Pollination Drop Proteome and Reproductive Organ Transcriptome Comparison in Gnetum Reveals Entomophilous Adaptation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pollination Drop Sampling
2.2. Gel Electrophoresis
2.3. Trypsin Digestion and LC-MS/MS
2.4. Proteomic Analyses and Bioinformatics
2.5. Reproductive Organ Collection and RNA Sequencing
2.6. Bioinformatics and Transcriptome Comparisons
2.7. Integrated Proteome and Transcriptome Analyses
3. Results
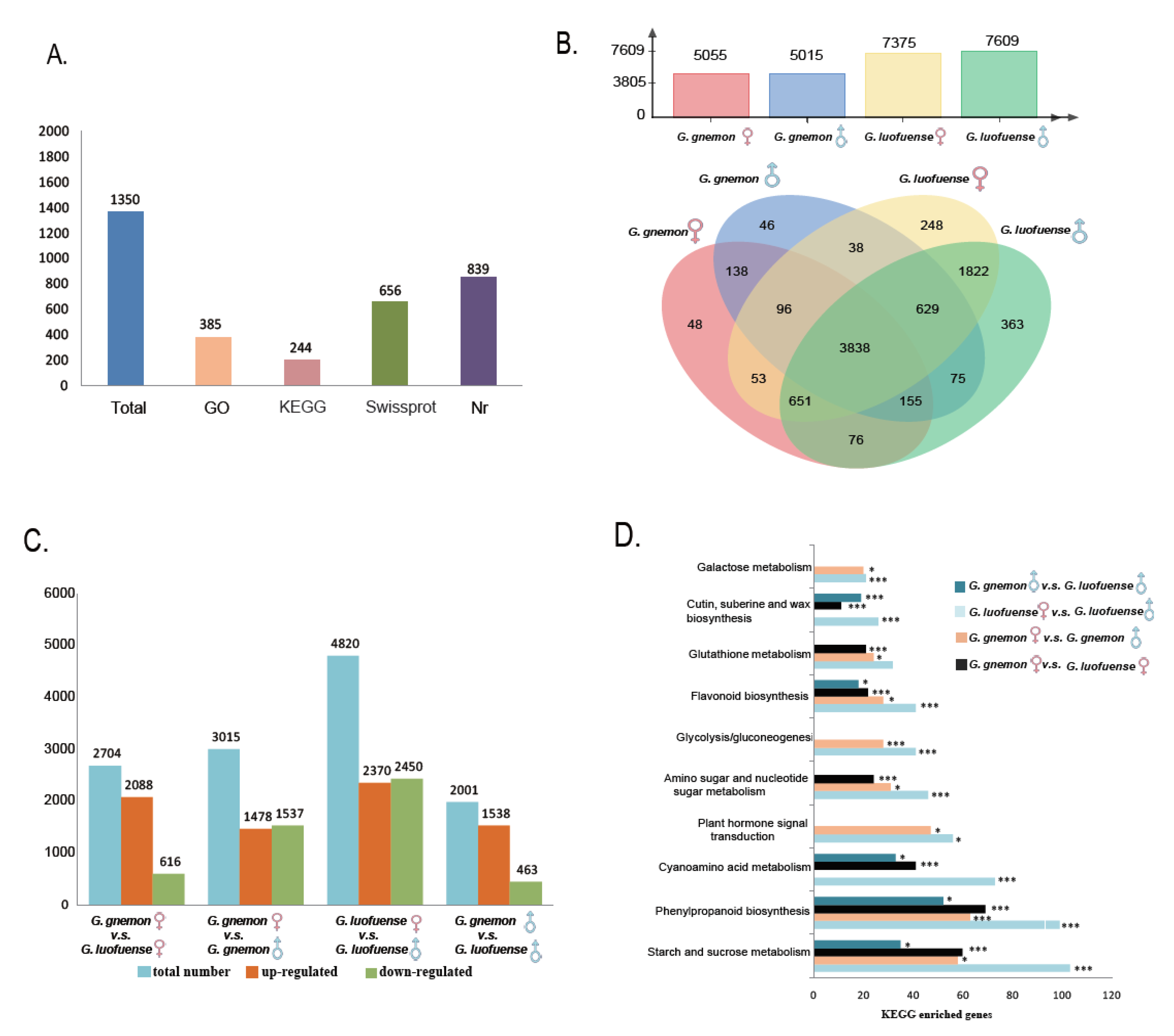
3.1. Proteomic Diversity
3.2. Differentially Abundant Proteinsand KEGG Enrichment Analyses
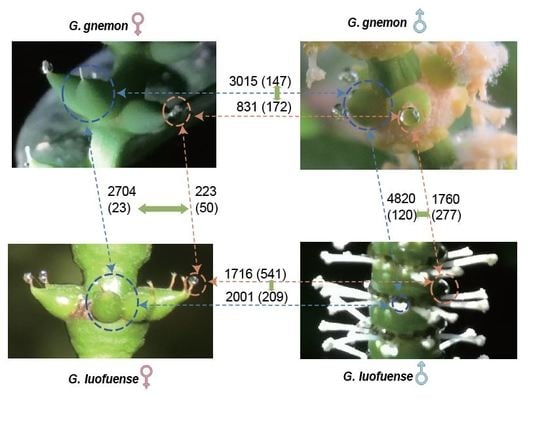
3.3. Transcripts of Reproductive Organs
3.4. Differentially Expressed Genes and KEGG Enrichment Analyses
3.5. Integrated Proteome and Transcriptome Analyses
4. Discussion
4.1. Proteomic Diversity in the Pollination Drops of Gnetum
4.2. Infraspecific Variation of Protein Profiles
4.3. Interspecific Variation of Protein Profiles
4.4. Transcriptome Data and Newly Detected Genes
4.5. Infraspecific Variation of Transcriptome Data
4.6. Interspecific Variation Intranscriptome Data
4.7. Inferred Reproductive Evolution of Gnetum
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ollerton, J. Pollinator diversity: Distribution, ecological function, and conservation. Annu. Rev. Ecol. Evol. Syst. 2017, 48, 353–376. [Google Scholar] [CrossRef]
- Ollerton, J.; Winfree, R.; Tarrant, S. How many flowering plants are pollinated by animals? Oikos 2011, 120, 321–326. [Google Scholar] [CrossRef]
- Gervasi, D.D.L.; Schiestl, F.P. Real-time divergent evolution in plants driven by pollinators. Nat. Commun. 2017, 8, 14691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norstog, K. Cycads and the origin of insect pollination. Am. Sci. 1987, 75, 270–279. [Google Scholar]
- Terry, I.; Walter, G.H.; Moore, C.; Roemer, R.; Hull, C. Odor-mediated push-pull pollination in cycads. Science 2007, 318, 70. [Google Scholar] [CrossRef] [PubMed]
- Tang, W. Insect pollination in the cycad Zamia pumila (Zamiaceae). Am. J. Bot. 1987, 74, 90–99. [Google Scholar] [CrossRef]
- Ickert-Bond, S.M.; Renner, S.S. The Gnetales: Recent insights on their morphology, reproductive biology, chromosome numbers, biogeography, and divergence times. J. Syst. Evol. 2015, 54, 1–16. [Google Scholar] [CrossRef]
- Endress, P.K. Structure and function of female and bisexual organ complexes in Gnetales. Int. J. Plant Sci. 1996, 157, 113–125. [Google Scholar] [CrossRef]
- Bolinder, K.; Humphreys, A.M.; Ehrlén, J.; Alexandersson, R.; Ickert-Bond, S.M.; Rydin, C. From near extinction to diversification by means of a shift in pollination mechanism in the gymnosperm relict Ephedra (Ephedraceae, Gnetales). Bot. J. Linn. Soc. 2016, 4, 461–477. [Google Scholar] [CrossRef]
- Bolinder, K.; Niklas, K.J.; Rydin, C. Aerodynamics and pollen ultrastructure in Ephedra. Am. J. Bot. 2015, 102, 457–470. [Google Scholar] [CrossRef]
