Spatial Transcriptomic and miRNA Analyses Revealed Genes Involved in the Mesometrial-Biased Implantation in Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. Histological Analysis
2.3. RNA-seq and miRNA-seq Data Analysis
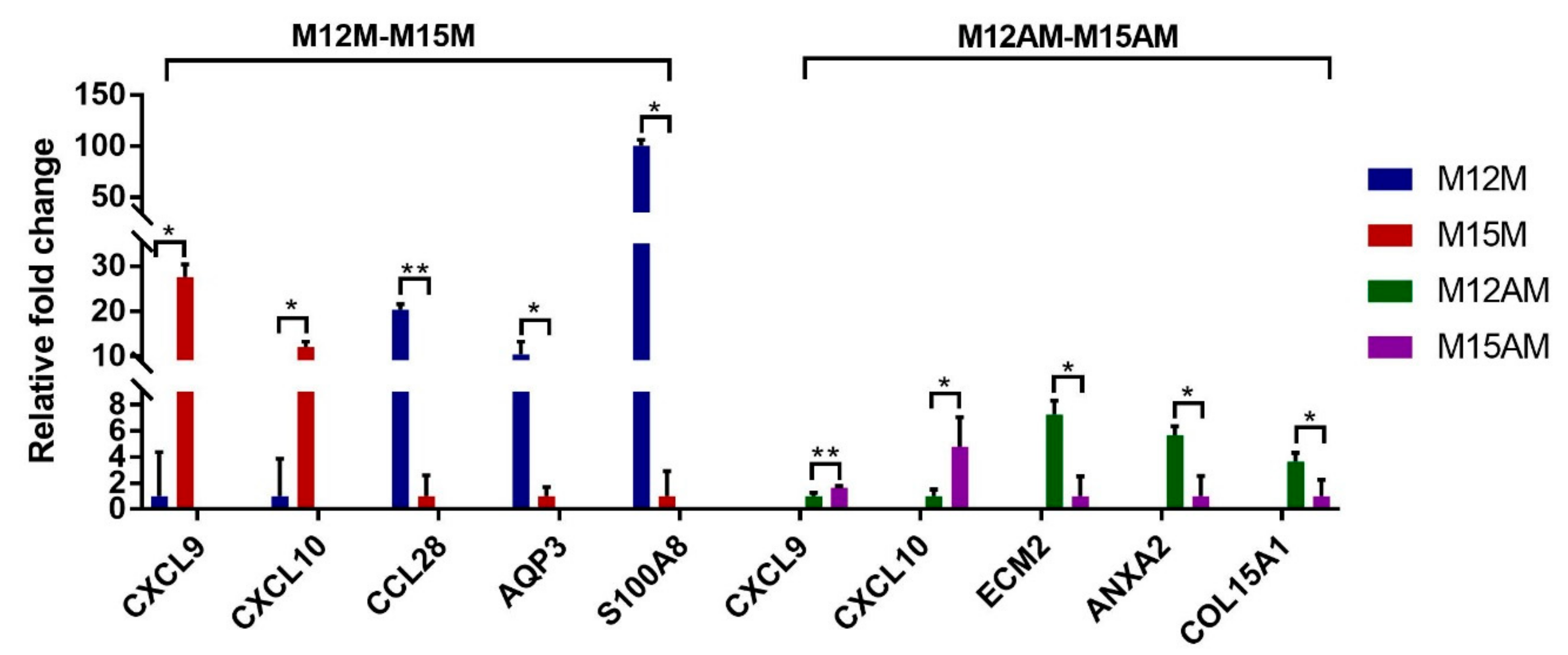
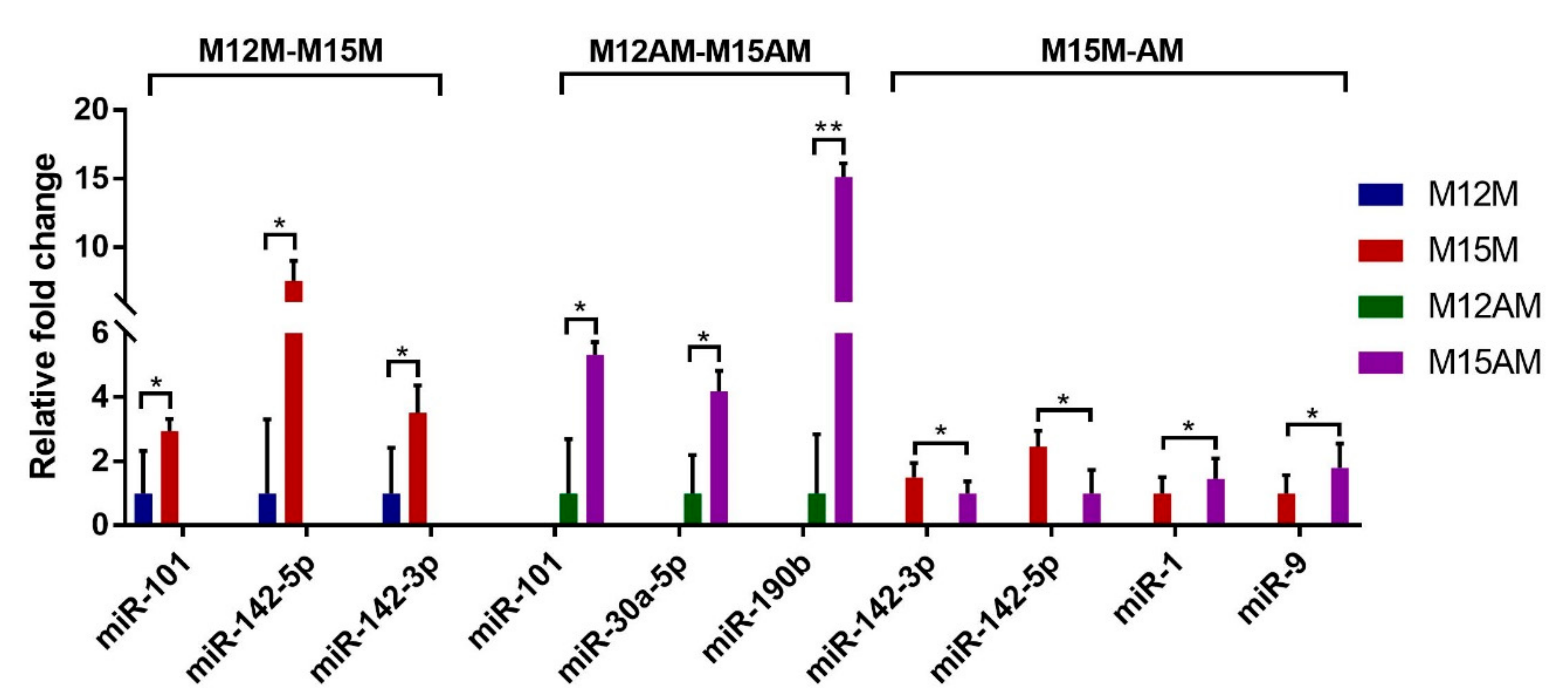
2.4. Real-Time Polymerase Chain Reaction (RT-PCR) for Genes and miRNAs
2.5. Dual Luciferase Reporter Assays
3. Results
3.1. Analysis of the Histological Features of the Whole Cross-Sectional View of the Uterus during Implantation in Pigs
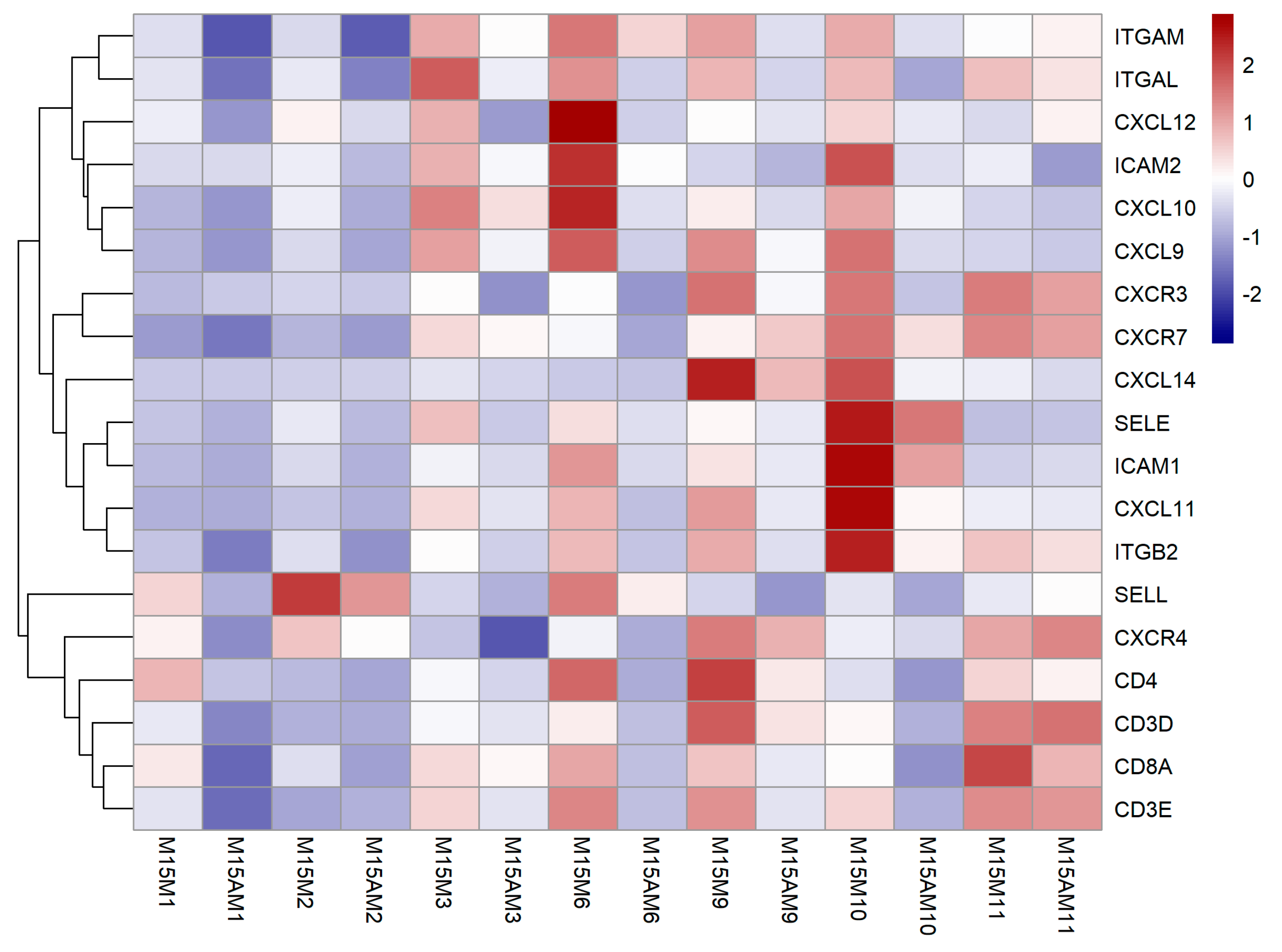
3.2. Gene Expression Patterns before and after Conceptus Attachment Differed between the Mesometrial and Anti-Mesometrial Endometrium in the Uterus
3.3. Endometrial Genes Exhibit Spatial Expression Patterns in the Uterus
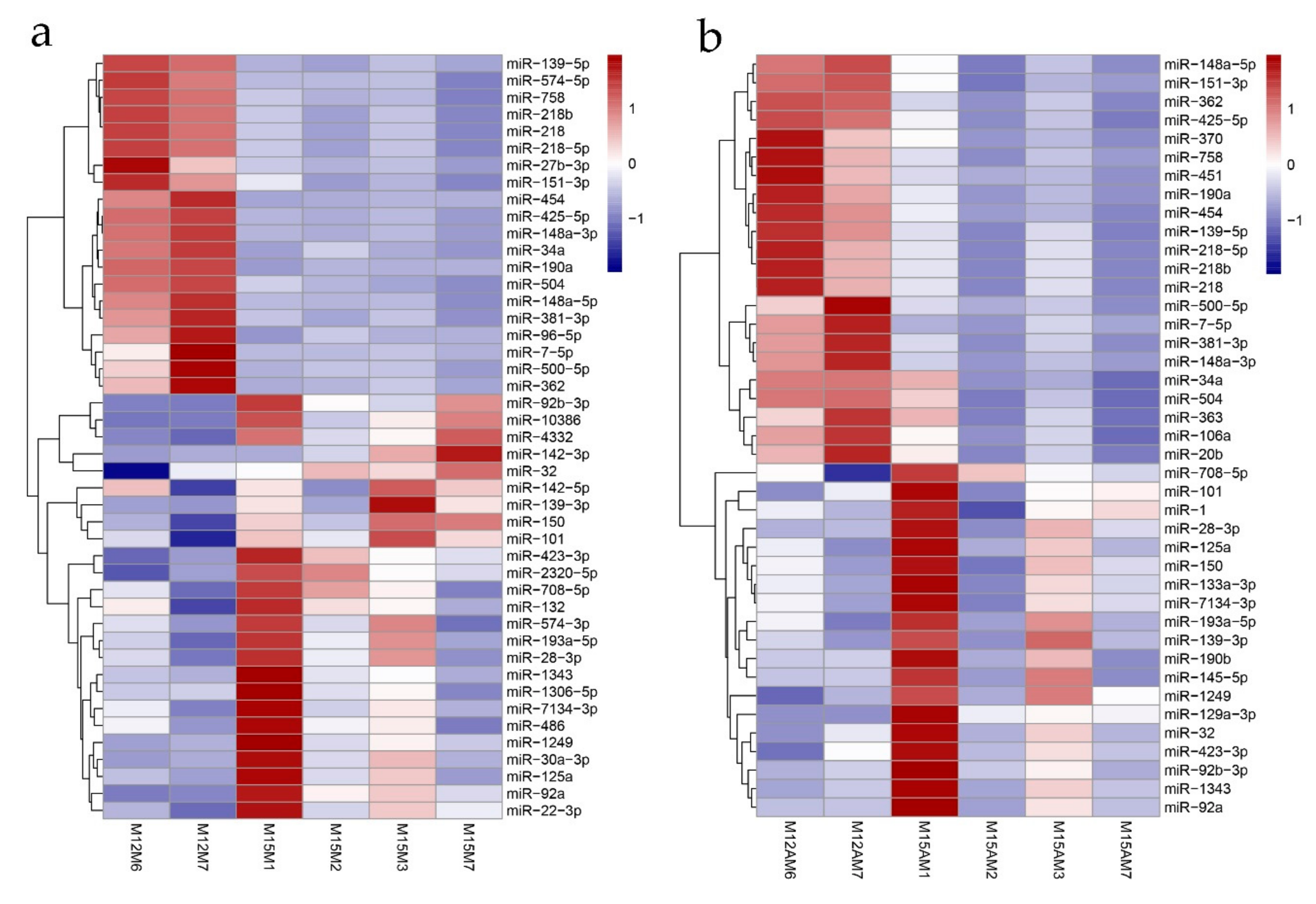
3.4. Identification of Spatiotemporal Endometrial miRNA Expression Profiles in the Uterus during Implantation
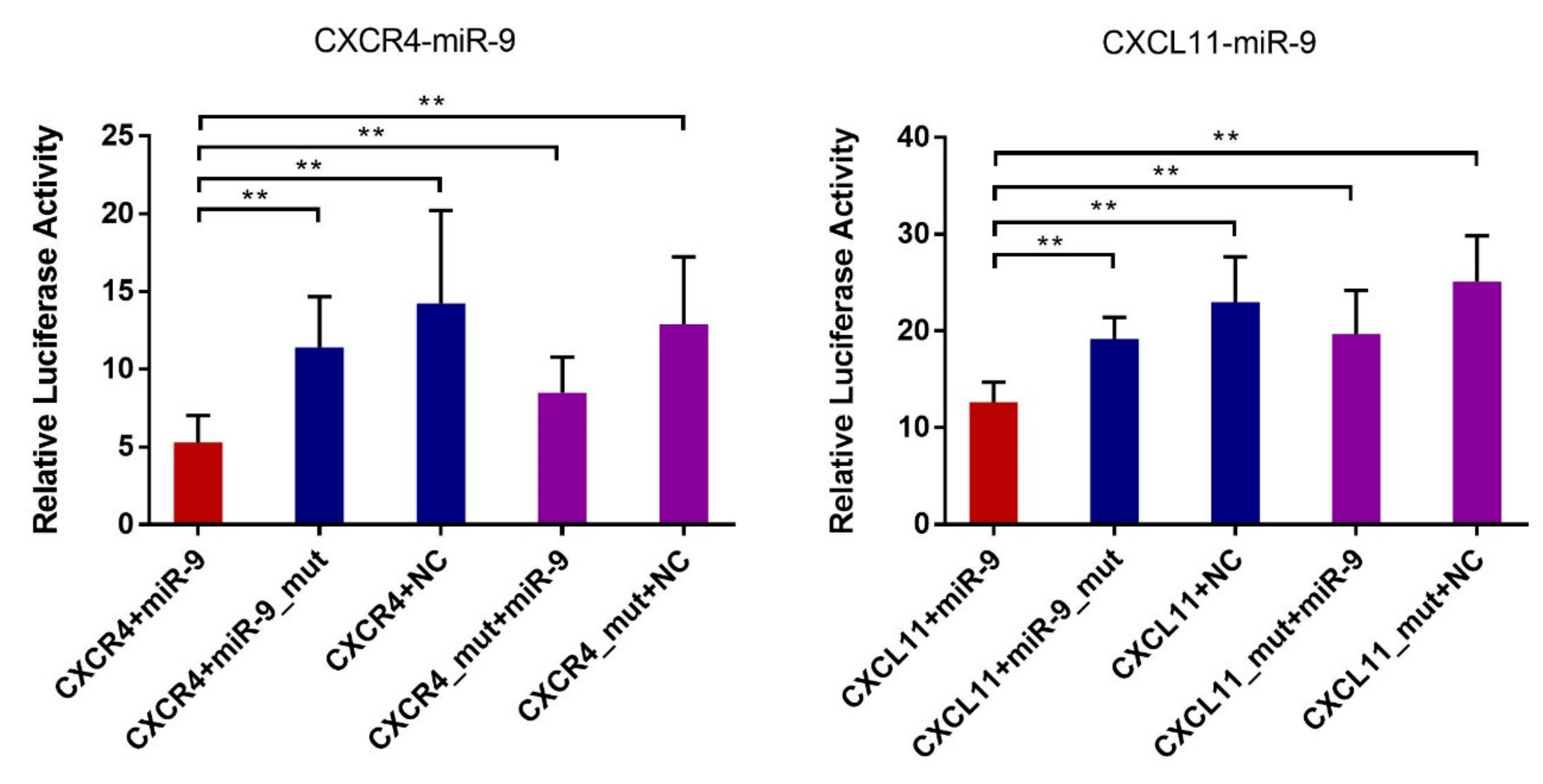
3.5. Validation of the Predicted Interaction of miRNA-Target Pairs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bazer, F.W. Pregnancy recognition signaling mechanisms in ruminants and pigs. J. Anim. Sci. Biotechnol. 2013, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Johnson, G.A. Pig blastocyst-uterine interactions. Differentiation 2014, 87, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Dantzer, V. Electron microscopy of the initial stages of placentation in the pig. Anat. Embryol. 