i6mA-DNCP: Computational Identification of DNA N6-Methyladenine Sites in the Rice Genome Using Optimized Dinucleotide-Based Features
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset
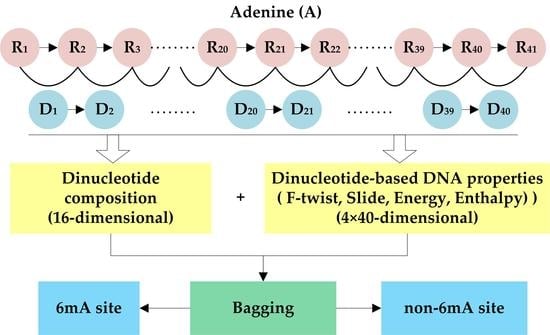
2.2. Feature Extraction
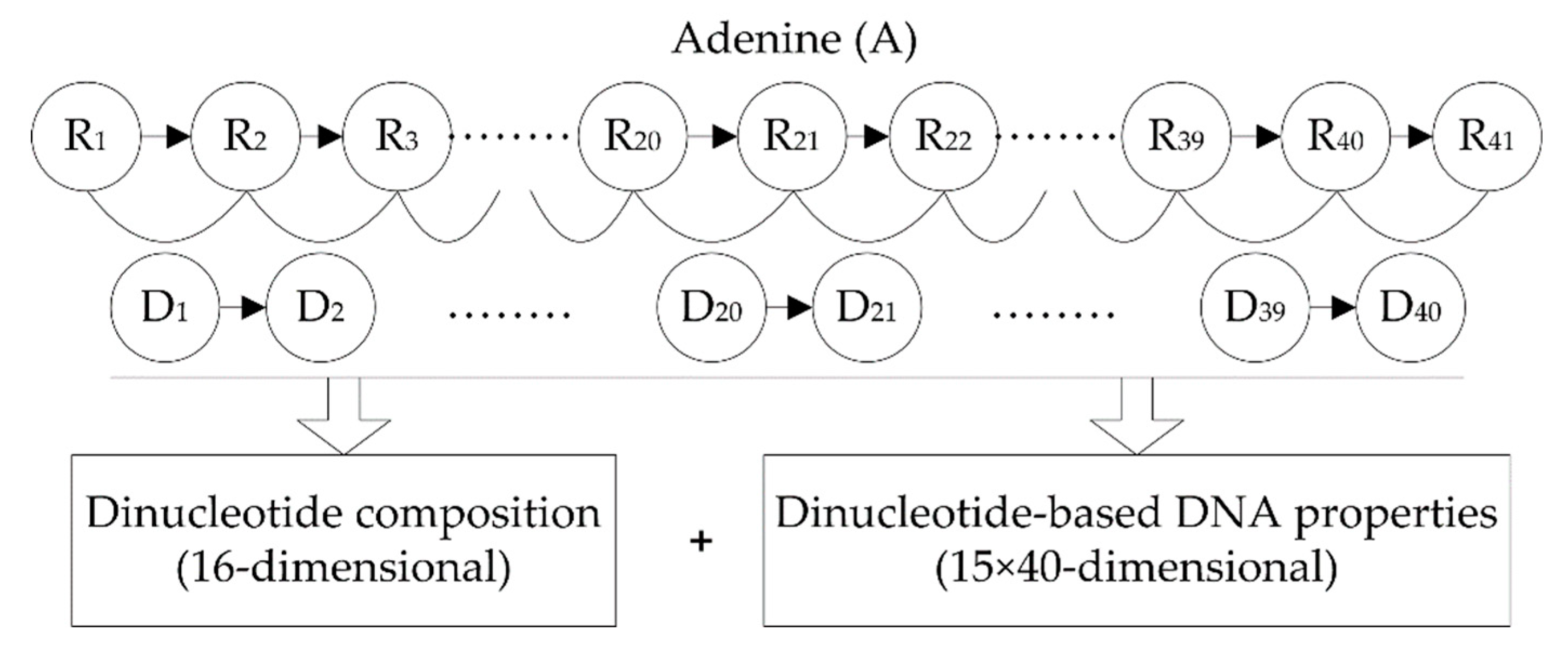
2.2.1. Dinucleotide Composition
2.2.2. Dinucleotide-Based DNA Properties
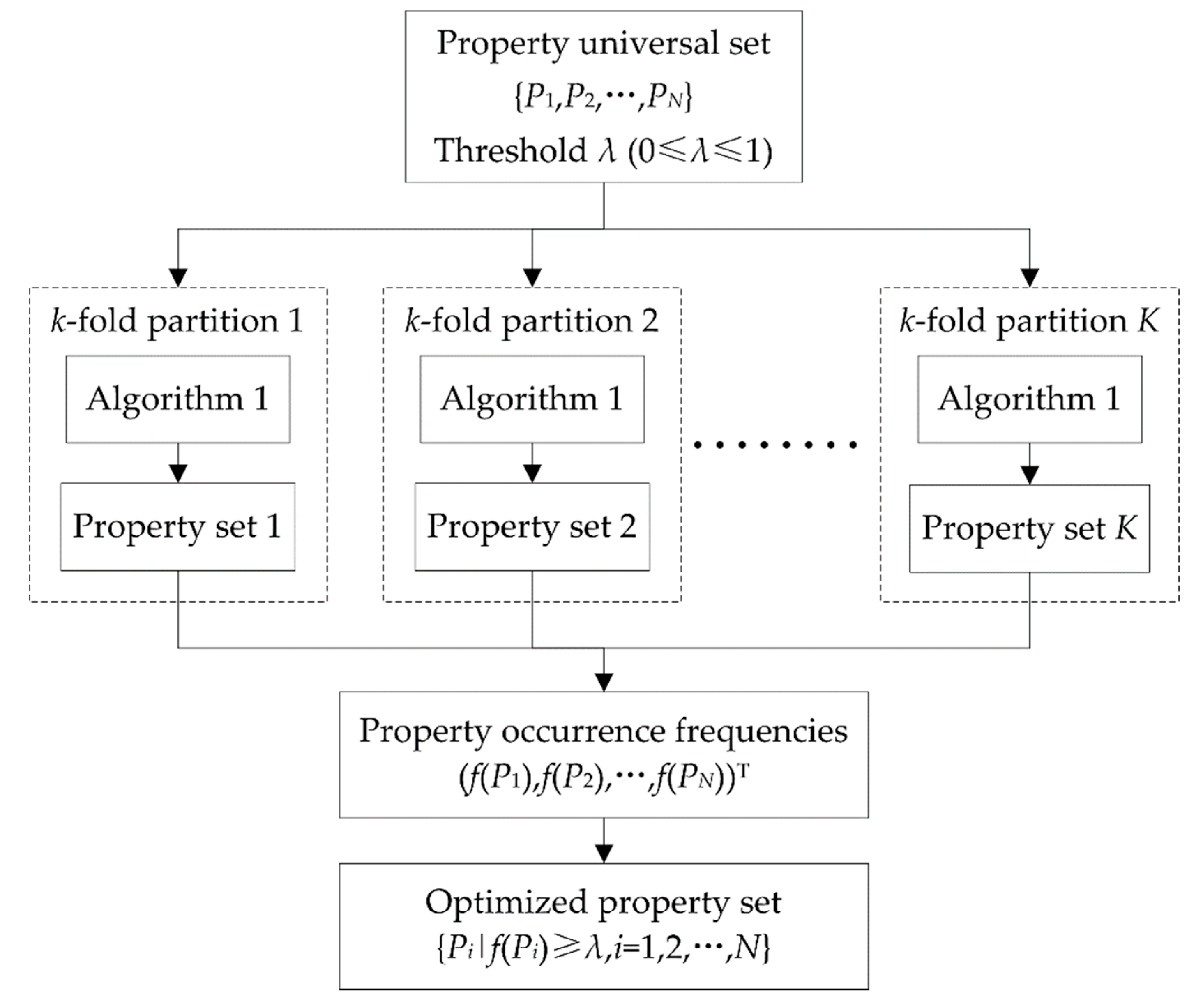
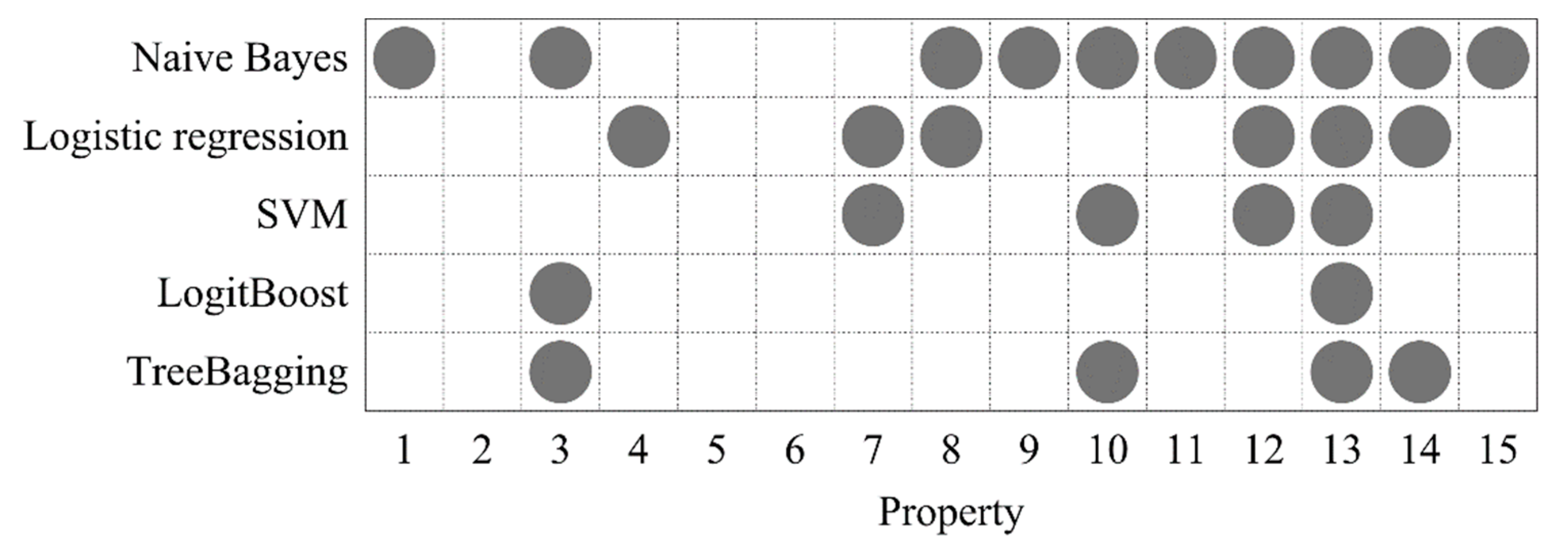
2.3. DNA Property Selection
| Algorithm 1. Heuristic DNA property selection. |
| Input: Universal set Output: Optimized property set S |
| 1. 2. is the accuracy corresponding to S 3. while do 4. for each property do 5. generating a candidate set 6. calculating 7. end for 8. 9. if do 10. 11. 12. else 13. break while 14. end if 15. end while 16. return S |
2.4. Classification Algorithms
2.5. Performance Evaluation
3. Results and Discussion
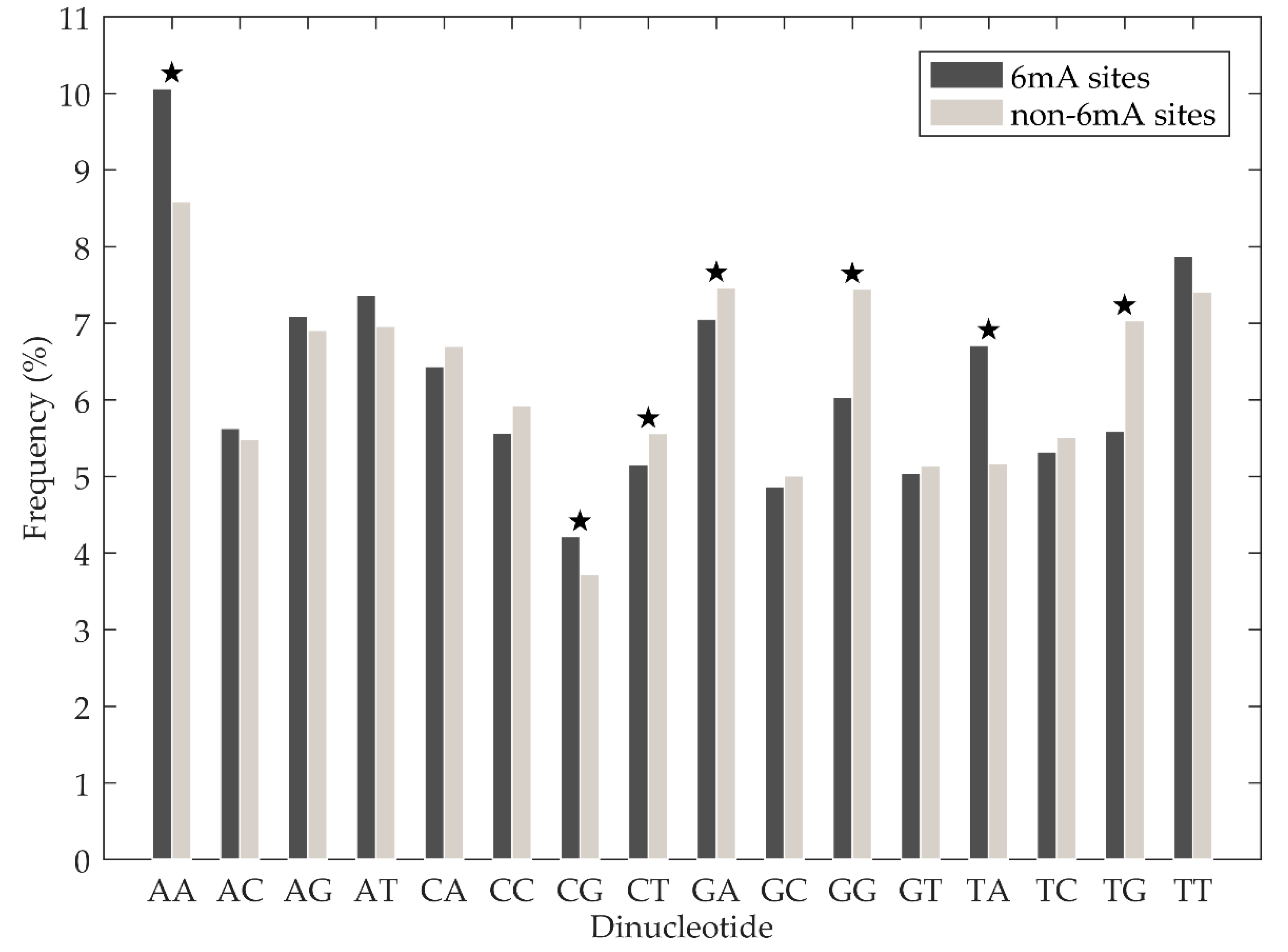
3.1. Sequence Analysis
3.2. Performance Evaluation Using 10-Fold Cross-Validation Tests
3.3. The Effect of Optimized DNA Properties on the Model Performance
3.4. Comparison with Other Methods
3.5. Validation on Independent Datasets
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ratel, D.; Ravanat, J.L.; Berger, F.; Wion, D. N6-methyladenine: The other methylated base of DNA. BioEssays 2006, 28, 309–315. [Google Scholar] [CrossRef]