- Rydin, C.; Bolinder, K. Moonlight pollination in the gymnosperm Ephedra (Gnetales). Biol. Lett. 2015, 11, 20140993. [Google Scholar] [CrossRef] [PubMed]
- Wetschnig, W.; Depisch, B. Pollination biology of Welwitschia mirabilis Hook. f. (Welwitschiaceae, Gnetopsida). Phyton 1999, 39, 167–184. [Google Scholar]
- Kato, M.; Inoue, T. Origin of insect pollination. Nature 1994, 368, 195. [Google Scholar] [CrossRef]
- Kato, M.; Inoue, T.; Nagamitsu, T. Pollination biology of Gnetum (Gnetaceae) in a lowland mixed dipterocarp forest in Sarawak. Am. J. Bot. 1995, 82, 862–868. [Google Scholar] [CrossRef]
- Corlett, R.T. Pollination in a degraded tropical landscape: A Hong Kong case study. J. Trop. Ecol. 2001, 17, 155–161. [Google Scholar] [CrossRef]
- Karsten, G. Untersuchungen über die Gattung Gnetum. Ann. Jard. Bot. Buitenzorg 1892, 2, 195–217. [Google Scholar]
- Karsten, G. Beitrag zur Entwickelungsgeschichte einiger Gnetum-Arten. Bot. Gaz. 1893, 50, 205–215. [Google Scholar]
- Niklas, K.J. A biophysical perspective on the pollination biology of Ephedra nevadensis and E. trifurca. Bot. Rev. 2015, 81, 28–41. [Google Scholar] [CrossRef]
- Jörgensen, A.; Rydin, C. Reproductive morphology in the Gnetumcuspidatum group (Gnetales) and its implications for pollination biology in the Gnetales. Plant Ecol. Evol. 2015, 148, 387–396. [Google Scholar] [CrossRef]
- Rydin, C.; Hoorn, C. The Gnetales: Past and present. Grana 2016, 55, 1–4. [Google Scholar] [CrossRef]
- Gong, Y.B.; Yang, M.; Vamosi, J.C.; Yang, H.M.; Mu, W.X.; Li, J.K.; Wan, T. Wind or insect pollination? Ambophily in a subtropical gymnosperm Gnetum parvifolium (Gnetales). Plant Spec. Biol. 2016, 31, 272–279. [Google Scholar] [CrossRef]
- Owens, J.N.; Takaso, T.; Runions, C.J. Pollination in conifers. Trends Plant Sci. 1998, 3, 479–485. [Google Scholar] [CrossRef]
- Tomlinson, P.; Braggins, J.; Rattenbury, J. Contrasted pollen capture mechanisms in Phyllocladaceae and certain Podocarpaceae (Coniferales). Am. J. Bot. 1997, 84, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Gelbart, G.; von Aderkas, P. Ovular secretions as part of pollination mechanisms in conifers. Ann. For. Sci. 2002, 59, 345–357. [Google Scholar] [CrossRef] [Green Version]
- Nepi, M.; von Aderkas, P.; Wagner, R.; Mugnaini, S.; Coulter, A.; Pacini, E. Nectar and pollination drops: How different are they? Ann. Bot. 2009, 104, 205–219. [Google Scholar] [CrossRef]
- Nepi, M.; von Aderkas, P.; Pacini, E. Sugary exudates in plant pollination. In Secretions and Exudates in Biological Systems; Vivanco, J.M., Baluška, F., Eds.; Springer: Berlin, Germany, 2012; Volume 12, pp. 155–185. [Google Scholar]
- Nepi, M.; Little, S.; Guarnieri, M.; Nocentini, D.; Prior, N.; Gill, J.; Barry Tomlinson, P.; Ickert-Bond, S.M.; Pirone, C.; Pacini, E.; et al. Phylogenetic and functional signals in gymnosperm ovular secretions. Ann. Bot. 2017, 120, 923–936. [Google Scholar] [CrossRef] [Green Version]