1985, 172, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xi, Y.; Xue, S.; Wang, Y.; Wu, L.; Liu, H.; Lei, M. Sequence analysis of microRNAs during pre-implantation between Meishan and Yorkshire pigs. Gene 2018, 646, 20–27. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, T.; Wu, L.; Liu, X.; Xue, S.; Lei, M. Identification of non-coding and coding RNAs in porcine endometrium. Genomics 2017, 109, 43–50. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, S.; Liu, X.; Liu, H.; Hu, T.; Qiu, X.; Zhang, J.; Lei, M. Analyses of Long Non-Coding RNA and mRNA profiling using RNA sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 2016, 6, 20238. [Google Scholar] [CrossRef]
- Chen, X.; Li, A.; Chen, W.; Wei, J.; Fu, J.; Wang, A. Differential gene expression in uterine endometrium during implantation in pigs. Biol. Reprod. 2015, 92, 52. [Google Scholar] [CrossRef]
- Krawczynski, K.; Bauersachs, S.; Reliszko, Z.P.; Graf, A.; Kaczmarek, M.M. Expression of microRNAs and isomiRs in the porcine endometrium: Implications for gene regulation at the maternal-conceptus interface. BMC Genomics 2015, 16, 906. [Google Scholar] [CrossRef]
- Zeng, S.; Bick, J.; Ulbrich, S.E.; Bauersachs, S. Cell type-specific analysis of transcriptome changes in the porcine endometrium on Day 12 of pregnancy. BMC Genomics 2018, 19, 459. [Google Scholar] [CrossRef] [PubMed]
