Molecular Cloning and Characterization of Small Heat Shock Protein Genes in the Invasive Leaf Miner Fly, Liriomyza trifolii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Insects of Different Developmental Stages
2.2. Temperature Treatments
2.3. RNA Isolation and Cloning Experiments
2.4. Reverse Transcription Quantitative PCR
2.5. Sequence Alignment of sHSPs and Data Analysis
3. Results
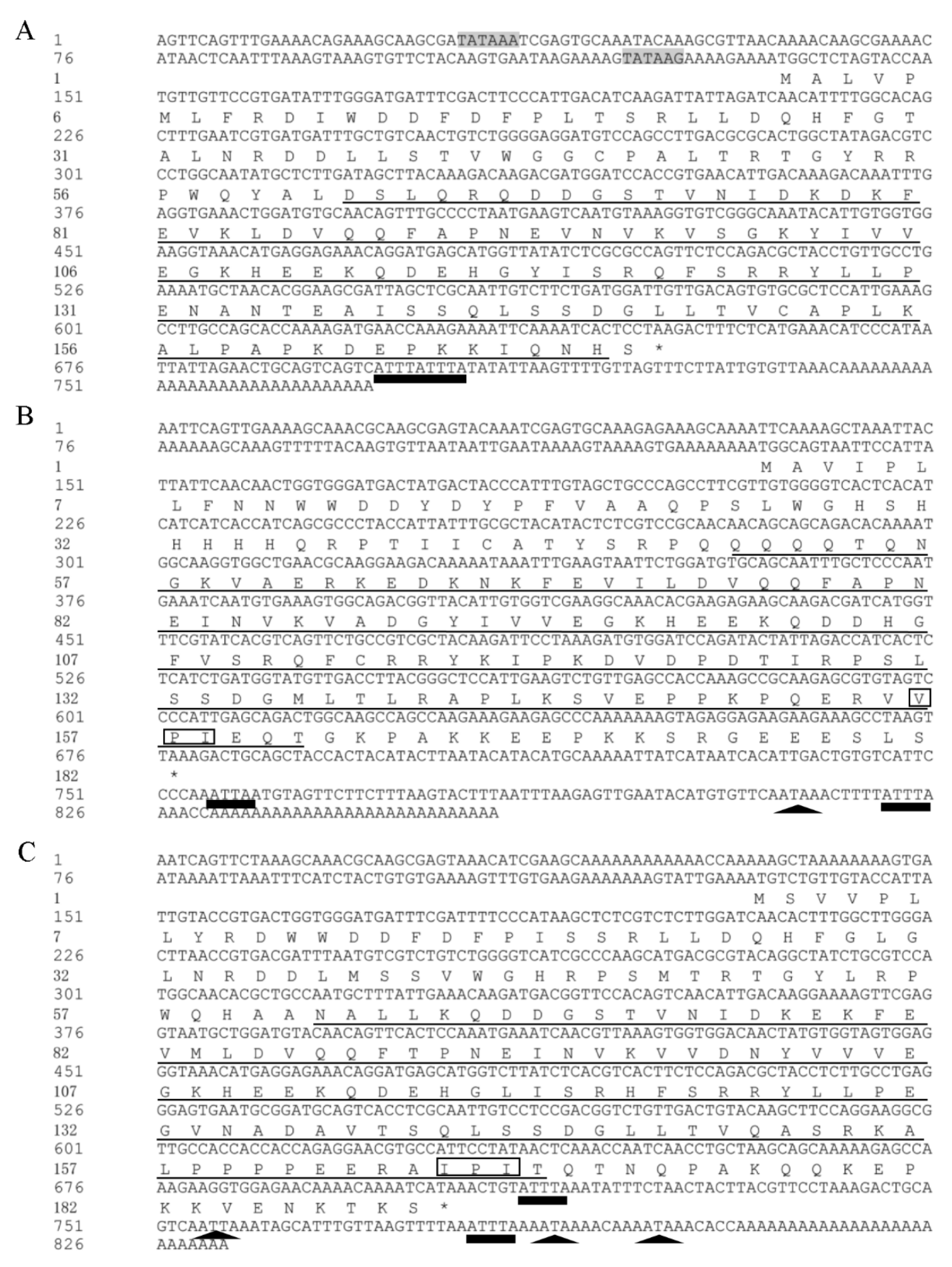
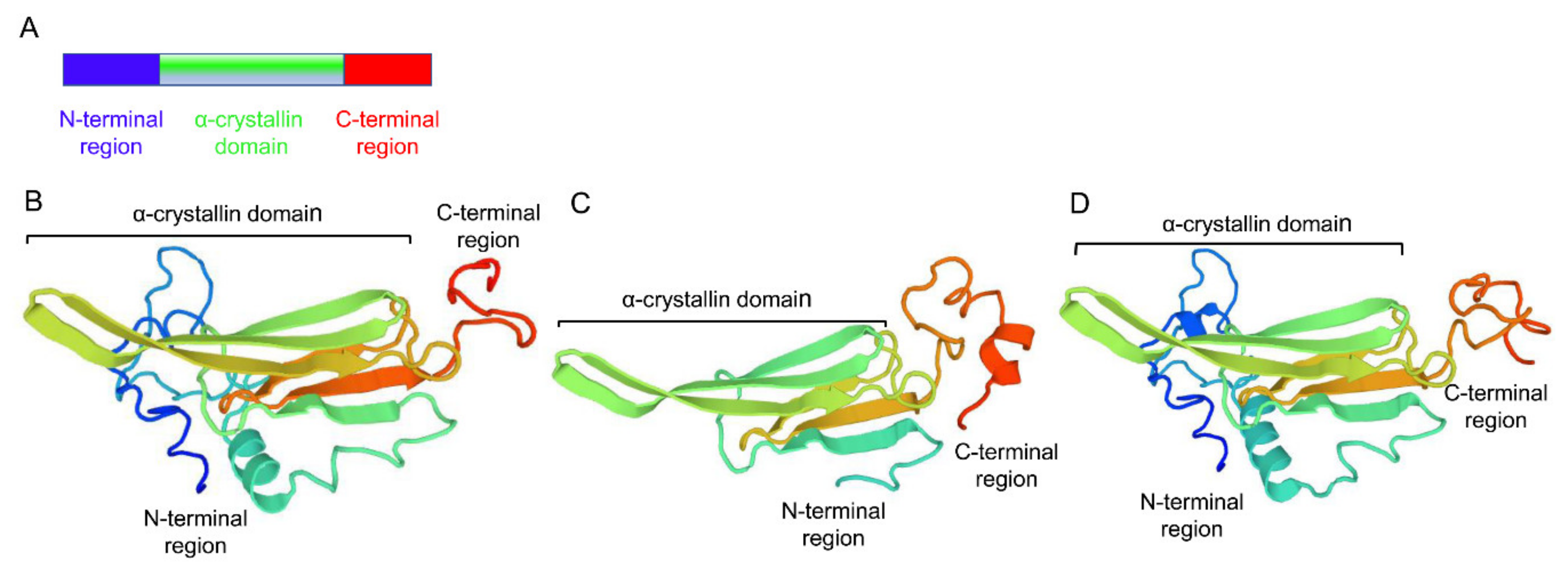
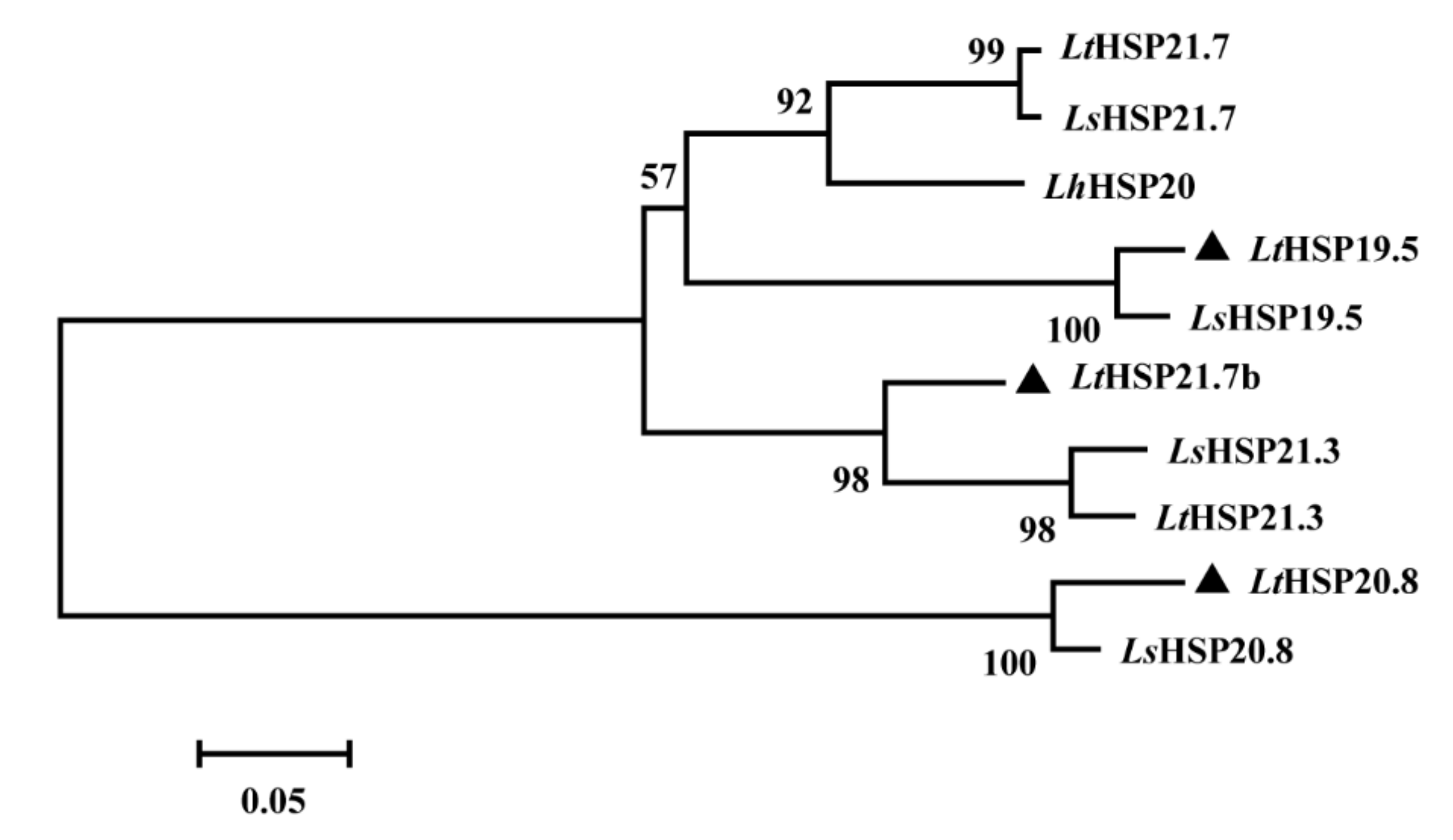
3.1. Cloning, Characterization and Phylogenetic Analysis of Three sHSP Genes from Liriomyza trifolii
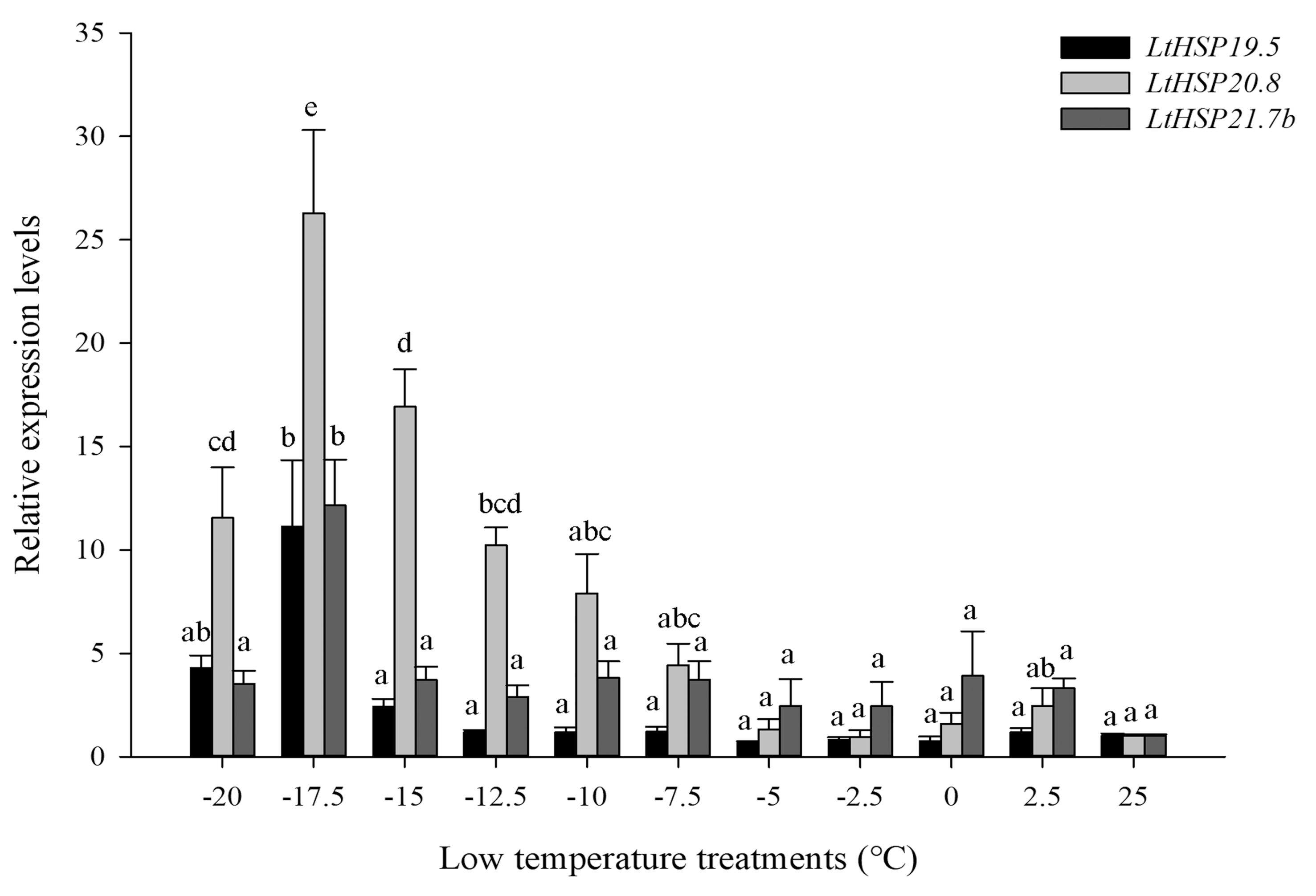
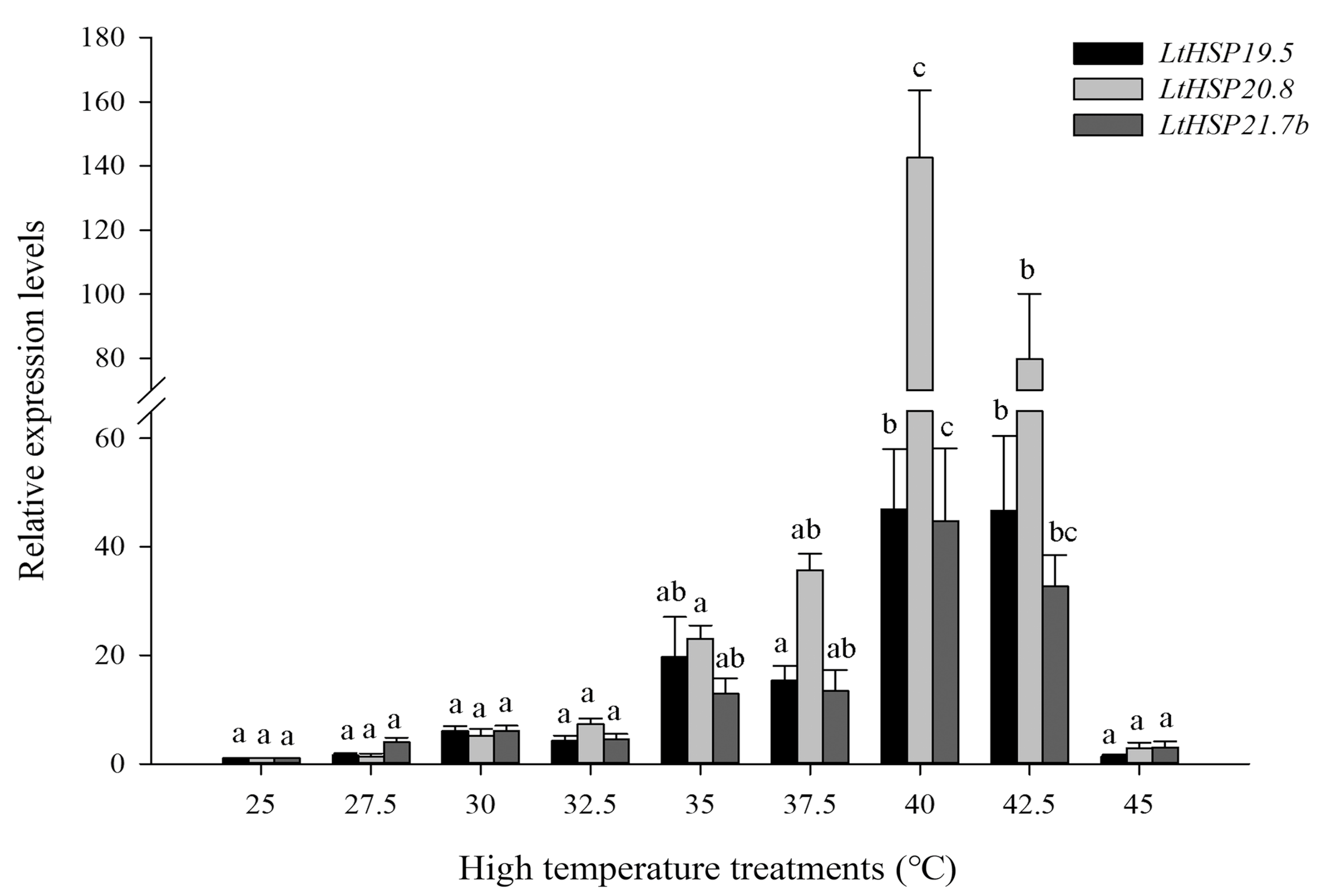
3.2. Expression of Three LtsHSPs in Response to Temperature Treatments
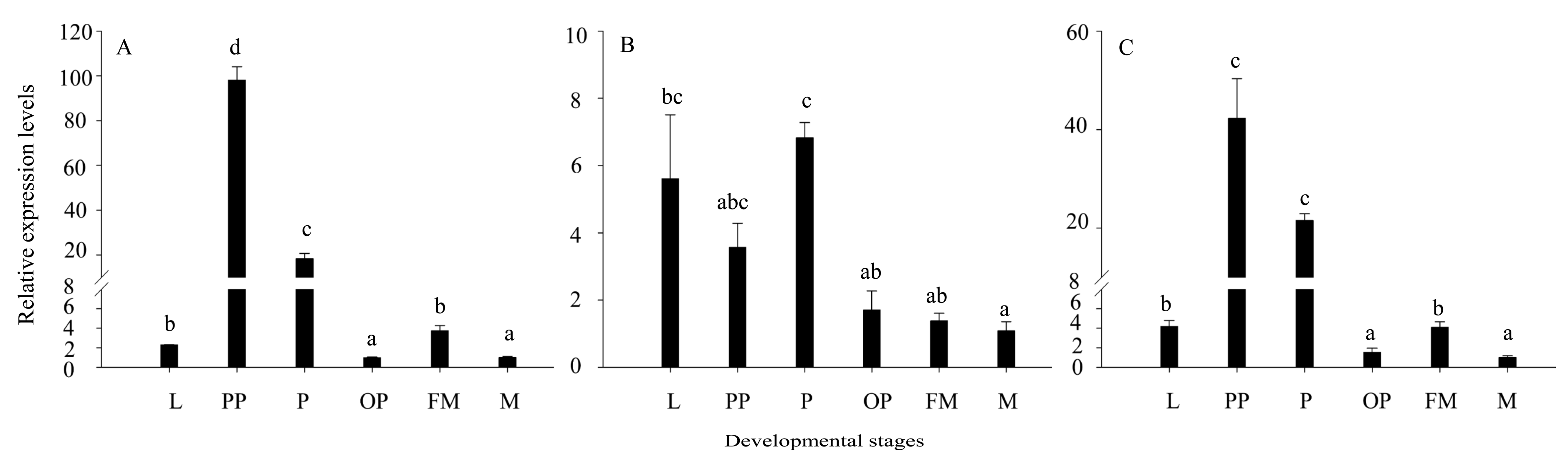
3.3. Expression of Three LtsHSPs at the Developmental Stages
3.4. Comparative Characteristics and Expression Pattern of Five LtsHSPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Spencer, K.A. Agromyzidae (Diptera) of Economic Importance; 9: Series Entomologica; The Hague Publishers: Bath, UK, 1973; pp. 19–28. [Google Scholar]
- Johnson, M.W.; Welter, S.C.; Toscano, N.C.; Tingi, P.; Trumble, J.T. Reduction of tomato leaflet photosynthesis rates by mining activity of Liriomyza sativae (Diptera: Agromyzidae). J. Econ. Entomol. 1983, 76, 1061–1063. [Google Scholar] [CrossRef]
- Parrella, M.P.; Jones, V.P.; Youngman, R.R.; Lebeck, L.M. Effect of leaf mining and leaf stippling of Liriomyza spp. on photosynthetic rates of chrysanthemum. Ann. Entomol. Soc. A 1985, 78, 90–93. [Google Scholar] [CrossRef]
- Reitz, S.R.; Kund, G.S.; Carson, W.G.; Phillips, P.A.; Trumble, J.T. Economics of reducing insecticide use on celery through low-input pest management strategies. Agric. Ecosyst. Environ. 1999, 73, 185–197. [Google Scholar] [CrossRef]