- Luo, G.Z.; Blanco, M.A.; Greer, E.L.; He, C.; Shi, Y. DNA N6-methyladenine: A new epigenetic mark in eukaryotes? Nat. Rev. Mol. Cell Bio. 2015, 16, 705–710. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, C.; Liu, H.; Zhou, Q.; Liu, Q.; Guo, Y.; Peng, T.; Song, J.; Zhang, J.; Chen, L.; et al. Identification and analysis of adenine N6-methylation sites in the rice genome. Nat. Plants. 2018, 4, 554–563. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, G.Z.; Chen, K.; Deng, X.; Yu, M.; Han, D.; Hao, Z.; Liu, J.; Lu, X.; Dore, L.C.; et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 2015, 161, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizabal-Corrales, D.; Hsu, C.H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Koziol, M.J.; Bradshaw, C.R.; Allen, G.E.; Costa, A.S.H.; Frezza, C.; Gurdon, J.B. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 2016, 23, 24–30. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, Y.; Luo, G.Z.; Wang, X.; Yue, Y.; Wang, X.; Zong, X.; Chen, K.; Yin, H.; Fu, Y.; et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 2016, 7, 13052. [Google Scholar] [CrossRef]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Cheng, Y.; Wang, Z.; Li, Y.; Chen, L.; Huang, L.; Zhang, W.; Chen, D.; Wu, H.; Tang, B.; et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 2017, 8, 1122. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Shen, L.; Cui, X.; Bao, S.; Geng, Y.; Yu, G.; Liang, F.; Xie, S.; Lu, T.; Gu, X.; et al. DNA N6-Adenine Methylation in Arabidopsis thaliana. Dev. Cell 2018, 45, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liang, Z.; Cui, X.; Ji, C.; Li, Y.; Zhang, P.; Liu, J.; Riaz, A.; Yao, P.; Liu, M.; et al. N6-Methyladenine DNA methylation in Japonica and Indica rice genomes and its association with gene expression, plant development, and stress responses. Mol. Plant 2018, 11, 1492–1508. [Google Scholar] [CrossRef]