- Samborski, A.; Graf, A.; Krebs, S.; Kessler, B.; Bauersachs, S. Deep sequencing of the porcine endometrial transcriptome on day 14 of pregnancy. Biol. Reprod. 2013, 88, 84. [Google Scholar] [CrossRef] [PubMed]
- Samborski, A.; Graf, A.; Krebs, S.; Kessler, B.; Reichenbach, M.; Reichenbach, H.D.; Ulbrich, S.E.; Bauersachs, S. Transcriptome changes in the porcine endometrium during the preattachment phase. Biol. Reprod. 2013, 89, 134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, S.; Liu, M.; Zhang, A.; Wu, Z.; Zhang, Z.; Li, J. Differential gene expression in the endometrium on gestation day 12 provides insight into sow prolificacy. BMC Genomics 2013, 14, 45. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Zhu, M.-J.; Schroyen, M.; Qu, L.; Nettleton, D.; Kuhar, D.; Lunney, J.K.; Ross, J.W.; Zhao, S.-h.; Tuggle, C.K. Endometrial gene expression profiling in pregnant Meishan and Yorkshire pigs on day 12 of gestation. BMC Genomics 2014, 15, 156. [Google Scholar] [CrossRef] [PubMed]
- Kiewisz, J.; Krawczynski, K.; Lisowski, P.; Blitek, A.; Zwierzchowski, L.; Ziecik, A.J.; Kaczmarek, M.M. Global gene expression profiling of porcine endometria on Days 12 and 16 of the estrous cycle and pregnancy. Theriogenology 2014, 82, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hua, R.; Xue, S.; Li, W.; Wu, L.; Kang, T.; Lei, M. mRNA/lncRNA expression patterns and the function of fibrinogen-like protein 2 in Meishan pig endometrium during the preimplantation phases. Mol. Reprod. Dev. 2019, 86, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.; Bartos, A.; Park, C.; Sun, X.; Li, Y.; Cha, S.W.; Ajima, R.; Ho, H.Y.; Yamaguchi, T.P.; Dey, S.K. Appropriate crypt formation in the uterus for embryo homing and implantation requires Wnt5a-ROR signaling. Cell Rep. 2014, 8, 382–392. [Google Scholar] [CrossRef]
- Goad, J.; Ko, Y.A.; Kumar, M.; Syed, S.M.; Tanwar, P.S. Differential Wnt signaling activity limits epithelial gland development to the anti-mesometrial side of the mouse uterus. Dev. Biol. 2017, 423, 138–151. [Google Scholar] [CrossRef]
- Kridli, R.T.; Khalaj, K.; Bidarimath, M.; Tayade, C. Placentation, maternal-fetal interface, and conceptus loss in swine. Theriogenology 2016, 85, 135–144. [Google Scholar] [CrossRef]
- White, F.J.; Ross, J.W.; Joyce, M.M.; Geisert, R.D.; Burghardt, R.C.; Johnson, G.A. Steroid regulation of cell specific secreted phosphoprotein 1 (osteopontin) expression in the pregnant porcine uterus. Biol. Reprod. 2005, 73, 1294–1301. [Google Scholar] [CrossRef]
- Johnson, G.A.; Burghardt, R.C.; Bazer, F.W. Osteopontin: A leading candidate adhesion molecule for implantation in pigs and sheep. J. Anim. Sci. Biotechnol. 2014, 5, 56. [Google Scholar] [CrossRef]
