The Selection and Validation of Reference Genes for mRNA and microRNA Expression Studies in Human Liver Slices Using RT-qPCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Ethics Statement
2.3. Human Liver
2.4. Preparation of Precision-Cut Liver Slice and Experimental Treatment
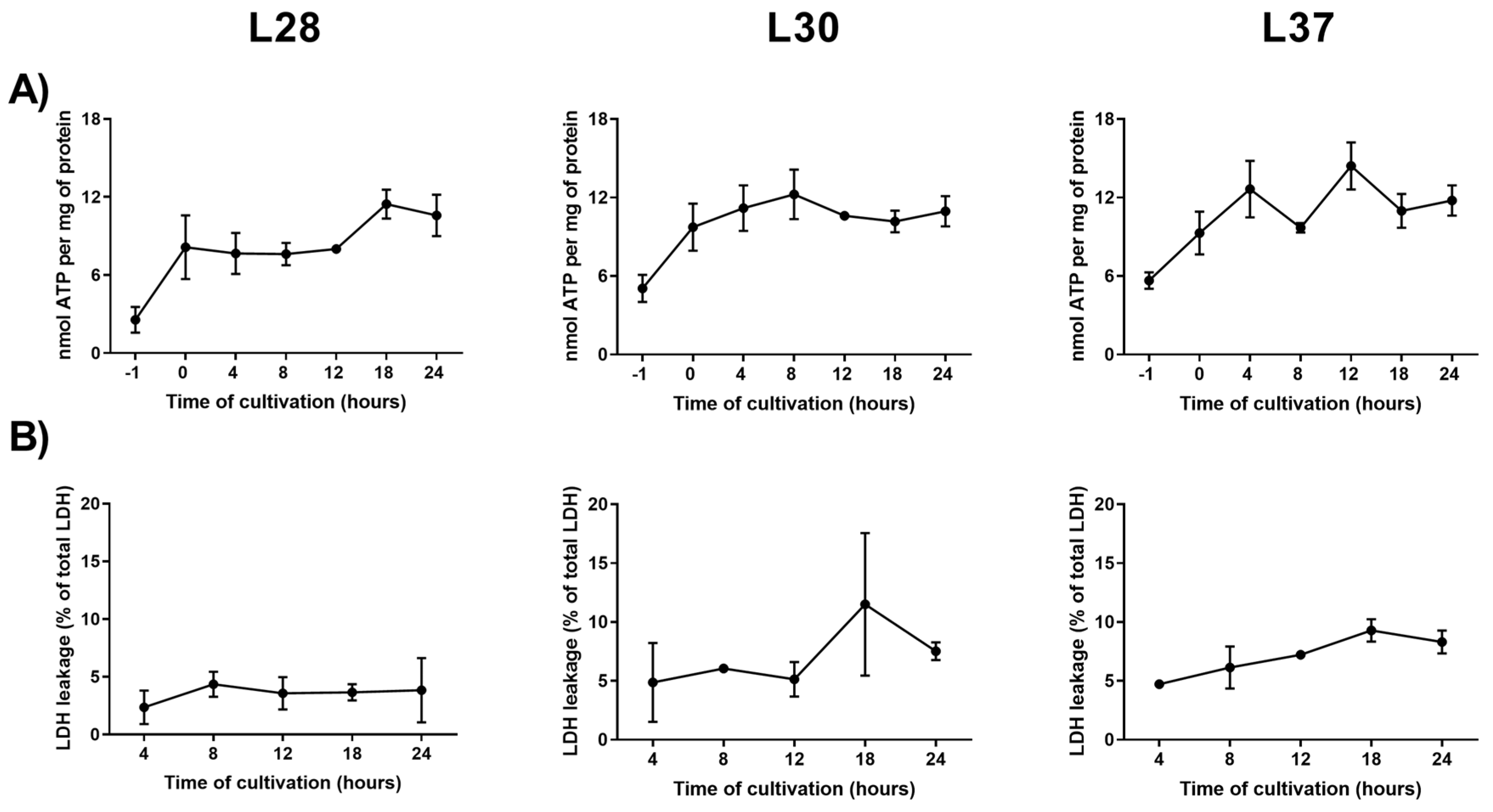
2.5. Viability
2.6. Tissue RNA Extraction
2.7. cDNA Synthesis
2.8. Primer Design, Quantitative Real-Time PCR
2.9. Data Analysis
3. Results
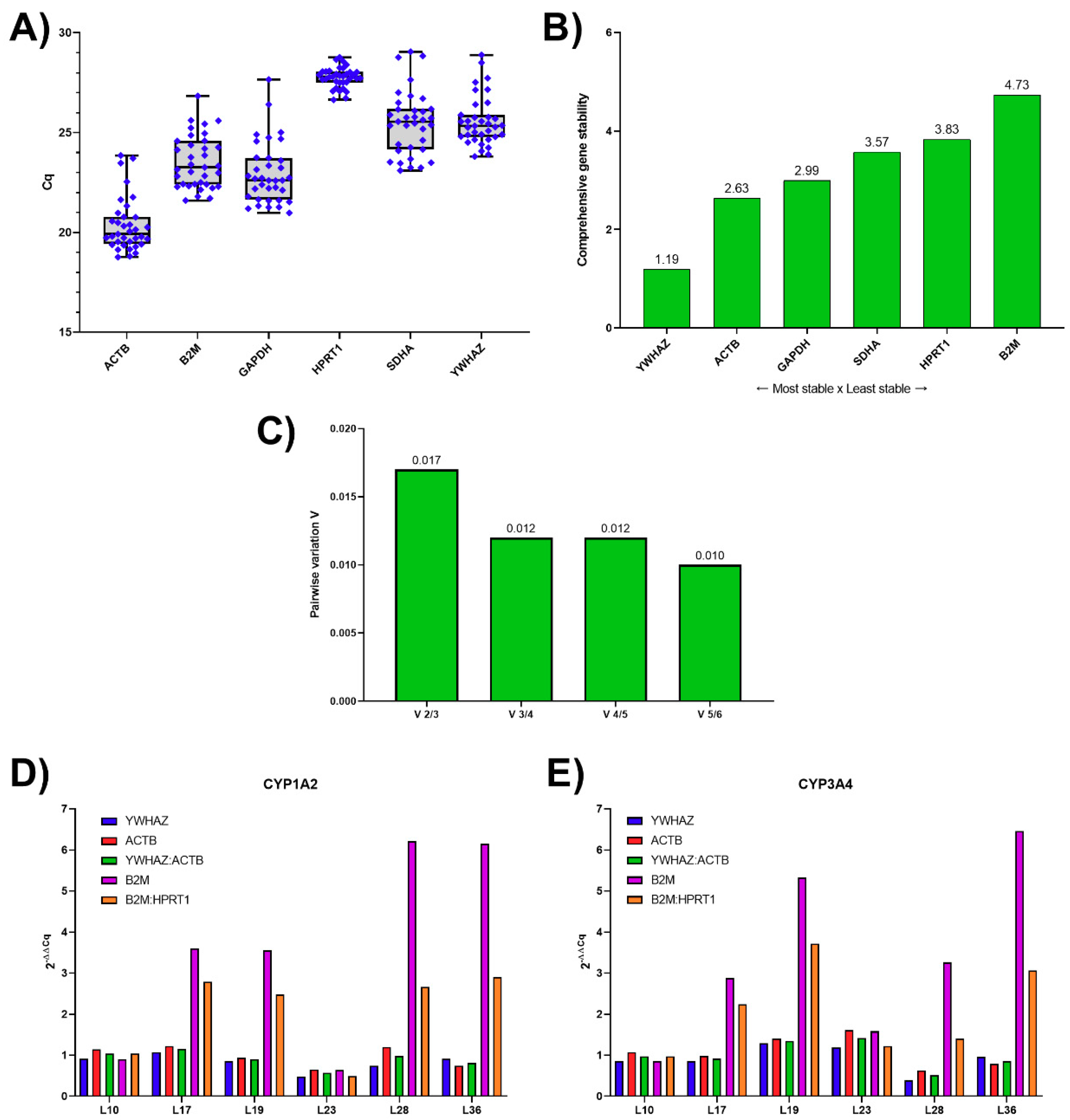
3.1. Analysis of Candidate RGs Expression Stability for mRNA Normalization and Their Validation in Human Liver Tissue
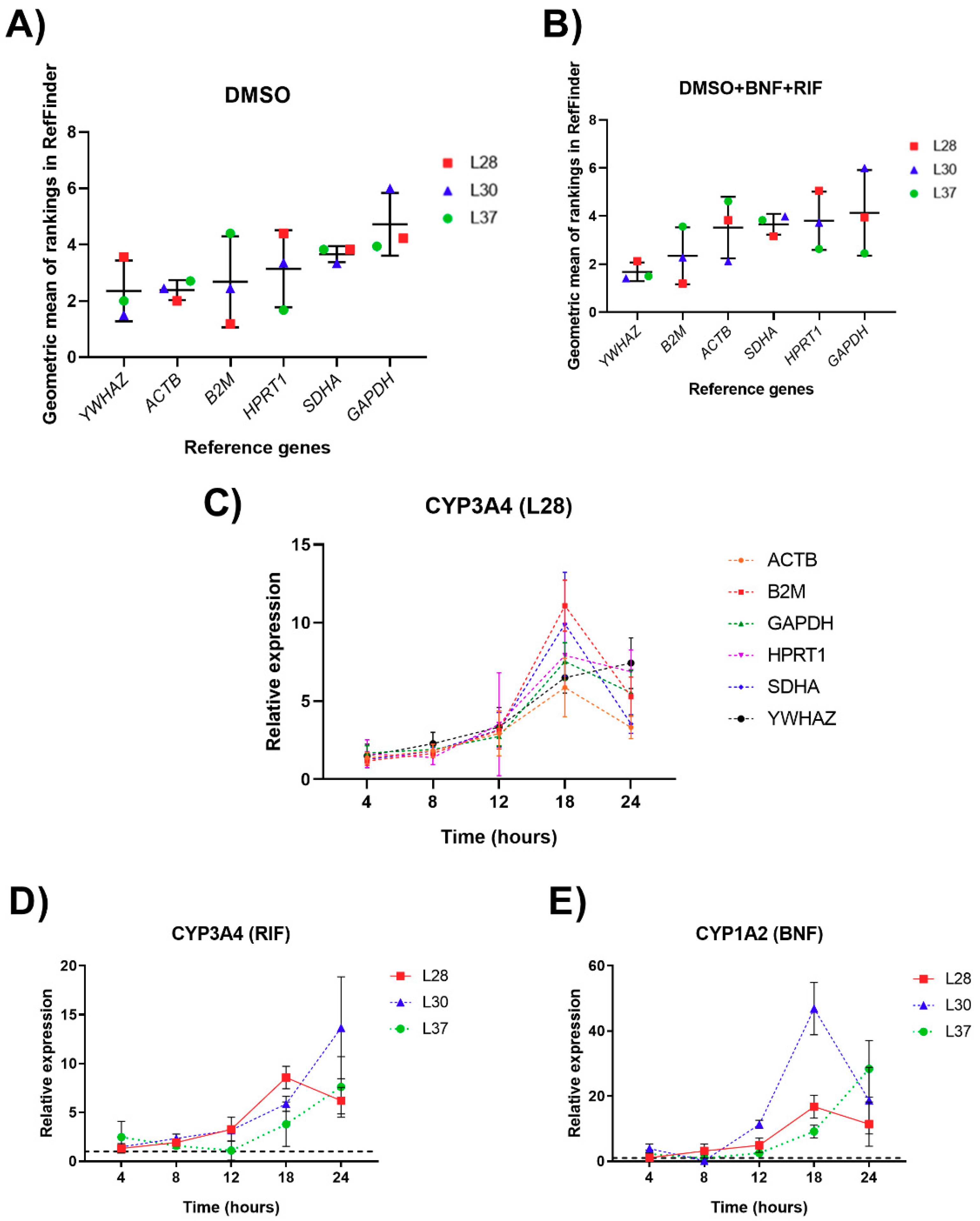
3.2. Analysis of Candidate RGs Expression Stability for mRNA Normalization in Human PCLS
3.3. Reference Genes Ranking According to Their Expression Stability
3.4. Validation of Reference Genes for mRNA Normalization
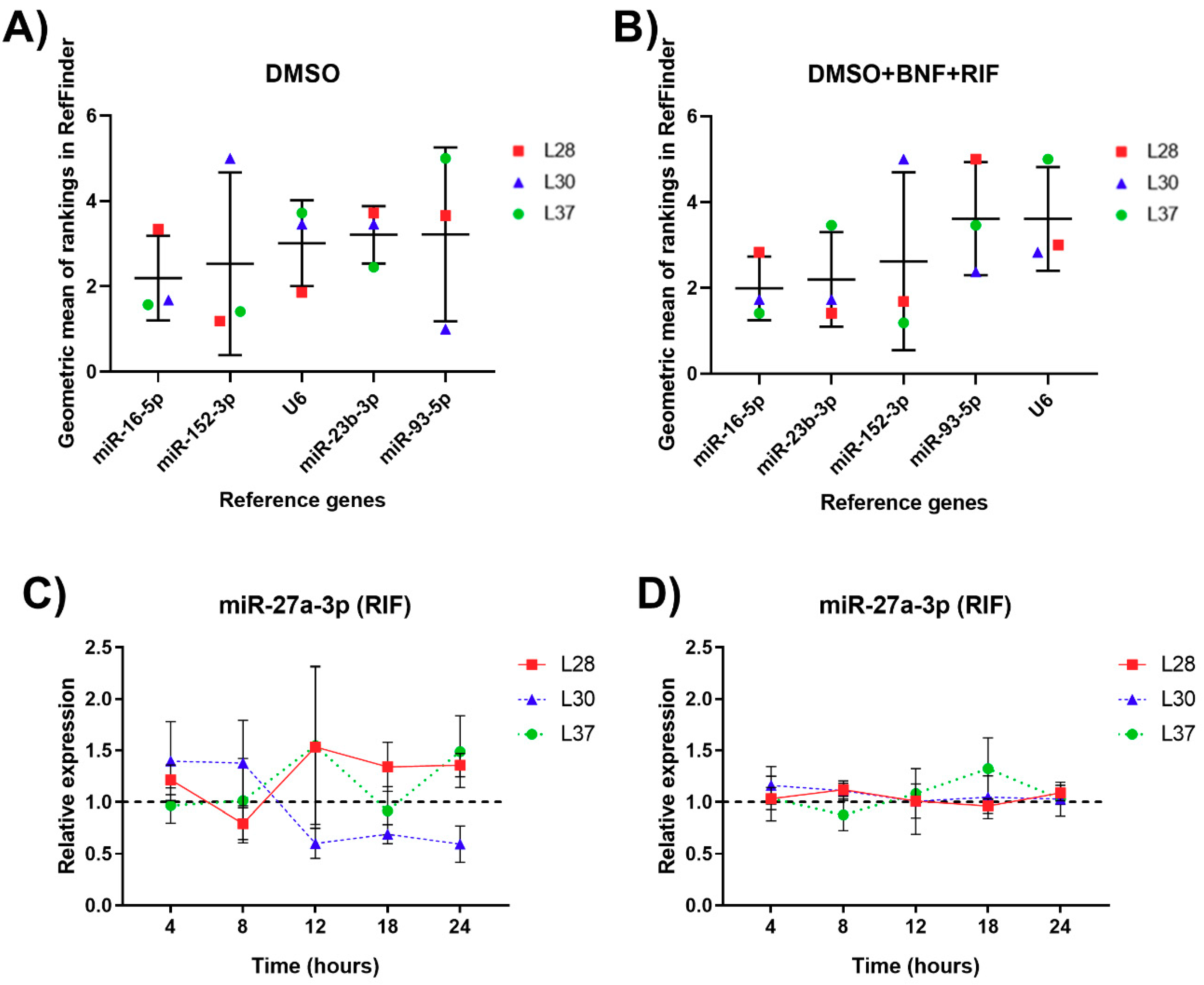
3.5. Analysis of Expression Stability and Validation of RGs for miRNA Normalization in Human PCLS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fisher, R.L.; Vickers, A.E.M. Preparation and culture of precision-cut organ slices from human and animal. Xenobiotica. 2013, 43, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Olinga, P.; Hof, I.H.; Merema, M.T.; Smit, M.; de Jager, M.H.; Swart, P.J.; Slooff, M.J.H.; Meijer, D.K.F.; Groothuis, G.M.M. The applicability of rat and human liver slices to the study of mechanisms of hepatic drug uptake. J. Pharmacol. Toxicol. Methods. 2001, 45, 55–63. [Google Scholar] [CrossRef]
- Vickers, A.E.M.; Ulyanov, A.V.; Fisher, R.L. Liver Effects of Clinical Drugs Differentiated in Human Liver Slices. Int. J. Mol. Sci. 2017, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Zarybnicky, T.; Matouskova, P.; Lancosova, B.; Subrt, Z.; Skalova, L.; Bousova, I. Inter-Individual Variability in Acute Toxicity of R-Pulegone and R-Menthofuran in Human Liver Slices and Their Influence on miRNA Expression Changes in Comparison to Acetaminophen. Int. J. Mol. Sci. 2018, 19, 17. [Google Scholar] [CrossRef] [PubMed]