- Von Aderkas, P.; Prior, N.A.; Little, S.A. The evolution of sexual fluids in gymnosperms from pollination drops to nectar. Front. Plant. Sci. 2018, 9, 1844. [Google Scholar] [CrossRef]
- Little, S.; Prior, N.; Pirone, C.; von Aderkas, P. Pollen-ovule interactions in gymnosperms. In Reproductive Biology of Plants; Ramawat, K.G., Mérillon, J.-M., Shivanna, K.R., Eds.; CRC Press, Taylor & Francis Group: New York, NY, USA, 2014; pp. 97–117. [Google Scholar]
- Poulis, B.A.; O’Leary, S.J.; Haddow, J.D.; von Aderkas, P. Identification of proteins present in the Douglas fir ovular secretion: An insight into conifer pollen selection and development. Int. J. Plant Sci. 2005, 166, 733–739. [Google Scholar] [CrossRef]
- Wagner, R.E.; Mugnaini, S.; Sniezko, R.; Hardie, D.; Poulis, B.; Nepi, M.; Pacini, E.; Aderkas, P. Proteomic evaluation of gymnosperm pollination drop proteins indicates highly conserved and complex biological functions. Plant Reprod. 2007, 20, 181–189. [Google Scholar] [CrossRef]
- O’Leary, S.J.; Poulis, B.A.; von Aderkas, P. Identification of two thaumatin-like proteins (TLPs) in the pollination drop of hybrid yew that may play a role in pathogen defense during pollen collection. Tree Physiol. 2007, 27, 1649–1659. [Google Scholar] [CrossRef]
- Coulter, A.; Poulis, B.A.; von Aderkas, P. Pollination drops as dynamic apoplastic secretions. Flora 2012, 207, 482–490. [Google Scholar] [CrossRef]
- Von Aderkas, P.; Prior, N.; Gagnon, S.; Little, S.; Cross, T.; Hardie, D.; Borchers, C.; Thornburg, R.; Hou, C.; Lunny, A. Degradome and secretome of pollination drops of Ephedra. Bot. Rev. 2015, 81, 1–27. [Google Scholar] [CrossRef]
- Prior, N.; Little, S.A.; Boyes, I.; Griffith, P.; Husby, C.; Pirone-Davies, C.; Stevenson, D.W.; Tomlinson, P.B.; von Aderkas, P. Complex reproductive secretions occur in all extant gymnosperm lineages: A proteomic survey of gymnosperm pollination drops. Plant Reprod. 2019, 32, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Cecchi Fiordi, A.; Papini, A.; Brighigna, L. Programmed cell death of the non-functional megaspores in Larix leptolepis (Sieb. Et Zucc.) Gordon (Pinaceae): Ultrastructural aspects. Phytomorphology 2002, 52, 187–195. [Google Scholar]
- Luigi, B.; Papini, A.; Milocani, E.; Vesprini, J.L. Programmed cell death in the nucellus of Tillandsia (Bromeliaceae). Caryologia 2006, 59, 334–339. [Google Scholar] [CrossRef]
- Ziegler, H. Über die Zusammensetzung des “Bestäubungstropfens” und den Mechanismus seiner Sekretion. Planta 1959, 52, 587–599. [Google Scholar] [CrossRef]
- Van der Pijl, L. On the flower biology of some plants from Java with general remarks on fly-traps (species of Annona, Artocarpus, Typhonium, Gnetum, Arisaema and Bogorienses). Ann. Bogor. 1953, 1, 77–99. [Google Scholar]
