expression level per gene over all tissues is indicated in bold. CL = cluster of several unigenes.
4. Discussion
We here report the first well-annotated transcriptome data of a high-altitude amphibian taxon from the Greater Himalaya, based on RNAseq of multiple tissues. Our work, based on a male Scutiger cf. sikimmensis from central Nepal, provides a transcriptome of a representative species from a branch of the amphibian tree of life (Pelobatoidea of some authors [49]; Mesobatrachia of others [50], currently Megophryidae [27]), from which no complete genomes have been sequenced. Although genomic approaches have been used [14,15], to our knowledge no annotated multi-tissue transcriptome has been published for the genus Scutiger. Our study is mostly descriptive, but it has yielded novel discoveries and represents an important turning point for genomic studies in megophryd anurans.
4.1. Indications for Potential Genetic Adaptation to High Elevations in Scutiger
Sun et al. [6] used comparative transcriptomic analyses to prove that amphibian and reptile populations, occurring at different altitudes around the Tibetan Plateau, show parallel evolution. They have provided evidence for convergent and continuous genetic adaptation to high elevations in taxa as distant as Anura (Ranidae) and Sauropsida (Agamidae), in which genes with related functions, especially DNA-repair and energy metabolism pathways, exhibit evidence for rapid change and continuous positive selection with increasing elevations. These data let us assume that a similar genomic high-elevation-selection-syndrome might be detectable in Scutiger, sampled in 3289 m above sea level (Methods). Indeed, in two out of 19 key genes [6], we have detected diversifying selection and thus potentially positive selection in the Scutiger transcriptome as well. In another three of these key genes (PGS: uncorr. p = 0.180, corr. p = 1.000; OLFM4: uncorr. p = 0.0818, corr. p = 0.6542; PPIL2: uncorr. p = 0.185, corr. p = 1.000) we obtained uncorrected p-values close to significance level, possibly indicating weak signs of adaptive molecular evolution. However, the analyses of only a single Scutiger-transcriptome (data were unobtainable from a whole radiation of species or an altitudinal gradient as available for Ranidae [6]) limits our approach and did not allow for further comparative tests. Therefore, future research with multiple species and a greater scale of altitudinal variation might yield additional evidence for genes adapted to the life in high altitudes in lazy toads. This seems especially justified since similar traits have been studied extensively in other vertebrates, such as mammals including humans [51].
4.2. Expression of Genes Involved in Sex Determination or Sex Differentiation
As a second application, we used the transcriptomes of Scutiger cf. sikimmensis to search for candidate genes, known for their role in sex determination or sex differentiation in other vertebrates (e.g., [45]). Most, if not all, anurans exhibit genetic sex determination [52,53,54] and generally exhibit sex-biased gene expression, depending on phenotypic sex [55]. As most anurans [56], to our knowledge, Scutiger exhibits homomorphic sex chromosomes and nothing is known about sex determination in Scutiger.
Of the 27 genes with potential sex roles, detected in Scutiger cf. sikimmensis, two tissues, the brain (32%) and the testes (22%), exhibited the highest numbers of highly expressed genes, as typical of sex determination or differentiation genes. About a quarter of the relevant 37 genes have remained uncharacterized, presumably, since their levels of expression was below the threshold to be assembled into unigenes or might be expressed only during earlier developmental stages or exclusively in females.
5. Conclusions
Overall, our data provide the first well-annotated multi-tissue transcriptomic resource for a Himalayan amphibian of the genus Scutiger. This transcriptome is available for further studies on evolution and adaptation in Himalayan high-altitude vertebrate species.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/11/873/s1, Figure S1: Frequency length distribution of unigenes in the Scutiger cf. sikimmensis transcriptome, Figure S2: Gene ontology classification of Scutiger cf. sikimmensis transcriptome unigenes, Figure S3: KOG functional classification of Scutiger cf. sikimmensis transcriptome unigenes, Figure S4: Functional distributions of Scutiger cf. sikimmensis transcriptome unigenes annotated by KEGG. Table S1: Annotation summary for the transcriptomes of Scutiger cf. sikimmensis, File S1: Annotation results of unigenes from five databases, File S2: Alignments and test statistics for 19 genes, potentially exhibiting diversifying selection in Scutiger cf. sikimmensis in comparison with ranid frogs [6].
Author Contributions
Conceptualization, S.H. and M.S.; methodology, S.H., C.B.B., H.K., and M.S.; formal analysis, S.H., H.K., and M.S.; resources, S.H., C.B.B., and M.S.; writing—original draft preparation, S.H.; writing—review and editing, C.B.B., H.K., and M.S.; visualization, S.H.; project administration, S.H.; funding acquisition, S.H. and M.S.