- Rius, B.; Titos, E.; Moran-Salvador, E.; Lopez-Vicario, C.; Garcia-Alonso, V.; Gonzalez-Periz, A.; Arroyo, V.; Claria, J. Resolvin D1 primes the resolution process initiated by calorie restriction in obesity-induced steatohepatitis. Faseb J. 2014, 28, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Rius, B.; Duran-Guell, M.; Flores-Costa, R.; Lopez-Vicario, C.; Lopategi, A.; Alcaraz-Quiles, J.; Casulleras, M.; Lozano, J.J.; Titos, E.; Claria, J. The specialized proresolving lipid mediator maresin 1 protects hepatocytes from lipotoxic and hypoxia-induced endoplasmic reticulum stress. Faseb J. 2017, 31, 5384–5398. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Roberto, J.B.; Knupp, A.; Kenerson, H.L.; Truong, C.D.; Yuen, S.Y.; Brempelis, K.J.; Tuefferd, M.; Chen, A.; Horton, H.; et al. Precision-cut human liver slice cultures as an immunological platform. J. Immunol. Methods. 2018, 455, 71–79. [Google Scholar] [CrossRef]
- Elferink, M.G.L.; Olinga, P.; Draaisma, A.L.; Merema, M.T.; Bauerschmidt, S.; Polman, J.; Schoonen, W.G.; Groothuis, G.M.M. Microarray analysis in rat liver slices correctly predicts in vivo hepatotoxicity. Toxicol. Appl. Pharmacol. 2008, 229, 300–309. [Google Scholar] [CrossRef]
- Melgert, B.N.; Olinga, P.; Van der Laan, J.M.S.; Weert, B.; Cho, J.; Schuppan, D.; Groothuis, G.M.M.; Meijer, D.K.F.; Poelstra, K. Targeting dexamethasone to Kupffer cells: Effects on liver inflammation and fibrosis in rats. Hepatology 2001, 34, 719–728. [Google Scholar] [CrossRef]
- van de Bovenkamp, M.; Groothuis, G.M.M.; Draaisma, A.L.; Merema, M.T.; Bezuijen, J.I.; van Gils, M.J.; Meijer, D.K.F.; Friedman, S.L.; Olinga, P. Precision-cut liver slices as a new model to study toxicity-induced hepatic stellate cell activation in a physiologic milieu. Toxicol. Sci. 2005, 85, 632–638. [Google Scholar] [CrossRef]
- Vickers, A.E.M.; Saulnier, M.; Cruz, E.; Merema, M.T.; Rose, K.; Bentley, P.; Olinga, P. Organ slice viability extended for pathway characterization: An in vitro model to investigate fibrosis. Toxicol. Sci. 2004, 82, 534–544. [Google Scholar] [CrossRef]