- Frelon, S.; Douki, T.; Ravanat, J.L.; Pouget, J.P.; Tornabene, C.; Cadet, J. High-performance liquid chromatography--tandem mass spectrometry measurement of radiation-induced base damage to isolated and cellular DNA. Chem. Res. Toxicol. 2000, 13, 1002–1010. [Google Scholar] [CrossRef]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 2010, 7, 461–465. [Google Scholar] [CrossRef] [Green Version]
- Feng, P.; Yang, H.; Ding, H.; Lin, H.; Chen, W.; Chou, K.C. iDNA6mA-PseKNC: Identifying DNA N6-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics 2019, 111, 96–102. [Google Scholar] [CrossRef]
- Chen, W.; Lv, H.; Nie, F.; Lin, H. i6mA-Pred: Identifying DNA N6-methyladenine sites in the rice genome. Bioinformatics 2019, 35, 2796–2800. [Google Scholar] [CrossRef]
- Tahir, M.; Tayara, H.; Chong, K.T. iDNA6mA (5-step rule): Identification of DNA N6-methyladenine sites in the rice genome by intelligent computational model via Chou’s 5-step rule. Chemometr. Intell. Lab. 2019, 189, 96–101. [Google Scholar] [CrossRef]
- Lv, H.; Dao, F.Y.; Guan, Z.X.; Zhang, D.; Tan, J.X.; Zhang, Y.; Chen, W.; Lin, H. iDNA6mA-Rice: A computational tool for detecting N6-methyladenine sites in rice. Front Genet. 2019, 10, 793. [Google Scholar] [CrossRef]