- Scheffer, S.J.; Lewis, M.L. Mitochondrial phylogeography of the vegetable pest Liriomyza trifolii (Diptera: Agromyzidae): Diverged clades and invasive populations. Ann. Entomol. Soc. Am. 2006, 99, 991–998. [Google Scholar] [CrossRef]
- Wang, Z.G.; Guan, W.; Chen, D.H. Preliminary report of the Liriomyza trifolii in Zhongshan area. Plant Quarantine 2007, 21, 19–20. [Google Scholar]
- Gao, Y.L.; Reitz, S.; Xing, Z.L.; Ferguson, S.; Lei, Z.R. A decade of a leafminer invasion in China: Lessons learned. Pest Manag. Sci. 2017, 73, 1775–1779. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Chen, B.; Wei, J.N.; Liu, T.X. Roles of thermal adaptation and chemical ecology in Liriomyza distribution and control. Annu. Rev. Entomol. 2009, 54, 127–145. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Cao, J.M.; Du, Y.Z. Survey and molecular identification of Liriomyza trifolii in Jiangsu, China. Plant Prot. 2010, 36, 108–111. [Google Scholar]
- Xiang, J.C.; Lei, Z.R.; Wang, H.H. Interspecific competition among three invasive Liriomyza species. Acta Ecol. Sin. 2012, 32, 1616–1622. [Google Scholar] [CrossRef]
- Wen, J.Z.; Lei, Z.R.; Wang, Y. Survey of Liriomyza huidobrensis in Yunnan Province and Guizhou Province, China. Plant Prot. 1998, 24, 18–20. [Google Scholar]
- Wen, J.Z.; Wang, Y.; Lei, Z.R. New record of Liriomyza sativae Blanchard (Diptera: Agromyzidae) from China. Entomotaxonomia 1996, 18, 311–312. [Google Scholar]
- Chang, Y.W.; Chen, J.Y.; Lu, M.X.; Gao, Y.; Tian, Z.H.; Gong, W.R.; Dong, C.S.; Du, Y.Z. Cloning and expression of genes encoding heat shock proteins in Liriomyza trifolii and comparison with two congener leafminer species. PLoS ONE 2017, 12, e0181355. [Google Scholar] [CrossRef] [PubMed]