- Starokozhko, V.; Vatakuti, S.; Schievink, B.; Merema, M.T.; Asplund, A.; Synnergren, J.; Aspegren, A.; Groothuis, G.M.M. Maintenance of drug metabolism and transport functions in human precision-cut liver slices during prolonged incubation for 5 days. Arch. Toxicol. 2017, 91, 2079–2092. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 12. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 17. [Google Scholar] [CrossRef]
- Vickers, A.E.M.; Fisher, R.L. Human Liver Slices to Investigate Injury and Repair. Appl. In Vitro Toxicol. 2018, 4, 280–287. [Google Scholar] [CrossRef]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.; Nolan, T. Talking the talk, but not walking the walk: RT-qPCR as a paradigm for the lack of reproducibility in molecular research. Eur. J. Clin. Invest. 2017, 47, 756–774. [Google Scholar] [CrossRef] [Green Version]
- Gerets, H.H.J.; Tilmant, K.; Gerin, B.; Chanteux, H.; Depelchin, B.O.; Dhalluin, S.; Atienzar, F.A. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol. Toxicol. 2012, 28, 69–87. [Google Scholar] [CrossRef] [Green Version]
- Hadi, M.; Westra, I.M.; Starokozhko, V.; Dragovic, S.; Merema, M.T.; Groothuis, G.M.M. Human Precision-Cut Liver Slices as an ex Vivo Model to Study Idiosyncratic Drug-Induced Liver Injury. Chem. Res. Toxicol. 2013, 26, 710–720. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Lardizábal, M.N.; Nocito, A.L.; Daniele, S.M.; Ornella, L.A.; Palatnik, J.F.; Veggi, L.M. Reference Genes for Real-Time PCR Quantification of MicroRNAs and Messenger RNAs in Rat Models of Hepatotoxicity. PLoS One. 2012, 7, e36323. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 9. [Google Scholar] [CrossRef]
- Xie, F.L.; Xiao, P.; Chen, D.L.; Xu, L.; Zhang, B.H. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol.Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pavo, N.; Raderer, M.; Goliasch, G.; Wurm, R.; Strunk, G.; Cho, A.; Novak, J.F.; Gisslinger, H.; Steger, G.G.; Hejna, M.; et al. Subclinical involvement of the liver is associated with prognosis in treatment naïve cancer patients. Oncotarget. 2017, 8, 81250–81260. [Google Scholar] [CrossRef] [PubMed]