- Ka, H.; Seo, H.; Choi, Y.; Yoo, I.; Han, J. Endometrial response to conceptus-derived estrogen and interleukin-1beta at the time of implantation in pigs. J. Anim. Sci. Biotechnol. 2018, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Tayade, C.; Black, G.P.; Fang, Y.; Croy, B.A. Differential Gene Expression in Endometrium, Endometrial Lymphocytes, and Trophoblasts during Successful and Abortive Embryo Implantation. J. Immunol. 2005, 176, 148–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, L.; Xu, X.; Huang, J.; Lei, M.; Xu, D.; Zhao, S.; Yu, M. Difference in expression patterns of placental cholesterol transporters, ABCA1 and SR-BI, in Meishan and Yorkshire pigs with different placental efficiency. Sci. Rep. 2016, 6, 20503. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Liu, R.; Wang, M.; Su, L.; Li, X.; Zhao, S.; Yu, M. The Expression Pattern of MicroRNAs and the Associated Pathways Involved in the Development of Porcine Placental Folds That Contribute to the Expansion of the Exchange Surface Area. Biol. Reprod. 2015, 93, 62. [Google Scholar] [CrossRef]
- Han, J.; Gu, M.J.; Yoo, I.; Choi, Y.; Jang, H.; Kim, M.; Yun, C.H.; Ka, H. Analysis of cysteine-X-cysteine motif chemokine ligands 9, 10, and 11, their receptor CXCR3, and their possible role on the recruitment of immune cells at the maternal-conceptus interface in pigs. Biol. Reprod. 