- Liu, G.; Xing, Y.; Cai, L. Using weighted features to predict recombination hotspots in Saccharomyces cerevisiae. J. Theor. Biol. 2015, 382, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.C.; Herman, G.; Modrich, P. Extent of equilibrium perturbation of the DNA helix upon enzymatic methylation of adenine residues. J. Biol. Chem. 1985, 260, 191–194. [Google Scholar] [PubMed]
- Richmond, T.J.; Davey, C.A. The structure of DNA in the nucleosome core. Nature 2003, 423, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Tolstorukov, M.Y.; Colasanti, A.V.; McCandlish, D.M.; Olson, W.K.; Zhurkin, V.B. A novel roll-and-slide mechanism of DNA folding in chromatin: Implications for nucleosome positioning. J. Mol. Biol. 2007, 371, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, Y.; Jin, X.; Wang, X.; Liu, B. iRSpot-DACC: A computational predictor for recombination hot/cold spots identification based on dinucleotide-based auto-cross covariance. Sci. Rep. 2016, 6, 33483. [Google Scholar] [CrossRef]
- Zhang, L.; Kong, L. iRSpot-ADPM: Identify recombination spots by incorporating the associated dinucleotide product model into Chou’s pseudo components. J. Theor. Biol. 2018, 441, 1–8. [Google Scholar] [CrossRef]
- Zhang, L.; Kong, L. iRSpot-PDI: Identification of recombination spots by incorporating dinucleotide property diversity information into Chou’s pseudo components. Genomics 2019, 111, 457–464. [Google Scholar] [CrossRef]
- Zhang, S.; Chang, M.; Zhou, Z.; Dai, X.; Xu, Z. pDHS-ELM: Computational predictor for plant DNase I hypersensitive sites based on extreme learning machines. Mol. Genet. Genomics 2018, 293, 1035–1049. [Google Scholar] [CrossRef]