Funding
This research was funded in part by the German Research Foundation (DFG), grant number Sto493/2-2.
Acknowledgments
We thank Nadine Possnien for RNA extraction and quantification; David M. Hillis and Yan-Bo Sun for sending 21 gene alignments for anurans and lizards for comparative selection analyses. We thank Santos Tamang and colleagues for help during the fieldwork.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Mittermeier, R.A.; Robles-Gil, P.; Hoffman, M.; Pilgrim, J.; Brooks, T.; Mittermeier, C.G.; Lamoreux, J.; da Fonseca, G.A.B. Hotspots Revisited; CEMEX: Mexico City, Mexico, 2004; p. 392. [Google Scholar]
- Hofmann, S.; Stoeck, M.; Zheng, Y.; Ficetola, F.G.; Li, J.T.; Scheidt, U.; Schmidt, J. Molecular Phylogenies Indicate a Paleo-Tibetan Origin of Himalayan Lazy Toads (Scutiger). Sci. Rep. 2017, 7, 3308. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Qi, Y.; Fu, J. Exploring the Genetic Basis of Adaption to High Elevations in Reptiles: A Comparative Transcriptome Analysis of Two Toad Headed Agamas (Genus Phrynocephalus). PLoS ONE 2014, 9, e112218. [Google Scholar]
- Wang, G.-D.; Zhang, B.-L.; Zhou, W.-W.; Li, Y.-X.; Jin, J.-Q.; Shao, Y.; Yang, H.-C.; Liu, Y.-H.; Yan, F.; Chen, H.-M.; et al. Selection and Environmental Adaptation Along a Path to Speciation in the Tibetan Frog Nanorana parkeri. Proc. Natl. Acad. Sci. USA 2018, 115, E5056–E5065. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Zhou, W.-W.; Hu, J.-S.; Yan, F.; Papenfuss, T.J.; Wake, D.B.; Zhang, Y.-P. Spiny frogs (Paini) illuminate the history of the Himalayan region and Southeast Asia. Proc. Natl. Acad. Sci. USA 2010, 107, 13765–13770. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.B.; Fu, T.T.; Jin, J.Q.; Murphy, R.W.; Hillis, D.M.; Zhang, Y.P.; Che, J. Species Groups Distributed across Elevational Gradients Reveal Convergent and Continuous Genetic Adaptation to High Elevations. Proc. Natl. Acad. Sci. USA 2018, 115, E10634–E10641. [Google Scholar] [CrossRef]
- Yang, W.J.; Qi, Y.; Bi, K.; Fu, J. Toward Understanding the Genetic Basis of Adaption to High-Elevation Life in Poikilothermic Species: A Comparative Transcriptomic Analysis of Two Ranid Frogs, Rana chensinensis and R. kukunoris. BMC Genom. 2012, 13, 588. [Google Scholar] [CrossRef]
- Yang, W.; Qi, Y.; Fu, J. Genetic Signals of High-Altitude Adaption in Amphibians: A Comparative Transcriptome Analysis. BMC Genet. 2016, 17, 134. [Google Scholar] [CrossRef]
- Mosbrugger, V.; Favre, L.; Müllner-Riehl, A.; Päckert, M.; Mulch, A. Cenozoic Evolution of Geo-Biodiversity in the Tibeto-Himalayan Region. In Mountains, Climate and Biodiversity; Hoorn, C., Perrigo, A., Antonelli, A., Eds.; Wiley-Blackwell: Oxford, UK, 2018; pp. 429–449. [Google Scholar]
- Bolch, T.; Kulkarni, A.; Kääb, A.; Huggel, C.; Paul, F.; Cogley, J.G.; Fey, H.; Kargel, J.S.; Fujita, K.; Scheel, M.; et al. The State and the Fate of Himalayan Glaciers. Science 2012, 336, 310–314. [Google Scholar] [CrossRef]
- Mittermeier, R.A.; Turner, W.R.; Larsen, F.W.; Brooks, T.M.; Gascon, C. Global Biodiversity Conservation: The Critical Role of Hotspots. In Biodiversity Hotspots; Zachos, F.E., Habel, J.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 3–22. [Google Scholar]
- Pandit, M.K.; Navjot, S.S.; Koh, L.P.; Bhaskar, A.; Brook, B.W. Unreported yet Massive Deforestation Driving Loss of Endemic Biodiversity in Indian Himalaya. Biodivers. Conserv. 2017, 16, 153–163. [Google Scholar] [CrossRef]