- Nygard, A.B.; Jorgensen, C.B.; Cirera, S.; Fredholm, M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol. Biol. 2007, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.; Huh, J.W.; Park, S.J.; Kim, D.S.; Ha, H.S.; Kim, Y.J.; Lee, J.R.; Chang, K.T.; Kim, H.S. Selection of internal reference genes for SYBR green qRT-PCR studies of rhesus monkey (Macaca mulatta) tissues. BMC Mol. Biol. 2008, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Granitzny, A.; Knebel, J.; Schaudien, D.; Braun, A.; Steinberg, P.; Dasenbrock, C.; Hansen, T. Maintenance of high quality rat precision cut liver slices during culture to study hepatotoxic responses: Acetaminophen as a model compound. Toxicol. In Vitro 2017, 42, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.L.; Schelcher, C.; Gashi, S.; Schreiber, S.; Thasler, R.M.K.; Jauch, K.-W.; Thasler, W.E.J.M.B. RNA Stability in Human Liver: Comparison of Different Processing Times, Temperatures and Methods. Mol. Biotechnol. 2013, 53, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vickers, A.E.; Fisher, R.L.; Sinclair, J.R. Glutathione Modulation and Oxidative Stress in Human Liver Slices. Curr. Drug Discov. Technol. 2010, 7, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matoušková, P. microRNAs and Reference Gene Methodology. In Handbook of Nutrition, Diet, and Epigenetics, 1st ed.; Patel, V., Preedy, V., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–17. [Google Scholar]
- Wang, J.; Lu, M.; Qiu, C.; Cui, Q. TransmiR: A transcription factor–microRNA regulation database. Nucleic Acids Res. 2010, 38, D119–D122. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef]
- Matoušková, P.; Bártíková, H.; Boušová, I.; Hanušová, V.; Szotáková, B.; Skálová, L. Reference Genes for Real-Time PCR Quantification of Messenger RNAs and MicroRNAs in Mouse Model of Obesity. PLoS ONE 2014, 9, e86033. [Google Scholar] [CrossRef]