- Kubitzki, K. Gnetaceae. In The families and Genera of Vascular Plants; Kramer, K.U., Green, P.S., Eds.; Springer: Berlin, Germany, 1990; pp. 383–386. [Google Scholar]
- Hou, C.; Humphreys, A.M.; Thureborn, O.; Rydin, C. New insights into the evolutionary history of Gnetum (Gnetales). Taxon 2015, 64, 239–253. [Google Scholar] [CrossRef]
- Price, R.A. Systematics of the Gnetales: A review of morphological and molecular evidence. Int. J. Plant Sci. 1996, 157, 40–49. [Google Scholar] [CrossRef]
- Won, H.; Renner, S.S. Dating dispersal and radiation in the gymnosperm Gnetum (Gnetales)—Clock calibration when outgroup relationships are uncertain. Syst. Biol. 2006, 55, 610–622. [Google Scholar] [CrossRef]
- Won, H.; Renner, S.S. Horizontal gene transfer from flowering plants to Gnetum. Proc. Natl. Acad. Sci. USA 2003, 100, 10824–18029. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gong, Y.-B.; Wan, T. Early pollination in the dark: Simple volatiles attract more effective pollinators in gnetophytes. 2019; in preparation. [Google Scholar]
- Markgraf, F. Monographie der Gattung Gnetum Ser. 3. Bull. Jard. Bot. Buitenzorg 1930, 10, 407–511. [Google Scholar]
- Lan, Q.; Liu, J.F.; Shi, S.Q.; Deng, N.; Jiang, Z.P.; Chang, E.M. Anatomy, microstructure and endogenous hormone changes in Gnetum parvifolium during anthesis. J. Syst. Evol. 2018, 56, 14–24. [Google Scholar] [CrossRef]
- Markgraf, F. Gnetaceae. In Flora Malesiana Ser. 1; Steenis, C.G.G.J., Ed.; Djakarta: Noordhoff-Kolff: Batavia, Dutch East Indies, 1951; Volume 4, pp. 336–347. [Google Scholar]
- Hou, C.; Wikström, N.; Strijk, J.; Rydin, C. Resolving phylogenetic relationships and species delimitations in closely related gymnosperms using high-throughput NGS, Sanger sequencing and morphology. Plant Syst. Evol. 2016, 302, 1345–1365. [Google Scholar] [CrossRef]
- Markgraf, F. New discoveries of Gnetum in Tropical America. Ann. Missouri. Bot. Gard. 1965, 52, 379–386. [Google Scholar] [CrossRef]
- Pearson, H.H.W. On the microsporangium and microspore of Gnetum, with some notes on the structure of the inflorescence. Ann. Bot. 1912, 26, 603–620. [Google Scholar] [CrossRef]
- Pearson, H.H.W. Notes on the morphology of certain structures concerned in reproduction, in the genus Gnetum. Trans. Linn. Soc. Lond. 1915, 8, 311–332. [Google Scholar]
- Biye, E.H.; Balkwill, K.; Cron, G.V. A clarification of Gnetum L. (Gnetaceae) in Africa and the description of two new species. Plant Syst. Evol. 2014, 300, 263–272. [Google Scholar] [CrossRef]
- Biye, E.H.; Cron, G.V.; Balkwill, K. Morphometric delimitation of Gnetum species in Africa. Plant Syst. Evol. 2016, 302, 1067–1082. [Google Scholar] [CrossRef]
- Berridge, E.M. On some points of resemblance between gnetalean and Bennettitean seeds. New Phytol. 1911, 10, 140–144. [Google Scholar] [CrossRef]
- Takaso, T.; Bouman, F. Ovule and seed ontogeny in Gnetum gnemon L. J. Plant Res. 1986, 99, 241–266. [Google Scholar]