- Zhang, S.; Zhuang, W.; Xu, Z. Prediction of DNase I hypersensitive sites in plant genome using multiple modes of pseudo components. Anal. Biochem. 2018, 549, 149–156. [Google Scholar] [CrossRef]
- Zhang, S.; Lin, J.; Su, L.; Zhou, Z. pDHS-DSET: Prediction of DNase I hypersensitive sites in plant genome using DS evidence theory. Anal. Biochem. 2019, 564-565, 54–63. [Google Scholar] [CrossRef]
- Chen, W.; Xing, P.; Zou, Q. Detecting N6-methyladenosine sites from RNA transcriptomes using ensemble Support Vector Machines. Sci. Rep. 2017, 7, 40242. [Google Scholar] [CrossRef]
- He, W.; Jia, C.; Zou, Q. 4mCPred: Machine learning methods for DNA N4-methylcytosine sites prediction. Bioinformatics 2019, 35, 593–601. [Google Scholar] [CrossRef]
- Zhou, Y.; Zeng, P.; Li, Y.H.; Zhang, Z.; Cui, Q. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016, 44, e91. [Google Scholar] [CrossRef]
- Wei, L.; Su, R.; Wang, B.; Li, X.; Zou, Q.; Gao, X. Integration of deep feature representations and handcrafted features to improve the prediction of N6-methyladenosine sites. Neurocomputing 2019, 324, 3–9. [Google Scholar] [CrossRef]
- Chen, W.; Feng, P.; Ding, H.; Lin, H.; Chou, K.C. iRNA-Methyl: Identifying N6-methyladenosine sites using pseudo nucleotide composition. Anal. Biochem. 2015, 490, 26–33. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 389–396. [Google Scholar] [CrossRef]
- Chen, W.; Feng, P.M.; Lin, H.; Chou, K.C. iRSpot-PseDNC: Identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res. 2013, 41, e68. [Google Scholar] [CrossRef]