- Reitz, S.R.; Trumble, J.T. Interspecific and intraspecific differences in two Liriomyza leafminer species in California. Entomol. Exp. Appl. 2002, 102, 101–113. [Google Scholar] [CrossRef]
- Abe, Y.; Tokumaru, S. Displacement in two invasive species of leafminer fly in different localities. Biol. Invasions 2008, 10, 951–953. [Google Scholar] [CrossRef]
- Wang, H.H.; Reitz, S.R.; Xiang, J.C.; Smagghe, G.; Lei, Z.R. Does Temperature-Mediated Reproductive Success Drive the Direction of Species Displacement in Two Invasive Species of Leafminer Fly? PLoS ONE 2014, 9, e98761. [Google Scholar] [CrossRef] [PubMed]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef] [PubMed]
- Moseley, P.L. Heat shock proteins and heat adaptation of the whole organism. J. Appl. Physiol. 1997, 83, 1413–1417. [Google Scholar] [CrossRef] [PubMed]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- Sørensen, J.G.; Kristensen, T.N.; Loeschcke, V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003, 6, 1025–1037. [Google Scholar] [CrossRef]
- Horwitz, J. α-Crystalline can function as a molecular chaperone. Proc. Natl. Acad. Sci. USA 1992, 89, 10449–10453. [Google Scholar] [CrossRef]
- Gusev, N.B.; Bogatcheva, N.V.; Marston, S.B. Structure and properties of the small heat shock proteins (shsps) and their interaction with cytoskeleton proteins. Biochemistry (Mosc.) 2002, 67, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Franck, E.; Madsen, O.; van-Rheede, T.; Ricard, G.; Huynen, M.A.; de-Jong, W.W. Evolutionary diversity of vertebrate small heat shock proteins. J. Mol. Evol. 2004, 59, 792–805. [Google Scholar] [CrossRef]