- Thoday, M.G. Anatomy of the ovule and seed in Gnetum gnemon, with notes on Gnetum funiculare. Ann. Bot. 1921, 35, 37–53. [Google Scholar] [CrossRef]
- Thoday, M.G. The female inflorescence and ovules of Gnetum africanum with notes on Gnetum scandens. Ann. Bot. 1911, 25, 1101–1135. [Google Scholar] [CrossRef]
- Becker, A.; Kaufmann, K.; Freialdenhoven, A.; Vincent, C.; Li, M.A.; Saedler, H.; Theissen, G. A novel MADS-box gene subfamily with a sister-group relationship to class B floral homeotic genes. Mol. Genet. Genom. 2002, 266, 942–950. [Google Scholar]
- Becker, A.; Saedler, H.; Theissen, G. Distinct MADS-box gene expression patterns in the reproductive cones of the gymnosperm Gnetum gnemon. Dev. Genes Evol. 2003, 213, 567–572. [Google Scholar] [CrossRef]
- Shindo, S.; Ito, M.; Ueda, K.; Kato, M.; Hasebe, M. Characterization of MADS genes in the gymnosperm Gnetum parvifolium and its implication on the evolution of reproductive organs in seed plants. Evol. Dev. 1999, 1, 180–190. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Melzer, R.; Theissen, G. Molecular interactions of orthologues of floral homeotic proteins from the gymnosperm Gnetum gnemon provide a clue to the evolutionary origin of ‘floral quartets’. Plant J. 2010, 64, 177–190. [Google Scholar] [CrossRef]
- Pirone-Davies, C.; Prior, N.; von Aderkas, P.; Smith, D.; Hardie, D.; Friedman, W.E.; Mathews, S. Insights from the pollination drop proteome and the ovule transcriptome of Cephalotaxus at the time of pollination drop production. Ann. Bot. 2016, 117, 973–984. [Google Scholar] [CrossRef]
- Prior, N.; Little, S.A.; Pirone, C.; Gill, J.E.; Smith, D.; Han, J.; Hardie, D.; O’Leary, S.J.; Wagner, R.E.; Cross, T. Application of proteomics to the study of pollination drops. Appl. Plant Sci. 2013, 1, 1300008. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Wan, T.; Liu, Z.M.; Li, L.F.; Leitch, A.R.; Leitch, I.J.; Lohaus, R.; Liu, Z.J.; Xin, H.P.; Gong, Y.B.; Liu, Y.; et al. A genome for gnetophytes and early evolution of seed plants. Nat. Plants 2018, 4, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Park, G.W.; Hwang, H.; Kim, K.H.; Lee, J.Y.; Lee, H.K.; Park, J.Y.; Ji, E.S.; Park, S.K.R.; Yates, J.R.; Kwon, K.H.; et al. Integrated proteomic pipeline using multiple search engines for a proteogenomic study with a controlled protein false discovery rate. J. Proteome Res. 2016, 15, 4082–4090. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Meth. 2015, 12, 357. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing Version 3.2.0; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org (accessed on 2 October 2019).
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Zhu, W.; Smith, J.W.; Huang, C. Mass spectrometry-based label-free quantitative proteomics. Biomed. Res. Int. 2009, 2010, 840518. [Google Scholar] [CrossRef] [PubMed]
- Shivanna, K.R. Pollen Biology and Biotechnology; Science Publishers Inc.: Enfield, UK, 2003. [Google Scholar]