2017, 97, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Jeong, W.; Gu, M.J.; Yoo, I.; Yun, C.H.; Kim, J.; Ka, H. Cysteine-X-cysteine motif chemokine ligand 12 and its receptor CXCR4: Expression, regulation, and possible function at the maternal-conceptus interface during early pregnancy in pigs. Biol. Reprod. 2018, 99, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Lecce, L.; Kaneko, Y.; Madawala, R.J.; Murphy, C.R. ICAM1 and fibrinogen-γ are increased in uterine epithelial cells at the time of implantation in rats. Mol. Reprod. Dev. 2011, 78, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Seo, H.; Choi, Y.; Yoo, I.; Seo, M.; Lee, C.K.; Kim, H.; Ka, H. Analysis of Stage-Specific Gene Expression Profiles in the Uterine Endometrium during Pregnancy in Pigs. PLoS ONE 2015, 10, e0143436. [Google Scholar] [CrossRef]
- Bidarimath, M.; Tayade, C. Pregnancy and spontaneous fetal loss: A pig perspective. Mol. Reprod. Dev. 2017, 84, 856–869. [Google Scholar] [CrossRef] [Green Version]
- Engelhardt, H.; Croy, B.A.; King, G.J. Conceptus Influences the Distribution of Uterine Leukocytes During Early Porcine Pregnancy. Biol. Reprod. 2002, 66, 1875–1880. [Google Scholar] [CrossRef]
- Engelhardt, H.; Croy, B.A.; King, G.J. Evaluation of Natural Killer Cell Recruitment to Embryonic Attachment Sites during Early Porcine Pregnancy. Biol. Reprod. 2002, 66, 1185–1192. [Google Scholar] [CrossRef]