- Haslbeck, M.; Franzmann, T.; Weinfurtner, D.; Buchner, J. Some like it hot: The structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 2005, 12, 842–846. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.T.; Chen, B.; Li, Z.H. Thermal plasticity is related to the hardening response of heat shock protein expression in two Bactrocera fruit flies. J. Insect. Physiol. 2014, 67, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef]
- Basha, E.; O’Neill, H.; Vierling, E. Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.; Macrae, T.H. Insect heat shock proteins during stress and diapause. Annu. Rev. Entomol. 2015, 60, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.J.; Wehner, R. Heat shock protein synthesis and thermotolerance in Cataglyphis, an ant from the Sahara Desert. Proc. Natl. Acad. Sci. USA 1995, 92, 2994–2998. [Google Scholar] [CrossRef]
- Lu, M.X.; Hua, J.; Cui, Y.D.; Du, Y.Z. Five small heat shock protein genes from Chilo suppressalis: Characteristics of gene, genomic organization, structural analysis, and transcription profiles. Cell Stress Chaperon. 2014, 19, 91–104. [Google Scholar] [CrossRef]
- Pan, D.D.; Lu, M.X.; Li, Q.Y.; Du, Y.Z. Characteristics and expression of genes encoding two small heat shock protein genes lacking introns from Chilo suppressalis. Cell Stress Chaperon. 2017, 23, 1–10. [Google Scholar] [CrossRef]
- Wang, H.H.; Rreitz, S.; Wang, L.X.; Wang, S.Y.; Xue, L.I.; Lei, Z.R. The mRNA expression proifles of five heat shock protein genes from Frankliniella occidentalis at different stages and their responses to temperatures and insecticides. J. Integr. Agric. 2014, 13, 2196–2210. [Google Scholar] [CrossRef]