- Chen, W.; Lei, T.Y.; Jin, D.C.; Lin, H.; Chou, K.C. PseKNC: A flexible web server for generating pseudo K-tuple nucleotide composition. Anal. Biochem. 2014, 456, 53–60. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, X.; Brooker, J.; Lin, H.; Zhang, L.; Chou, K.C. PseKNC-General: A cross-platform package for generating various modes of pseudo nucleotide compositions. Bioinformatics 2015, 31, 119–120. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Xing, J.F.; Chen, W.; Luan, M.W.; Xie, R.; Huang, J.; Xie, S.Q.; Xiao, C.L. MDR: An integrative DNA N6-methyladenine and N4-methylcytosine modification database for Rosaceae. Hortic Res. 2019, 6, 78. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
| Classifier | Sn (%) | Sp (%) | Acc (%) | MCC | AUC |
|---|---|---|---|---|---|
| Naive Bayes | 81.02 | 79.63 | 80.32 | 0.607 | 0.879 |
| Logistic regression | 82.23 | 80.76 | 81.49 | 0.630 | 0.897 |
| SVM | 84.60 | 82.89 | 83.74 | 0.675 | 0.914 |
| LogitBoost | 84.16 | 84.77 | 84.47 | 0.689 | 0.916 |
| TreeBagging | 84.32 | 86.36 | 85.34 | 0.707 | 0.921 |
| Classifier | Sn (%) | Sp (%) | Acc (%) | MCC | AUC |
|---|---|---|---|---|---|
| Naive Bayes | 82.67 | 80.84 | 81.76 | 0.635 | 0.889 |
| Logistic regression | 83.42 | 81.85 | 82.64 | 0.653 | 0.901 |
| SVM | 85.47 | 83.61 | 84.54 | 0.691 | 0.915 |
| LogitBoost | 84.86 | 85.43 | 85.15 | 0.703 | 0.917 |
| TreeBagging | 84.09 | 88.07 | 86.08 | 0.722 | 0.926 |
| Method | Sn (%) | Sp (%) | Acc (%) | MCC | AUC |