- Lamba, V.; Ghodke-Puranik, Y.; Guan, W.; Lamba, J.K. Identification of suitable reference genes for hepatic microRNA quantitation. BMC Res. Notes 2014, 7, 129. [Google Scholar] [CrossRef]
- Zhang, J.; Le, T.D.; Liu, L.; Liu, B.; He, J.; Goodall, G.J.; Li, J. Identifying direct miRNA–mRNA causal regulatory relationships in heterogeneous data. J. Biomed. Inform. 2014, 52, 438–447. [Google Scholar] [CrossRef]
- Tang, X.; Chen, S. Epigenetic Regulation of Cytochrome P450 Enzymes and Clinical Implication. Curr. Drug Metab. 2015, 16, 86–96. [Google Scholar] [CrossRef]
- Ramamoorthy, A.; Liu, Y.; Philips, S.; Desta, Z.; Lin, H.; Goswami, C.; Gaedigk, A.; Li, L.; Flockhart, D.A.; Skaar, T.C. Regulation of MicroRNA Expression by Rifampin in Human Hepatocytes. Drug Metab. Dispos. 2013, 41, 1763–1768. [Google Scholar] [CrossRef]
- Takahashi, K.; Tatsumi, N.; Fukami, T.; Yokoi, T.; Nakajima, M. Integrated Analysis of Rifampicin-induced MicroRNA and Gene Expression Changes in Human Hepatocytes. Drug Metab. Pharmacokinet. 2014, 29, 333–340. [Google Scholar] [CrossRef] [Green Version]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.W.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef]

| Human Sample | Gender (Age) | Reason of Surgery | Long-Term Pharmacotherapy | PCLS Preparation |
|---|---|---|---|---|
| L1 | Female (37) | CRC 1 | Insulin | No |
| L3 | Male (58) | CCC 2 | Allopurinol, felodipine, ramipril, indapamide, atorvastatin, citalopram, pregabalin | No |
| L4 | Male (35) | Adenoma | Mesalazine, omeprazole, escitalopram, budesonide | No |
| L5 | Male (63) | CRC | Ramipril, atorvastatin, metformin, allopurinol | Yes |
| L6 | Male (69) | CRC | Hydrochlorothiazide | Yes |
| L7 | Male (69) | CRC | Nitrendipine, acetylsalicylic acid | Yes |
| L8 | Female (69) | CRC | Verapamil, trandolapril | No |
| L9 | Male (81) | CRC | Betaxolol | Yes |
| L10 | Female (66) | HCC 3 | Pantoprazole | No |