- Quan, G.X.; Duan, J.; Ladd, T.; Krell, P.J. Identification and expression analysis of multiple small heat shock protein genes in spruce budworm, Choristoneura fumiferana, (L.). Cell Stress Chaperon. 2017, 23, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Dou, W.; Tian, Y.; Liu, H.; Shi, Y.; Smagghe, G.; Wang, J.J. Characteristics of six small heat shock protein genes from Bactrocera dorsalis: Diverse expression under conditions of thermal stress and normal growth. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2017, 213, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Rinehart, J.P.; Li, A.; Yocum, G.D.; Robich, R.M.; Hayward, S.A.; Denlinger, D.L. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc. Natl. Acad. Sci. USA 2007, 104, 11130–11137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joanisse, D.R.; Michaud, S.; Inaguma, Y.; Tanguay, R.M. Small heat shock proteins of Drosophila: Developmental expression and functions. J. Biosci. 1998, 23, 369–376. [Google Scholar] [CrossRef]
- Haass, C.H.; Klein, U.L.; Kloetzel, P.M. Developmental expression of Drosophila melanogaster small heat-shock proteins. J. Cell Sci. 1990, 96, 413–418. [Google Scholar] [PubMed]
- Saravanakumar, R.; Ponnuvel, K.M.; Qadri, S.M.H. Expression of metabolic enzyme genes and heatshock protein genes during embryonic development in diapause and non-diapause egg of multivoltine silkworm Bombyx mori. Biologia 2008, 63, 737–744. [Google Scholar] [CrossRef]
- Shen, Y.; Gu, J.; Huang, L.H.; Zheng, S.C.; Liu, L.; Xu, W.H.; Feng, Q.L.; Kang, L. Cloning and expression analysis of six small heat shock protein genes in the common cutworm, Spodoptera litura. J. Insect Physiol. 2011, 57, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Huang, L.X.; Shen, Y.; Huang, L.H.; Feng, Q.L. Hsp70 and small Hsps are the major heat shock protein members involved in midgut metamorphosis in the common cutworm, Spodoptera litura. Insect Mol. Biol. 2012, 21, 534–543. [Google Scholar] [CrossRef]