- Price, T.D.; Hooper, D.M.; Buchanan, C.D.; Johansson, U.S.; Tietze, D.T.; Alström, P.; Olsson, U.; Ghosh-Harihar, M.; Ishtiaq, F.; Gupta, S.K.; et al. Niche Filling Slows the Diversification of Himalayan Songbirds. Nature 2014, 509, 222–225. [Google Scholar] [CrossRef]
- Huang, L.; Li, J.; Anboukaria, H.; Luo, Z.; Zhao, M.; Wu, H. Comparative Transcriptome Analyses of Seven Anurans Reveal Functions and Adaptations of Amphibian Skin. Sci. Rep. 2016, 6, 24069. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Liu, L.; Wang, X.; Gao, X.; Jiang, J.; Wang, B. Transcriptomics Reveals the Molecular Processes of Light-Induced Rapid Darkening of the Non-Obligate Cave Dweller Oreolalax Rhodostigmatus (Megophryidae, Anura) and Their Genetic Basis of Pigmentation Strategy. BMC Genom. 2018, 19, 422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Guo, Y.; Li, J.; Huang, L.; Kazitsa, E.G.; Wu, H. Transcriptome Analysis Reveals the Genetic Basis Underlying the Seasonal Development of Keratinized Nuptial Spines in Leptobrachium boringii. BMC Genom. 2016, 17, 978. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A.; Wiens, J.J. A Large-Scale Phylogeny of Amphibia Including over 2800 Species, and a Revised Classification of Extant Frogs, Salamanders, and Caecilians. Mol. Phylogenet. Evol. 2011, 61, 543–583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liang, D.; Mao, R.L.; Hillis, D.M.; Wake, D.B.; Cannatella, D. Efficient Sequencing of Anuran Mtdnas and a Mitogenomic Exploration of the Phylogeny and Evolution of Frogs. Mol. Biol. Evol. 2013, 30, 1899–1915. [Google Scholar] [CrossRef] [PubMed]
- Irisarri, I.; Vences, M.; San Mauro, D.; Glaw, F.; Zardoya, R. Reversal to Air-Driven Sound Production Revealed by a Molecular Phylogeny of Tongueless Frogs, Family Pipidaex. BMC Evol. Biol. 2011, 11, 114–123. [Google Scholar] [CrossRef]
- Frost, D.R.; Grant, T.; Faivovich, J.N.; Bain, R.H.; Haas, A.; Haddad, C.F.B.; De Sa, R.O.; Channing, A.; Wilkinson, M.; Donnellan, S.C.; et al. The Amphibian Tree of Life. Bull. Am. Mus. Nat. Hist. 2006, 2006, 1–291. [Google Scholar] [CrossRef]
- Roelants, K.; Bossuyt, F. Archaeobatrachian Paraphyly and Pangaean Diversification of Crown-Group Frogs. Syst. Biol. 2005, 54, 111–126. [Google Scholar] [CrossRef]
- Roelants, K.; Gower, D.J.; Wilkinson, M.; Loader, S.P.; Biju, S.D.; Guillaume, K.; Moriau, L.; Bossuyt, F. Global Patterns of Diversification in the History of Modern Amphibians. Proc. Natl. Acad. Sci. USA 2007, 104, 887–892. [Google Scholar] [CrossRef]
- Wiens, J.J. Global Patterns of Diversification and Species Richness in Amphibians. Am. Nat. 2007, 170, 86–106. [Google Scholar] [CrossRef]
- Ohler, A.; Wollenberg, K.C.; Grosjean, S.; Hendrix, R.; Vences, M.; Ziegler, T.; Dubois, A. Sorting out Lalos: Description of New Species and Additional Taxonomic Data on Megophryid Frogs from Northern Indochina (Genus Leptolalax, Megophryidae, Anura). Zootaxa 2011, 3147, 1–83. [Google Scholar] [CrossRef]
- Yang, J.-H.; Huang, X.-Y. A New Species of Scutiger (Anura: Megophryidae) from the Gaoligongshan Mountain Range, China. Copeia 2019, 107, 10–21. [Google Scholar] [CrossRef]
- Fei, L. Atlas of Amphibians of China; Henan Science and Technology Press: Zhengzhou, China, 1999; p. 432. [Google Scholar]
- Frost, D.R. Amphibian Species of the World: An Online Reference. Version 6.0. Electronic Database. American Museum of Natural History. Available online: http://research.amnh.org/herpetology/amphibia/index.html (accessed on 4 September 2019).