| L11 | Female (57) | CRC, liver metastases | None | Yes |
| L12 | Female (73) | CRC | Lerkanidipine, furosemide, perindopril, nadroparin | No |
| L13 | Female (67) | CCC | Nadroparin | No |
| L14 | Female (45) | BFNH 4 | None | Yes |
| L15 | Female (69) | CRC | Nebivolol, simvastatin, digoxin, irbesartan, hydrochlorothiazide, nadroparin | No |
| L16 | Female (59) | CRC | None | Yes |
| L17 | Male (39) | CRC | None | No |
| L18 | Male (83) | HCC | Lacidipine, solifenacin, tamsulosin | No |
| L19 | Female (65) | CRC | Amlodipine | No |
| L20 | Female (84) | Abscess | None | No |
| L21 | Male (34) | Jejunal adenocarcinoma | None | No |
| L22 | Female (84) | CRC | Telmisartan, nitrendipine, formoterol | No |
| L23 | Male (83) | CRC | Furosemide, amlodipine, acetylsalicylic acid, telmisartan, salmeterol, fluticasone | No |
| L24 | Male (77) | HCC | Metoprolol, felodipin, ramipril, metformin, gliclazid, finasteride, warfarin,nadroparin | No |
| L25 | Male (70) | CRC | Perindopril, betaxolol, metformin, atorvastatin, allopurinol, insulin | No |
| L26 | Male (72) | CRC | None | No |
| L27 | Male (70) | CRC | Insulin, atorvastatin | No |
| L28 | Female (26) | BFNH | None | Yes |
| L29 | Male (59) | Renal cell carcinoma, liver metastases | None | No |
| L30 | Female (81) | CRC, liver metastases | Hydrochlorothiazide, betaxolol, acetylsalicylic acid, zolpidem, insulin | Yes |
| L33 | Female (72) | CCC | Lerkanidipine, irbesartan, hydrochlorothiazide, lansoprazole, levothyroxine, acetylsalicylic acid, fenofibrate, nebivolol | No |
| L34 | Male (62) | CRC | Ramipril, felodipine, metoprolol, rosuvastatin, acetylsalicylic acid | No |
| L35 | Male (72) | CRC, liver metastases | Tamsulosin, metoprolol, omeprazole | No |
| L36 | Female (78) | CCC | Simvastatin, bisoprolol, furosemide, ramipril, enoxaparin, zolpidem | Yes |
| L37 | Male (50) | Neuroendocrine tumor, liver metastases | Insulin | Yes |
| L38 | Male (59) | CCC | None | Yes |
| Gene Symbol | Gene Name | GeneBank or miRbase Accession Number | Gene Function | Primer Sequences 5´-3´ | Tm1 (°C) | E2 (%) |