- Huang, L.H.; Kang, L. Cloning and interspecific altered expression of heat shock protein genes in two leafminer species in response to thermal stress. Insect Mol. Biol. 2007, 16, 491–500. [Google Scholar] [CrossRef]
- Huang, L.H.; Wang, C.Z.; Kang, L. Cloning and expression of five heat shock protein genes in relation to cold hardening and development in the leafminer, Liriomyza sativa. J. Insect Physiol. 2009, 55, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.W.; Chen, J.Y.; Lu, M.X.; Gao, Y.; Tian, Z.H.; Gong, W.R.; Zhu, W.; Du, Y.Z. Selection and validation of reference genes for quantitative real time PCR analysis under different experimental conditions in the leafminer Liriomyza trifolii (Diptera: Agromyzidae). PLoS ONE 2017, 12, e0181862. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Kang, L. Cold hardiness and supercoo ling capacity in the pea leafminer Liriomyza huidobrensis. Cryo Letters 2002, 23, 173–182. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Tabaska, J.E.; Zhang, M.Q. Detection of polyadenylation signals in human DNA sequences. Gene 1999, 231, 77–86. [Google Scholar] [CrossRef] [Green Version]
- Lindquist, S.; Petersen, R. Selective translation and degradation of heat-shock messenger RNAs in Drosophila. Enzyme 1990, 44, 147–166. [Google Scholar] [CrossRef]
- Colgan, D.F.; Manley, J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997, 11, 2755–2766. [Google Scholar] [CrossRef] [Green Version]
- Rubenstein, R.C.; Lyons, B.M. Sodium 4-phenylbutyrate downregulates HSC70 expression by facilitating mRNA degradation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001, 281, L43–L51. [Google Scholar] [CrossRef]