|---|---|---|---|---|---|
| i6mA-Pred | 82.95 | 83.30 | 83.13 | 0.660 | 0.886 |
| PseDNC | 63.52 | 65.57 | 64.55 | 0.290 | 0.636 |
| iDNA6mA | 86.70 | 86.59 | 86.64 | 0.730 | 0.931 |
| iDNA6mA-Rice | 83.86 | 83.41 | 83.63 | 0.670 | 0.910 |
| i6mA-DNCP | 84.43 | 88.86 | 86.65 | 0.734 | 0.926 |
| SVM-based method | 86.25 | 83.86 | 85.06 | 0.701 | 0.915 |
| Genome | Number of Samples | Number of Corrected Prediction | Success Rate (%) |
|---|---|---|---|
| Arabidopsis thaliana | 27,751 | 25,394 | 91.51 |
| Fragaria vesca | 8983 | 8680 | 96.63 |
| Rosa chinensis | 1479 | 1359 | 91.89 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, L.; Zhang, L. i6mA-DNCP: Computational Identification of DNA N6-Methyladenine Sites in the Rice Genome Using Optimized Dinucleotide-Based Features. Genes 2019, 10, 828. https://doi.org/10.3390/genes10100828
Kong L, Zhang L. i6mA-DNCP: Computational Identification of DNA N6-Methyladenine Sites in the Rice Genome Using Optimized Dinucleotide-Based Features. Genes. 2019; 10(10):828. https://doi.org/10.3390/genes10100828
Chicago/Turabian StyleKong, Liang, and Lichao Zhang. 2019. "i6mA-DNCP: Computational Identification of DNA N6-Methyladenine Sites in the Rice Genome Using Optimized Dinucleotide-Based Features" Genes 10, no. 10: 828. https://doi.org/10.3390/genes10100828
APA StyleKong, L., & Zhang, L. (2019). i6mA-DNCP: Computational Identification of DNA N6-Methyladenine Sites in the Rice Genome Using Optimized Dinucleotide-Based Features. Genes, 10(10), 828. https://doi.org/10.3390/genes10100828