- Achari, A.; Marshall, S.; Muirhead, H.; Palmieri, R.; Noltmann, E. Glucose-6-phosphate isomerase. Philos. Trans. R. Soc. B. 1981, 293, 145–157. [Google Scholar] [CrossRef]
- Kanner, J.; Elmaleh, H.; Reuveni, O.; Ben-Gera, I. Invertase (beta-fructofuranosidase) activity in three date cultivars. J. Agric. Food Chem. 1978, 26, 1238–1240. [Google Scholar] [CrossRef]
- Le Roy, K.; Lammens, W.; Van Laere, A.; Van den Ende, W. Influencing the binding configuration of sucrose in the active sites of chicory fructan 1-exohydrolase and sugar beet fructan 6-exohydrolase. New Phytol. 2008, 178, 572–580. [Google Scholar] [CrossRef]
- Zareie, R.; Melanson, D.L.; Murphy, P.J. Isolation of fungal cell wall degrading proteins from barley (Hordeum vulgare L.) leaves infected with Rhynchosporium secalis. Mol. Plant Microbe Interact. 2002, 15, 1031–1039. [Google Scholar] [CrossRef]
- Whitaker, C.; Pammenter, N.; Berjak, P. Infection of the cones and seeds of Welwitschia mirabilis by Aspergillus niger var. phoenicis in the Namib-Naukluft Park. S. Afr. J. Bot. 2008, 74, 41–50. [Google Scholar] [CrossRef]
- Gardener, M.C.; Gillman, M.P. The taste of nectar—A neglected area of pollination ecology. Oikos 2002, 98, 552–557. [Google Scholar] [CrossRef]
- Nicolson, S.W.; Thornburg, R.W. Nectar chemistry. In Nectaries and Nectar; Nicolson, S.W., Thornburg, R.W., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 215–264. [Google Scholar]
- Hansen, K.; Wacht, S.; Seebauer, H.; Schnuch, M. New aspects of chemoreception in flies. Ann. N. Y. Acad. Sci. 1998, 855, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.; Shafir, S.; Yehonatan, L.; Palmer, R.G.; Thornburg, R. A novel role for proline in plant floral nectars. Sci. Nat. 2006, 93, 72–79. [Google Scholar] [CrossRef]
- Rao, N.A.; Ambili, M.; Jala, V.R.; Subramanya, H.; Savithri, H. Structure-function relationship in serine hydroxymethyltransferase. BBA-Proteins Proteom. 2003, 1647, 24–29. [Google Scholar]
- Smith, M.E.; Greenberg, D.M. Characterization of an enzyme reducing pyrroline-5-carboxylate to proline. Nature 1956, 177, 1130. [Google Scholar] [CrossRef]
- Carafa, A.; Carratù, G.; Pizzolongo, P. Anatomical observations on the nucellar apex of Wellwitschia mirabilis and the chemical composition of the micropylar drop. Plant Reprod. 1992, 5, 275–279. [Google Scholar] [CrossRef]
- Winter, K.U.; Becker, A.; Munster, T.; Kim, J.T.; Saedler, H.; Theissen, G. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc. Natl. Acad. Sci. USA 1999, 96, 7342–7347. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Maroto, F.; Carmona, M.J.; Garrido, J.A.; Vilches-Ferron, M.; Rodriguez-Ruiz, J.; Alonso, D.L. New roles for MADS-box genes in higher plants. Biol. Plantarum 2003, 46, 321–330. [Google Scholar] [CrossRef]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef]