- Hunt, C.; Morimoto, R.I. Conserved features of eukaryotic hsp70 genes revealed by comparison with the nucleotide sequence of Human hsp70. Proc. Natl. Acad. Sci. USA 1985, 82, 6455–6459. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Madabusi, L.; Nishioka, H.; Emanuel, P.; Sypes, M.; Arkhipova, I.; Gilmour, D.S. Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol. Cell. Biol. 2001, 21, 1593–1602. [Google Scholar] [CrossRef]
- Grace, M.L.; Chandrasekharan, M.B.; Hall, T.C.; Crowe, A.J. Sequence and spacing of TATA box elements are critical for accurate initiation from the beta-phaseolin promoter. J. Biol. Chem. 2004, 279, 8102–8110. [Google Scholar] [CrossRef] [PubMed]
- Martín-Folgar, R.; Fuente, M.; Morcillo, G.; Martínez-Guitarte, J. Characterization of six small HSP genes from Chironomus riparius (Diptera, Chironomidae): Differential expression under conditions of normal growth and heat-induced stress. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 188, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Lu, M.X.; Tang, X.T.; Du, Y.Z. Characterization and Expression of Genes Encoding Three Small Heat Shock Proteins in Sesamia inferens (Lepidoptera: Noctuidae). Int. J. Mol. Sci. 2014, 15, 23196–23211. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.T.; Sun, M.; Lu, M.X.; Du, Y.Z. Expression patterns of five heat shock proteins in Sesamia inferens, (Lepidoptera: Noctuidae) during heat stress. J. Asia-Pac. Entomol. 2015, 18, 529–533. [Google Scholar] [CrossRef]
- Lu, M.X.; Li, H.B.; Zheng, Y.T.; Shi, L.; Du, Y.Z. Identification, genomic organization and expression profiles of four heat shock protein genes in the western flower thrips, Frankliniella occidentalis. J. Therm. Biol. 2016, 57, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.W.; Zhang, X.X.; Chen, J.Y.; Lu, M.X.; Gong, W.R.; Du, Y.Z. Characterization