|---|---|---|---|---|---|---|
| Candidate reference genes for mRNA normalization | ||||||
| ACTB | Actin beta | NM_001101.4 | Structural protein of cytoskeleton | F3: TCCCTGGAGAAGAGCTACGAG R4: CAGGAAGGAAGGCTGGAAGAG | 86.5 | 102 |
| B2M | Beta-2-microglobulin | NM_004048.2 | Beta-chain of major histocompatibility complex | F: TGCTGTCTCCATGTTTGATGTATC R: TCTCTGCTCCCCACCTCTAAG | 83 | 99 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | NM_002046 | Enzyme of glycolysis pathway | F: GAGTCCACTGGCGTCTTCAC R: GAGGCATTGCTGATGATCTTGAG | 86 | 101 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | NM_000194.2 | Metabolism of purines | F: TGGTCAGGCAGTATAATCCAAAGA R: TTCAAATCCAACAAAGTCTGGCT | 82 | 101 |
| SDHA | Succinate dehydrogenase complex, subunit A | NM_004168.3 | Critical function in mitochondrial respiratory chain | F: TGGGAACAAGAGGGCATCTG R: ACCACCACTGCATCAAATTCATG | 79.5 | 99 |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | NM_003406.3 | Important protein for many signal transduction pathways | F: TGATCCCCAATGCTTCACAAG R: GCCAAGTAACGGTAGTAATCTCC | 77.5 | 102 |
| Candidate reference genes for miRNA normalization | ||||||
| miR-16-5p | MicroRNA 16 (5p) | MIMAT0000069 | Regulation of apoptosis | RT5: GTCTCCTCTGGTGCAGGGTCCGAG GTATTCGCACCAGAGGAGACCGCC AA F: ACAGCCTAGCAGCACGTAAAT | 79 | 102 |
| miR-23b-3p | MicroRNA 23b (3p) | MIMAT0000418 | Associated with cell proliferation, invasion, and apoptosis | RT: GTCTCCTCTGGTGCAGGGTCCGAGGTA TTCGCACCAGAGGAGACGTGGTA F: ATCTGTATCACATTGCCAGGGA | 77.5 | 109 |
| miR-93-5p | MicroRNA 93 (5p) | MIMAT0000093 | OncomiR, plays an essential role in tumorigenesis and progression of various carcinomas | RT: GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGAC F: GTCAATCAAAGTGCTGTTCGTG | 78.5 | 105 |
| miR-152-3p | MicroRNA 152 (3p) | MIMAT0000438 | Regulates hepatic glycogenesis, tumor suppressor | RT: GTCTCCTCTGGTGCAGGGTCCGAGGTA TTCGCACCAGAGGAGACCCAAGT F: CGACGTTCAGTGCATGACAG | 78.5 | 100 |