- Becker, A.; Winter, K.U.; Meyer, B.; Saedler, H.; Theissen, G. MADS-box gene diversity in seed plants 300 million years ago. Mol. Biol. Evol. 2000, 17, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Rodin, R.J.; Kapil, R.N. Comparative anatomy of the seed coats of Gnetum and their probable evolution. Am. J. Bot. 1969, 56, 420–431. [Google Scholar] [CrossRef]
- Berridge, E.M. The structure of the female strobilus in Gnetum gnemon. Ann. Bot. 1912, 26, 987–992. [Google Scholar] [CrossRef]
- Barceló, A.R. Peroxidase and not laccase is the enzyme responsible for cell wall lignification in the secondary thickening of xylem vessels in Lupinus. Protoplasma 1995, 186, 41–44. [Google Scholar] [CrossRef]
- Peter, G.; Neale, D. Molecular basis for the evolution of xylem lignification. Curr. Opin. Plant Biol. 2004, 7, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cheng, X.F.; Leshkevich, J.; Umezawa, T.; Harding, S.A.; Chiang, V.L. The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 2001, 13, 1567–1586. [Google Scholar] [CrossRef] [PubMed]
- Lequeu, J.; Fauconnier, M.L.; Chammai, A.; Bronner, R.; Blee, E. Formation of plant cuticle: Evidence for the occurrence of the peroxygenase pathway. Plant J. 2003, 36, 155–164. [Google Scholar] [CrossRef]
- Pellmyr, O. Evolution of insect pollination and angiosperm diversification. Trends Ecol. Evolut. 1992, 7, 46–49. [Google Scholar] [CrossRef]
- Gorelick, R. Did insect pollination cause increased seed plant diversity? Biol. J. Linn. Soc. 2001, 74, 407–427. [Google Scholar] [CrossRef]
- Labandeira, C.C.; Kvacek, J.; Mostovski, M.B. Pollination drops, pollen, and insect pollination of Mesozoic gymnosperms. Taxon 2007, 56, 663–695. [Google Scholar] [CrossRef]
- Burleigh, J.G.; Mathews, S. Phylogenetic signal in nucleotide data from seed plants: Implications for resolving the seed plant tree of life. Am. J. Bot. 2004, 91, 1599–1613. [Google Scholar] [CrossRef] [PubMed]
- Ran, J.H.; Shen, T.T.; Wang, M.M.; Wang, X.Q. Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc. R. Soc. B Biol. Sci. 2018, 285, 20181012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rydin, C.; Källersjö, M.; Friis, E.M. Seed plant relationships and the systematic position of Gnetales based on nuclear and chloroplast DNA: Conflicting data, rooting problems, and the monophyly of conifers. Int. J. Plant Sci. 2002, 163, 197–214. [Google Scholar] [CrossRef]



| Groups | Cycads | Ginkgo | Conifers | Gnetales | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteins | CH | ZF | CR | GB | CS | CK | CL | JC | JO | PM | TM | ED | EF | EM | ET | EL | EM | EP | WM(F) | WM(M) | GG(F) | GG(M) | GG(F)* | GG(M)* | GL(F)* | GL(M)* | |
| Alpha(beta)-galactosidase | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||||
| Alpha-amylase | X | X | X | X | X | X | X | ||||||||||||||||||||
| Alpha-fucosidase | X | X | X | X | X | ||||||||||||||||||||||
| Arabidongalactan protein | X | X | X | X | X | X | |||||||||||||||||||||