- Chen, W.; Bi, K.; Fu, J. Frequent Mitochondrial Gene Introgression among High Elevation Tibetan Megophryid Frogs Revealed by Conflicting Genegenealogies. Mol. Ecol. 2019, 18, 2856–2876. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Li, X.; Qiao, P. Population Structure, Historical Biogeography and Demographic History of the Alpine Toad Scutiger ningshanensis in the Tsinling Mountains of Central China. PLoS ONE 2014, 9, e100729. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Weipert, J. Biodiversität Und Naturausstattung Im Himalaya V; Verein der Freunde und Förderer des Naturkundemuseums Erfurt e.V.: Erfurt, Germany, 2015. [Google Scholar]
- Hartmann, M.; Weipert, J.; Weigel, A. Die Zoologischen Nepal-Expeditionen Des Naturkundemuseums Erfurt [the Zoological Expeditions of the Natural History Museum of Erfurt 1992 to 1997]. Veröffentlichungen Naturkundemuseums Erfurt 1998, 17, 15–30. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a Full-Length Transcriptome without a Genome from RNA-Seq Data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. Tigr Gene Indices Clustering Tools (Tgicl): A Software System for Fast Clustering of Large Est Datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using Diamond. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Aligment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S. Blast2go: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. Interproscan: Protein Domains Identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: Hmmer3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. Emboss: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. Rsem: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple Alignment of Nucleotide Sequences Guided by Amino Acid Translations. Nucleic Acids Res. 2010, 38, 7–13. [Google Scholar] [CrossRef]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773777. [Google Scholar] [CrossRef]
- Delport, W.; Poon, A.F.; Frost, S.D.W.; Kosakovsky Pond, S.L. Datamonkey 2010: A Suite of Phylogenetic Analysis Tools for Evolutionary Biology. Bioinformatics 2010, 26, 2455–2457. [Google Scholar] [CrossRef]
- Gerchen, J.F.; Reichert, S.J.; Röhr, J.T.; Dieterich, C.; Kloas, W.; Stöck, M. A Single Transcriptome of a Green Toad (Bufo viridis) Yields Candidate Genes for Sex Determination and -Differentiation and Non-Anonymous Population Genetic Markers. PLoS ONE 2016, 11, e0156419. [Google Scholar] [CrossRef]
- James-Zorn, C.; Ponferrada, V.G.; Burns, K.A.; Fortriede, J.D.; Lotay, V.S.; Liu, Y.; Brad Karpinka, J.; Karimi, K.; Zorn, A.M.; Vize, P.D. Xenbase: Core Features, Data Acquisition, and Data Processing. Genesis 2015, 53, 486–497. [Google Scholar] [CrossRef]
- Kent, W.J. Blat--the Blast-Like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped Blast and Psi-Blast: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Dubois, A. Amphibia Mundi. 1.1. An Ergotaxonomy of Recent Amphibians. Alytes 2005, 23, 1–24. [Google Scholar]
- San Mauro, D.; Vences, M.; Alcobendas, M.; Zardoya, R.; Meyer, A. Initial Diversification of Living Amphibians Predated the Breakup of Pangaea. Am. Nat. 2005, 165, 590–599. [Google Scholar] [CrossRef]
- Witt, K.E.; Huerta-Sánchez, E. Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180235. [Google Scholar] [CrossRef]
- Miura, I. An Evolutionary Witness: The Frog Rana rugosa Underwent Change of Heterogametic Sex from Xy Male to Zw Female. Sex. Dev. 2007, 1, 323–331. [Google Scholar] [CrossRef]
- Furman, B.L.S.; Evans, B.J. Sequential Turnovers of Sex Chromosomes in African Clawed Frogs (Xenopus) Suggest Some Genomic Regions Are Good at Sex Determination. G3 Genes Genomes Genet. 2016, 6, 3625–3633. [Google Scholar] [CrossRef]
- Beukeboom, L.W.; Perrin, N. The Evolution of Sex Determination; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Ma, W.J.; Veltsos, P.; Sermier, R.; Parker, D.J.; Perrin, N. Evolutionary and Developmental Dynamics of Sex-Biased Gene Expression in Common Frogs with Proto-Y Chromosomes. Genome Biol. 2018, 19, 156. [Google Scholar] [CrossRef]
- Eggert, C. Sex Determination: The Amphibian Models. Reprod. Nutr. Dev. 2004, 44, 539–549. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).