of three heat shock protein 70 genes from Liriomyza trifolii and expression during thermal stress and insect development. B. Entomol. Res. 2019, 109, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Liu, X.N.; Lu, M.X.; Du, Y.Z. Characterization of genes encoding small heat shock proteins from Bemisia tabaci and expression under thermal stress. PeerJ 2019, 7, e6992. [Google Scholar] [CrossRef]
| Name of Genes | Molecular Weight | Isoelectric Point | Developmental Stages (Highest Expression) | High Temperature (Tmax) | Low Temperature (Tmax) |
|---|---|---|---|---|---|
| LtHSP19.5 | 19.46 kDa | 5.56 | prepupae | 40 °C | −17.5 °C |
| LtHSP20.8 | 20.89 kDa | 6.71 | two-day-old pupae | 40 °C | −17.5 °C |
| LtHSP21.7b | 21.72 kDa | 6.07 | two-day-old pupae | 40 °C | −17.5 °C |
| LtHSP21.3 | 21.23 kDa * | 6.38 * | prepupae | 40 °C * | −17.5 °C * |
| LtHSP21.7 | 21.66 kDa * | 7.03 * | prepupae * | 42.5 °C | −17.5 °C |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.-W.; Zhang, X.-X.; Lu, M.-X.; Du, Y.-Z.; Zhu-Salzman, K. Molecular Cloning and Characterization of Small Heat Shock Protein Genes in the Invasive Leaf Miner Fly, Liriomyza trifolii. Genes 2019, 10, 775. https://doi.org/10.3390/genes10100775
Chang Y-W, Zhang X-X, Lu M-X, Du Y-Z, Zhu-Salzman K. Molecular Cloning and Characterization of Small Heat Shock Protein Genes in the Invasive Leaf Miner Fly, Liriomyza trifolii. Genes. 2019; 10(10):775. https://doi.org/10.3390/genes10100775
Chicago/Turabian StyleChang, Ya-Wen, Xiao-Xiang Zhang, Ming-Xing Lu, Yu-Zhou Du, and Keyan Zhu-Salzman. 2019. "Molecular Cloning and Characterization of Small Heat Shock Protein Genes in the Invasive Leaf Miner Fly, Liriomyza trifolii" Genes 10, no. 10: 775. https://doi.org/10.3390/genes10100775