| U6 | Small nuclear RNA U6 | NR_003027 | RNA splicing | R: AACGCTTCACGAATTTGCGTG F: GCTCGCTTCGGCAGCACA | 80.5 | 99 |
| universal | R: GAGGTATTCGCACCAGAGGA | |||||
| Genes of interest | ||||||
| CYP1A2 | Cytochrome P450 family 1 subfamily A member 2 | NM_000761 | Phase I biotransformation | F: CTTCCCTGAGAGTAGCGATGAGA R: GCAGTCTCCACGAACTCATGAG | 85.5 | 101 |
| CYP3A4 | Cytochrome P450 family 3 subfamily A member 4 | NM_017460.5 | Phase I biotransformation | F: CCCCTGAAATTAAGCTTAGGAGG R: CTGGTGTTCTCAGGCACAGA | 82.5 | 99 |
| miR-27a-3p | MicroRNA 27a (3p) | MIMAT0000084 | Direct regulation of CYP3A4 | RT: GTCTCCTCTGGTGCAGGGTCCGAGGTA TTCGCACCAGAGGAGACGCGGAA F: CGGCGTTTCACAGTGGCTAA | 80.5 | 106 |
| miR-203a-3p | MicroRNA 203a (3p) | MIMAT0000264 | Indirect regulation of CYP1A2 via PXR receptor | RT: GTCTCCTCTGGTGCAGGGTCCGAGGTA TTCGCACCAGAGGAGACCTAGTG F: CGGCGTGTGAAATGTTTAGGA | 78.5 | 105 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zárybnický, T.; Matoušková, P.; Ambrož, M.; Šubrt, Z.; Skálová, L.; Boušová, I. The Selection and Validation of Reference Genes for mRNA and microRNA Expression Studies in Human Liver Slices Using RT-qPCR. Genes 2019, 10, 763. https://doi.org/10.3390/genes10100763
Zárybnický T, Matoušková P, Ambrož M, Šubrt Z, Skálová L, Boušová I. The Selection and Validation of Reference Genes for mRNA and microRNA Expression Studies in Human Liver Slices Using RT-qPCR. Genes. 2019; 10(10):763. https://doi.org/10.3390/genes10100763
Chicago/Turabian StyleZárybnický, Tomáš, Petra Matoušková, Martin Ambrož, Zdeněk Šubrt, Lenka Skálová, and Iva Boušová. 2019. "The Selection and Validation of Reference Genes for mRNA and microRNA Expression Studies in Human Liver Slices Using RT-qPCR" Genes 10, no. 10: 763. https://doi.org/10.3390/genes10100763
APA StyleZárybnický, T., Matoušková, P., Ambrož, M., Šubrt, Z., Skálová, L., & Boušová, I. (2019). The Selection and Validation of Reference Genes for mRNA and microRNA Expression Studies in Human Liver Slices Using RT-qPCR. Genes, 10(10), 763. https://doi.org/10.3390/genes10100763