- Dixit, N.; Simon, S.I. Chemokines, selectins and intracellular calcium flux: Temporal and spatial cues for leukocyte arrest. Front. Immunol 2012, 3, 188. [Google Scholar] [CrossRef]
- Natarajan, A.; Nadarajah, V.; Felsovalyi, K.; Wang, W.; Jeyachandran, V.R.; Wasson, R.A.; Cardozo, T.; Bracken, C.; Krogsgaard, M. Structural Model of the Extracellular Assembly of the TCR-CD3 Complex. Cell Rep. 2016, 14, 2833–2845. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Cooper, D.K.C.; Cai, Z.; Mou, L. Expression and Regulation Profile of Mature MicroRNA in the Pig: Relevance to Xenotransplantation. Biomed Res. Int. 2018, 2018, 2983908. [Google Scholar] [CrossRef]
- Kumar, V.; Maurya, V.K.; Joshi, A.; Meeran, S.M.; Jha, R.K. Integrin β 8 (ITGB8) regulates embryo implantation potentially via controlling the activity of TGF-B1 in mice. Biol. Reprod. 2015, 92, 109. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Soni, U.K.; Maurya, V.K.; Singh, K.; Jha, R.K. Integrin beta8 (ITGB8) activates VAV-RAC1 signaling via FAK in the acquisition of endometrial epithelial cell receptivity for blastocyst implantation. Sci. Rep. 2017, 7, 1885. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yang, H.; Han, K.; Zhu, D.; Lun, P.; Zhao, Y. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem. Biophys. Res. Commun. 