| Aspartyl protease | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||||||
| ATPases | X | X | X | X | |||||||||||||||||||||||
| Beta-glucanse (or endoglucanase) | X | X | X | X | X | ||||||||||||||||||||||
| Beta-glucosidase | X | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||
| Beta-hexosaminidase | X | X | X | X | X | X | |||||||||||||||||||||
| Calmodulin | X | X | X | X | X | ||||||||||||||||||||||
| Chitinase | X | X | X | X | X | X | X | X | X | ||||||||||||||||||
| Cystatin | X | X | X | X | X | ||||||||||||||||||||||
| Cysteine protease | X | X | X | X | X | X | X | ||||||||||||||||||||
| Dehydrogenases | X | X | X | X | X | X | |||||||||||||||||||||
| Elongation factors | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||
| Expansin | X | X | X | X | X | ||||||||||||||||||||||
| Fasciclin-like arabinogalactan | X | X | X | X | X | ||||||||||||||||||||||
| Glycosylhydrolase | X | X | X | X | X | X | X | X | X | ||||||||||||||||||
| GTP-binding nuclear protein | X | X | X | X | |||||||||||||||||||||||
| Heat shock protein | X | X | X | X | X | X | |||||||||||||||||||||
| Histones | X | X | X | X | X | X | X | ||||||||||||||||||||
| Invertase | X | X | |||||||||||||||||||||||||
| Peroxidase | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||||||
| Polygalacturonas(-like) protein | X | X | X | X | X | ||||||||||||||||||||||
| Ribosomal proteins | X | X | X | X | X | X | |||||||||||||||||||||
| Serine carboxypeptidase (-like) protein | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||
| Subtilisin-like proteinase (serine endopeptidase) | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||
| Thaumatin-like protein | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||||||
| Ubiquitins | X | X | X | X | |||||||||||||||||||||||
| Xylosidase | X | X | X | X | X | X | X | X | X | ||||||||||||||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, C.; Saunders, R.M.K.; Deng, N.; Wan, T.; Su, Y. Pollination Drop Proteome and Reproductive Organ Transcriptome Comparison in Gnetum Reveals Entomophilous Adaptation. Genes 2019, 10, 800. https://doi.org/10.3390/genes10100800
Hou C, Saunders RMK, Deng N, Wan T, Su Y. Pollination Drop Proteome and Reproductive Organ Transcriptome Comparison in Gnetum Reveals Entomophilous Adaptation. Genes. 2019; 10(10):800. https://doi.org/10.3390/genes10100800
Chicago/Turabian StyleHou, Chen, Richard M. K. Saunders, Nan Deng, Tao Wan, and Yingjuan Su. 2019. "Pollination Drop Proteome and Reproductive Organ Transcriptome Comparison in Gnetum Reveals Entomophilous Adaptation" Genes 10, no. 10: 800. https://doi.org/10.3390/genes10100800
APA StyleHou, C., Saunders, R. M. K., Deng, N., Wan, T., & Su, Y. (2019). Pollination Drop Proteome and Reproductive Organ Transcriptome Comparison in Gnetum Reveals Entomophilous Adaptation. Genes, 10(10), 800. https://doi.org/10.3390/genes10100800