2018, 498, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Luo, H.; Liu, X.; Peng, Y.; Zhang, B.; Wang, L.; Xu, X.; Peng, X.; Li, G.; Tian, W.; et al. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis 2014, 35, 554–563. [Google Scholar] [CrossRef]
- Yu, T.; Liu, K.; Wu, Y.; Fan, J.; Chen, J.; Li, C.; Yang, Q.; Wang, Z. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/β-catenin signaling pathway. Oncogene 2014, 33, 5017–5027. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Yang, Y.; Tian, M.; Deng, D.; Yu, M. Spatial Transcriptomic and miRNA Analyses Revealed Genes Involved in the Mesometrial-Biased Implantation in Pigs. Genes 2019, 10, 808. https://doi.org/10.3390/genes10100808
Huang J, Yang Y, Tian M, Deng D, Yu M. Spatial Transcriptomic and miRNA Analyses Revealed Genes Involved in the Mesometrial-Biased Implantation in Pigs. Genes. 2019; 10(10):808. https://doi.org/10.3390/genes10100808
Chicago/Turabian StyleHuang, Ji, Yifen Yang, Miao Tian, Dadong Deng, and Mei Yu. 2019. "Spatial Transcriptomic and miRNA Analyses Revealed Genes Involved in the Mesometrial-Biased Implantation in Pigs" Genes 10, no. 10: 808. https://doi.org/10.3390/genes10100808
APA StyleHuang, J., Yang, Y., Tian, M., Deng, D., & Yu, M. (2019). Spatial Transcriptomic and miRNA Analyses Revealed Genes Involved in the Mesometrial-Biased Implantation in Pigs. Genes, 10(10), 808. https://doi.org/10.3390/genes10100808



